Gene expression
Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product. These products are often proteins, but in non-protein coding genes such as rRNA genes or tRNA genes, the product is a functional RNA. The process of gene expression is used by all known life - eukaryotes (including multicellular organisms), prokaryotes (bacteria and archea) and viruses - to generate the macromolecular machinery for life. Several steps in the gene expression process may be modulated, including the transcription, RNA splicing, translation, and post-translational modification of a protein. Gene regulation gives the cell control over structure and function, and is the basis for cellular differentiation, morphogenesis and the versatility and adaptability of any organism. Gene regulation may also serve as a substrate for evolutionary change, since control of the timing, location, and amount of gene expression can have a profound effect on the functions (actions) of the gene in a cell or in a multicellular organism.
In genetics gene expression is the most fundamental level at which genotype gives rise to the phenotype. The genetic code is "interpreted" by gene expression, and the properties of the expression products give rise to the organism's phenotype.

Mechanism
Transcription

The gene itself is typically a long stretch of DNA and does not perform an active role. It is a blueprint for the production of RNA. The production of RNA copies of the DNA is called transcription, and is performed by RNA polymerase, which adds one RNA nucleotide at a time to a growing RNA strand. This RNA is complementary to the DNA nucleotide being transcribed; i.e. a T on the DNA means an A is added to the RNA.[1] However, in RNA the nitrogen-containing base Uracil is inserted instead of Thymine wherever there is an Adenine on the DNA strand. Therefore, the mRNA complement of a DNA strand reading "TAC" would be transcribed as "AUG".
RNA processing
Transcription of protein encoding genes creates a primary transcript of RNA at the place where the gene was located. This transcript can be altered before being translated, this is particularly common in eukaryotes. The most common RNA processing is splicing to remove introns. Introns are RNA segments which are not found in the mature RNA, although they can function as precursors, e.g. for snoRNAs, which are RNAs that direct modification of nucleotides in other RNAs.[2] Introns are common in eukaryotic genes but rare in prokaryotes.
RNA processing, also known as post-transcriptional modification, can start during transcription, as is the case for splicing, where the spliceosome removes introns from newly formed RNA.[3]
Extensive RNA processing may be an evolutionary advantage made possible by the nucleus of eukaryotes. In prokaryotes transcription and translation (see below) happen together whilst in eukaryotes the nuclear membrane separates the two processes giving time for RNA processing to occur.
non-coding RNA maturation
In most organisms non-coding genes (ncRNA) are transcribed as precursors which undergo further processing. In the case of ribosomal RNAs (rRNA), they are often transcribed as a pre-rRNA which contains one or more rRNAs, the pre-rRNA is cleaved and modified (2′-O-methylation and pseudouridine formation) at a specific sites by approximately 150 different small nucleolus-restricted RNA species, called small nucleolar RNAs(snoRNAs) , which like snRNAs, snoRNAs associate with proteins, forming snoRNPs. In eukaryotes, in particular a snoRNP, called RNase MRP cleaves the 45S pre-rRNA into the 28S, 5.8S, and 18S rRNAs. The rRNA and RNA processing factors are form large aggregates called the nucleolus[4].
In the case of transfer RNA (tRNA), for example, the 5' sequence is removed by RNase P,[5] whereas the 3' end is removed by the tRNase Z enzyme.[6]. In the case of micro RNA (miRNA), miRNAs are first transcribed as primary transcripts or pri-miRNA with a cap and poly-A tail and processed to short, 70-nucleotide stem-loop structures known as pre-miRNA in the cell nucleus by the enzymes Drosha and Pasha, after being exported, it is then processed to mature miRNAs in the cytoplasm by interaction with the endonuclease Dicer, which also initiates the formation of the RNA-induced silencing complex (RISC), composed of the Argonaute protein.
RNA export
In eukaryotes most mature RNA must be exported to the cytoplasm from the nucleus. While some RNAs function in the nucleus, many RNAs are transported through the nuclear pores and into the cytosol. Notably this includes all RNA types involved in protein synthesis.[7] In some cases RNAs are additionally transported to a specific part of the cytoplasm, such as a synapse; they are then towed by motor proteins that bind through linker proteins to specific sequences (called "zipcodes") on the RNA.[8]
Translation
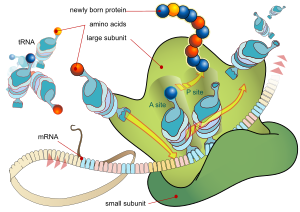
For some RNA (non-coding RNA) the mature RNA is the finished gene product.[9] In the case of messenger RNA (mRNA) the RNA is an information carrier coding for the synthesis of one or more proteins. mRNA carrying a single protein sequence (common in eukaryotes) is monocistronic whilst mRNA carrying multiple protein sequences (common in prokaryotes) is known as polycistronic.
Each triplet of nucleotides of the coding regions of a messenger RNA corresponds to a binding site for a transfer RNA. Transfer RNAs carry amino acids, and these are chained together by the ribosome. The ribosome helps transfer RNA to bind to messenger RNA and takes the amino acid from each transfer RNA and makes a structure-less protein out of it.[10][11] In prokaryotes translation generally occurs at the point of transcription, often using a messenger RNA which is still in the process of being created. In eukaryotes translation can occur in a variety of regions of the cell depending on where the protein being written is supposed to be. Major locations are the cytoplasm for soluble cytoplasmic proteins and the endoplasmic reticulum for proteins which are for export from the cell or insertion into a cell membrane. Proteins which are supposed to be expressed at the endoplasmic reticulum are recognised part-way through the translation process. This is governed by the signal recognition particle - a protein which binds to the ribosome and directs it to the endoplasmic reticulum when it finds a signal sequence on the growing (nascent) amino acid chain.[12]
Folding
Enzymes called chaperones assist the newly formed protein to attain (fold into) the 3-dimensional structure it needs to function.[13] Similarly, RNA chaperones help RNAs attain their functional shapes.[14] Assisting protein folding is one of the main roles of the endoplasmic reticulum in eukaryotes.
Protein transport
Many proteins are destined for other parts of the cell than the cytosol and a wide range of signalling sequences are used to direct proteins to where they are supposed to be. In prokaryotes this is normally a simple process due to limited compartmentalisation of the cell. However in eukaryotes there is a great variety of different targeting processes to ensure the protein arrives at the correct organelle.
Not all proteins remain within the cell and many are exported, for example digestive enzymes, hormones and extracellular matrix proteins. In eukaryotes the export pathway is well developed and the main mechanism for the export of these proteins is translocation to the endoplasmatic reticulum, followed by transport via the Golgi apparatus.[15][16]
Measurement
Measuring gene expression is an important part of many life sciences - the ability to quantify the level at which a particular gene is expressed within a cell, tissue or organism can give a huge amount of information. For example measuring gene expression can:
- Identify viral infection of a cell (viral protein expression)
- Determine an individual's susceptibility to cancer (oncogene expression)
- Find if a bacterium is resistant to penicillin (beta-lactamase expression)
Similarly the analysis of the location of expression protein is a powerful tool and this can be done on an organism or cellular scale. Investigation of localisation is particularly important for study of development in multicellular organisms and as an indicator of protein function in single cells. Ideally measurement of expression is done by detecting the final gene product (for many genes this is the protein) however it is often easier to detect one of the precursors, typically mRNA, and infer gene expression level.
mRNA quantification
Levels of mRNA can be quantitatively measured by Northern blotting which gives size and sequence information about the mRNA molecules. A sample of RNA is separated on an agarose gel and hybridized to a radio-labeled RNA probe that is complementary to the target sequence. The radio-labeled RNA is then detected by an autoradiograph. The main problems with Northern blotting stem from the use of radioactive reagents (which make the procedure time consuming and potentially dangerous) and lower quality quantification than more modern methods (due to the fact that quantification is done by measuring band strength in an image of a gel). Northern blotting is, however, still widely used as the additional mRNA size information allows the discrimination of alternately spliced transcripts.
A more modern low-throughput approach for measuring mRNA abundance is reverse transcription quantitative polymerase chain reaction (RT-PCR followed with qPCR). RT-PCR first generates a DNA template from the mRNA by reverse transcription. The DNA template is then used for qPCR where the change in fluorescence of a probe changes as the DNA amplification process progresses. With a carefully constructed standard curve qPCR can produce an absolute measurement such as number of copies of mRNA, typically in units of copies per nanolitre of homogenized tissue or copies per cell. qPCR is very sensitive (detection of a single mRNA molecule is possible), but can be expensive due to the fluorescent probes required.
Northern blots and RT-qPCR are good for detecting whether a single gene is being expressed, but it quickly becomes impractical if many genes within the sample are being studied. Using DNA microarrays transcript levels for many genes at once (expression profiling) can be measured. Recent advances in microarray technology allow for the quantification, on a single array, of transcript levels for every known gene in several organism's genomes, including humans.
Alternatively "tag based" technologies like Serial analysis of gene expression (SAGE), which can provide a relative measure of the cellular concentration of different messenger RNAs, can be used. The great advantage of tag-based methods is the "open architecture", allowing for the exact measurement of any transcript, with a known or unknown sequence.
Protein quantification
For genes encoding proteins the expression level can be directly assessed by a number of means with some clear analogies to the techniques for mRNA quantification.
The most commonly used method is to perform a Western blot against the protein of interest - this gives information on the size of the protein in addition to its identity. A sample (often cellular lysate) is separated on a polyacrylamide gel, transferred to a membrane and then probed with an antibody to the protein of interest. The antibody can either be conjugated to a fluorophore or to horseradish peroxidase for imaging and/or quantification. The gel-based nature of this assay makes quantification less accurate but it has the advantage of being able to identify later modifications to the protein, for example proteolysis or ubiquitination, from changes in size.
By replacing the gene with a new version fused a green fluorescent protein (or similar) marker expression may be directly quantified in live cells. This is done by imaging using a fluorescence microscope. It is very difficult to clone a GFP-fused protein into its native location in the genome without affecting expression levels so this method often cannot be used to measure endogenous gene expression. It is, however, widely used to measure the expression of a gene artificially introduced into the cell, for example via an expression vector. It is important to note that by fusing a target protein to a fluorescent reporter the protein's behavior, including its cellular localization and expression level, can be significantly changed.
The enzyme-linked immunosorbent assay works by using antibodies immobilised on a microtiter plate to capture proteins of interest from samples added to the well. Using a detection antibody conjugated to an enzyme or fluorophore the quantity of bound protein can be accurately measured by fluorometric or colourimetric detection. The detection process is very similar to that of a Western blot, but by avoiding the gel steps more accurate quantification can be achieved.
Localisation
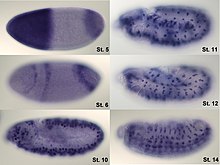
Analysis of expression is not limited to only quantification; localisation can also be determined. mRNA can be detected with a suitably labelled complementary mRNA strand and protein can be detected via labelled antibodies. The probed sample is then observed by microscopy to identify where the mRNA or protein is.
Regulation of gene expression

Regulation of gene expression refers to the control of the amount and timing of appearance of the functional product of a gene. Control of expression is vital to allow a cell to produce the gene products it needs when it needs them; in turn this gives cells the flexibility to adapt to a variable environment, external signals, damage to the cell, etc. Some simple examples of where gene expression is important are:
- Control of Insulin expression so it gives a signal for blood glucose regulation
- X chromosome inactivation in female mammals to prevent an "overdose" of the genes it contains.
- Cyclin expression levels control progression through the eukaryotic cell cycle
More generally gene regulation gives the cell control over all structure and function, and is the basis for cellular differentiation, morphogenesis and the versatility and adaptability of any organism.
Any step of gene expression may be modulated, from the DNA-RNA transcription step to post-translational modification of a protein. The stability of the final gene product, whether it is RNA or protein, also contributes to the expression level of the gene - an unstable product results in a low expression level. In general gene expression is regulated through changes[17] in the number and type of interactions between molecules[18] that collectively influence transcription of DNA[19] and translation of RNA.[20]
Transcription
Regulation of transcription can be broken down into three main routes of influence; genetic (direct interaction of a control factor with the gene), modulation (interaction of a control factor with the transcription machinery) and epigenetic (non-sequence changes in DNA structure which influence transcription).

Direct interaction with DNA is the simplest and most direct method a protein can change transcription levels and genes often have several protein binding sites around the coding region with the specific function of regulating transcription. There are many classes of regulatory DNA binding sites known as enhancers, insulators, repressors and silencers. The mechanisms for regulating transcription are very varied, from blocking key binding sites on the DNA for RNA polymerase to acting as an activator and promoting transcription by assisting RNA polymerase binding.
The activity of transcription factors is further modulated by intracellular signals causing protein post-translational modification including phosphorylated, acetylated, or glycosylated. These changes influence a transcription factor's ability to bind, directly or indirectly, to promoter DNA, to recruit RNA polymerase, or to favor elongation of a newly synthetized RNA molecule.
The nuclear membrane in eukaryotes allows further regulation of transcription factors by the duration of their presence in the nucleus which is regulated by reversible changes in their structure and by binding of other proteins.[21] Environmental stimuli or endocrine signals[22] may cause modification of regulatory proteins[23] eliciting cascades of intracellular signals,[24] which result in regulation of gene expression.
More recently it has become apparent that there is a huge influence of non-DNA-sequence specific effects on translation. These effects are referred to as epigenetic and involve the higher order structure of DNA, non-sequence specific DNA binding proteins and chemical modification of DNA. In general epigenetic effects alter the accessibility of DNA to proteins and so modulate transcription.

DNA methylation is a widespread mechanism for epigenetic influence on gene expression and is seen in bacteria and eukaryotes and has roles in heritable transcription silencing and transcription regulaton. In eukaryotes the structure of chromatin, controlled by the histone code, regulates access to DNA with significant impacts on the expression of genes in euchromatin and heterochromatin areas.
Nuclear export
In eukaryotes, where export of RNA is required before translation is possible, nuclear export is thought to provide additional control over gene expression. All transport in and out of the nucleus is via the nuclear pore and transport is controlled by a wide range of importin and exportin proteins.
RNA stability
Expression of a gene coding for a protein is only possible if the messenger RNA carrying the code survives long enough to be translated. In a typical cell an RNA molecule is only stable if specifically protected from degradation. RNA degradation has particular importance in regulation of expression in eukaryotic cells where mRNA has to travel significant distances before being translated. In eukaryotes RNA is stabilised by certain post-transcriptional modifications, particularly the 5' cap and poly-adenylated tail.
Intentional degradation of mRNA is used not just as a defence mechanism from foreign RNA (normally from viruses) but also as an route of mRNA destabilisation. If an mRNA molecule has a complementary sequence to a small interfering RNA then it is targeted for destruction via the RNA interference pathway.
Translation
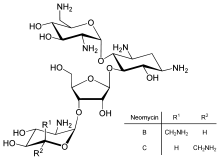
Direct regulation of translation is less prevalent than control of transcription or mRNA stability but is occasionally used. Inhbition of protein translation is a major target for toxins and antibiotics in order to kill a cell by overriding its normal gene expression control. Protein synthesis inhibitors include the antibiotic neomycin and the toxin ricin.
Protein degradation
Once protein synthesis is complete the level of expression of that protein can be reduced by protein degradation. There are major protein degradation pathways in all prokaryotes and eukaryotes of which the proteasome is a common component. An unneeded or damaged protein is often labelled for degradation by addition of ubiquitin.
Expression system
An expression system is a system specifically designed for the production of a gene product of choice. This is normally a protein although may also be RNA, such as tRNA or a ribozyme. An expression system consists of a gene, normally encoded by DNA, and the molecular machinery required to transcribe the DNA into mRNA and translate the mRNA into protein using the reagents provided. In the broadest sense this includes every living cell but the term is more normally used to refer to expression as a laboratory tool. An expression system is therefore often artificial in some manner. Expression systems are, however, a fundamentally natural process. Viruses are an excellent example where they replicate by using the host cell as an expression system for the viral proteins and genome.
In nature
In addition to these biological tools, certain naturally observed configurations of DNA (genes, promoters, enhancers, repressors) and the associated machinery itself are referred to as an expression system. This term is normally used in the case where a gene or set of genes is switched on under well defined conditions. For example the simple repressor 'switch' expression system in Lambda phage and the lac operator system in bacteria. Several natural expression systems are directly used or modified and used for artificial expression systems such as the Tet-on and Tet-off expression system.
Gene networks and expression
Genes have sometimes been regarded as nodes in a network, with inputs being proteins such as transcription factors, and outputs being the level of gene expression. The node itself performs a function, and the operation of these functions have been interpreted as performing a kind of information processing within cell and determine cellular behaviour.
Techniques and tools
The following experimental techniques are used to measure gene expression and are listed in roughly chronological order, starting with the older, more established technologies. They are divided into two groups based on their degree of multiplexity.
- Low-to-mid-plex techniques:
- Higher-plex techniques:
See also
- transcriptional noise
- transcriptional bursting
- Bookmarking
- Expression profiling
- Expressed sequence tag
- Paramutation
- Sequence profiling tool
- Genetically modified organism
- Genetic engineering
- Epigenetics
- List of human genes
- Oscillating gene
- Ridges
References
- ^ Brueckner F, Armache KJ, Cheung A; et al. (2009). "Structure-function studies of the RNA polymerase II elongation complex". Acta Crystallogr. D Biol. Crystallogr. 65 (Pt 2): 112–20. doi:10.1107/S0907444908039875. PMC 2631633. PMID 19171965.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Riccardo S, Tortoriello G, Giordano E, Turano M, Furia M (2007). "The coding/non-coding overlapping architecture of the gene encoding the Drosophila pseudouridine synthase". BMC Mol. Biol. 8: 15. doi:10.1186/1471-2199-8-15. PMC 1821038. PMID 17328797.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G (2004). "Multiple links between transcription and splicing". RNA. 10 (10): 1489–98. doi:10.1261/rna.7100104. PMC 1370635. PMID 15383674.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (2008). "Nucleolus: the fascinating nuclear body". Histochem. Cell Biol. 129 (1): 13–31. doi:10.1007/s00418-007-0359-6. PMC 2137947. PMID 18046571.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Frank DN, Pace NR (1998). "Ribonuclease P: unity and diversity in a tRNA processing ribozyme". Annu. Rev. Biochem. 67: 153–80. doi:10.1146/annurev.biochem.67.1.153. PMID 9759486.
- ^ Ceballos M, Vioque A (2007). "tRNase Z". Protein Pept. Lett. 14 (2): 137–45. PMID 17305600.
- ^ Köhler A, Hurt E (2007). "Exporting RNA from the nucleus to the cytoplasm". Nat. Rev. Mol. Cell Biol. 8 (10): 761–73. doi:10.1038/nrm2255. PMID 17786152.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Jambhekar A, Derisi JL (2007). "Cis-acting determinants of asymmetric, cytoplasmic RNA transport". RNA. 13 (5): 625–42. doi:10.1261/rna.262607. PMC 1852811. PMID 17449729.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Amaral PP, Dinger ME, Mercer TR, Mattick JS (2008). "The eukaryotic genome as an RNA machine". Science (journal). 319 (5871): 1787–9. doi:10.1126/science.1155472. PMID 18369136.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hansen TM, Baranov PV, Ivanov IP, Gesteland RF, Atkins JF (2003). "Maintenance of the correct open reading frame by the ribosome". EMBO Rep. 4 (5): 499–504. doi:10.1038/sj.embor.embor825. PMC 1319180. PMID 12717454.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Berk V, Cate JH (2007). "Insights into protein biosynthesis from structures of bacterial ribosomes". Curr. Opin. Struct. Biol. 17 (3): 302–9. doi:10.1016/j.sbi.2007.05.009. PMID 17574829.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Hegde RS, Kang SW (2008). "The concept of translocational regulation". J. Cell Biol. 182 (2): 225–32. doi:10.1083/jcb.200804157. PMC 2483521. PMID 18644895.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Hebert DN, Molinari M (2007). "In and out of the ER: protein folding, quality control, degradation, and related human diseases". Physiol. Rev. 87 (4): 1377–408. doi:10.1152/physrev.00050.2006. PMID 17928587.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Russell R (2008). "RNA misfolding and the action of chaperones". Front. Biosci. 13: 1–20. doi:10.2741/2557. PMC 2610265. PMID 17981525.
- ^ Moreau P, Brandizzi F, Hanton S; et al. (2007). "The plant ER-Golgi interface: a highly structured and dynamic membrane complex". J. Exp. Bot. 58 (1): 49–64. doi:10.1093/jxb/erl135. PMID 16990376.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Prudovsky I, Tarantini F, Landriscina M; et al. (2008). "Secretion without Golgi". J. Cell. Biochem. 103 (5): 1327–43. doi:10.1002/jcb.21513. PMC 2613191. PMID 17786931.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, Lian JB, van Wijnen AJ, Stein GS (2004). "Intranuclear trafficking: organization and assembly of regulatory machinery for combinatorial biological control". J. Biol. Chem. 279 (42): 43363–6. doi:10.1074/jbc.R400020200. PMID 15277516.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF (2009). "RNA regulation of epigenetic processes". Bioessays. 31 (1): 51–9. doi:10.1002/bies.080099. PMID 19154003.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Martinez NJ, Walhout AJ (2009). "The interplay between transcription factors and microRNAs in genome-scale regulatory networks". Bioessays. 31 (4): 435–45. doi:10.1002/bies.200800212. PMID 19274664.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Tomilin NV (2008). "Regulation of mammalian gene expression by retroelements and non-coding tandem repeats". Bioessays. 30 (4): 338–48. doi:10.1002/bies.20741. PMID 18348251.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Veitia RA (2008). "One thousand and one ways of making functionally similar transcriptional enhancers". Bioessays. 30 (11–12): 1052–7. doi:10.1002/bies.20849. PMID 18937349.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Nguyen T, Nioi P, Pickett CB (2009). "The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress". J. Biol. Chem. 284 (20): 13291–5. doi:10.1074/jbc.R900010200. PMID 19182219.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Paul S (2008). "Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches". Bioessays. 30 (11–12): 1172–84. doi:10.1002/bies.20852. PMID 18937370.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Los M, Maddika S, Erb B, Schulze-Osthoff K (2009). "Switching Akt: from survival signaling to deadly response". Bioessays. 31 (5): 492–5. doi:10.1002/bies.200900005. PMID 19319914.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Song Y, Wang W, Qu X, Sun S (2009). "Effects of hypoxia inducible factor-1alpha (HIF-1alpha) on the growth & adhesion in tongue squamous cell carcinoma cells". Indian J. Med. Res. 129 (2): 154–63. PMID 19293442.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hanriot L, Keime C, Gay N; et al. (2008). "A combination of LongSAGE with Solexa sequencing is well suited to explore the depth and the complexity of transcriptome". BMC Genomics. 9: 418. doi:10.1186/1471-2164-9-418. PMC 2562395. PMID 18796152.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Wheelan SJ, Martínez Murillo F, Boeke JD (2008). "The incredible shrinking world of DNA microarrays". Mol Biosyst. 4 (7): 726–32. doi:10.1039/b706237k. PMC 2535915. PMID 18563246.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Miyakoshi M, Nishida H, Shintani M, Yamane H, Nojiri H (2009). "High-resolution mapping of plasmid transcriptomes in different host bacteria". BMC Genomics. 10: 12. doi:10.1186/1471-2164-10-12. PMC 2642839. PMID 19134166.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Denoeud F, Aury JM, Da Silva C; et al. (2008). "Annotating genomes with massive-scale RNA sequencing". Genome Biol. 9 (12): R175. doi:10.1186/gb-2008-9-12-r175. PMC 2646279. PMID 19087247.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
External links
- "Genes & Gene Expression". The Virtual Library of Biochemistry and Cell Biology. BioChemWeb.org. 2005-12-04. Retrieved 2008-06-10.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
- John Kryk (2008-05-28). "DNA makes RNA". Retrieved 2008-06-10.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
- "Advancing Gene Expression Studies". Genetic Engineering & Biotechnology News. Mary Ann Liebert, Inc. 2008-08-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
- "Optimizing Transient Gene Expression". Genetic Engineering & Biotechnology News. Mary Ann Liebert, Inc. 2008-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
