Most recent common ancestor
| Part of a series on |
| Genetic genealogy |
|---|
| Concepts |
| Related topics |
In genetics, the most recent common ancestor (MRCA) of any set of organisms is the most recent individual from which all organisms in the group are directly descended. The term is often applied to human genealogy.
The MRCA of a set of individuals can sometimes be determined by referring to an established pedigree. However, in general, it is impossible to identify the specific MRCA of a large set of individuals, but an estimate of the time at which the MRCA lived can often be given. Such time to MRCA (TMRCA) estimates can be given based on DNA test results and established mutation rates as practiced in genetic genealogy, or by reference to a non-genetic, mathematical model or computer simulation.
The term MRCA is usually used to describe a common ancestor of individuals within a species. It can also be used to describe a common ancestor between species. To avoid confusion, last common ancestor (LCA) or the equivalent term concestor is sometimes used in place of MRCA when discussing ancestry between species.
The term MRCA may also be used to identify a common ancestor between a set of organisms via specific gene pathways. Mitochondrial Eve and Y-chromosomal Adam are examples of such MRCAs. Such genealogies in reality trace ancestry of individual genes, not organisms. As a result, TMRCA estimates for genetic MRCAs are necessarily greater than those for MRCAs of organisms.
MRCA of all living humans
Tracing one person's lineage back in time forms a binary tree of parents, grandparents, great-grandparents and so on. However, the number of individuals in such an ancestor tree grows exponentially and will eventually become impossibly high. For example, an individual human alive today would, over 30 generations, going back to about the High Middle Ages, have 230 or about 1.07 billion ancestors, more than the total world population at the time.[1]
In reality, an ancestor tree is not a binary tree. Rather, pedigree collapse changes the binary tree to a directed acyclic graph.
Consider the formation, one generation at a time, of the ancestor graph of all currently living humans with no descendants. Start with living people with no descendants at the bottom of the graph. Adding the parents of all those individuals at the top of the graph will connect (half-) siblings via one or two common ancestors, their parent(s). Adding the next generation will connect all first cousins. As each of the following generations of ancestors is added to the top of the graph, the relationship between more and more people is mapped (second cousins, third cousins and so on). Eventually a generation is reached where one or more of the many top-level ancestors is an MRCA from whom it is possible to trace a path of direct descendants all the way down to every living person at the bottom generations of the graph.
The MRCA of everyone alive today could thus have co-existed with a large human population, most of whom either have no living descendants today or else are ancestors of a subset of people alive today. The existence of an MRCA does therefore not imply the existence of a population bottleneck or first couple.
It is incorrect to assume that the MRCA passed all of his or her genes (or indeed any single gene) down to every person alive today. Because of sexual reproduction, at every generation, an ancestor only passes half of his or her genes to each particular descendant in the next generation. Save for inbreeding, the percentage of genes inherited from the MRCA becomes smaller and smaller in individuals at every successive generation, sometimes even decreasing to zero (at which point the Ship of Theseus situation arises[2]), as genes inherited from contemporaries of MRCA are interchanged via sexual reproduction.[3] [4]
It is estimated that the MRCA lived between 5,000 to 2,000 years ago.[5][4]
Identical ancestors point
The MRCA of all living humans had many contemporary companions of both sexes. Many of these contemporaries left direct descendants, but not all of them left an unbroken link of descendants all the way down to today's population. That is, some contemporaries of the MRCA are ancestors of no one in the current population. The rest of the contemporaries of the MRCA may claim ancestry over only a subset of current population.
Because ancestors of the MRCA are by definition also common ancestors, we can continue to find (less recent) common ancestors by pushing further back in time to more and more ancient common ancestors of all people alive today. Eventually we reach a point in the past where all humans can be divided into two groups: those who left no descendants today and those who are common ancestors of all living humans today. This point in time is termed the identical ancestors point. Even though each living person receives genes (in original or mutated forms) in dramatically different proportions from these ancestors from the identical ancestors point,[4] from this point back, all living people share exactly the same set of ancestors, all the way to the very first single-celled organism.[1]
The identical ancestors point for Homo sapiens has been estimated to between 15,000 and 5,000 years ago.[5][4]
MRCA of different species
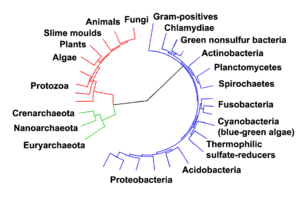
It is also possible to use the term MRCA to describe the common ancestor of two or more different species. In the past, the term MRCA was used interchangeably with last common ancestor (LCA) to denote both the common ancestor within a species and that between species. But MRCA is now more frequently used to describe common ancestors within a species. On the other hand, LCA now describes the common ancestor between two species.
The concept of the last common ancestor is described in Richard Dawkins' book, The Ancestor's Tale, in which he imagines a 'pilgrimage' backwards in time, during which we humans travel back through our own evolutionary history and as we do so are joined at each successive stage by all the other species of organism with which we share each respective common ancestor. Dawkins uses the word "concestor" (coined by Nicky Warren) as an alternative to LCA.
In The Ancestor's Tale, following the human evolutionary tree backwards, we first meet the concestor which we share with the species that are our closest relatives, the chimpanzee and bonobo. Dawkins estimates this to have occurred between 5 and 7 million years ago. Another way of looking at this is to say that our (approximately) 250,000-greats-grandparent was a creature from which all humans, chimpanzees and bonobos are directly descended. Further on in Dawkins' imaginary journey, we meet the concestor we share with the Gorilla, our next nearest relative, then the Orangutan, and so on, until we finally meet the concestor of all living organisms, known as the last universal ancestor.
A common mistake made by the public as well as some authors is to refer to a proposed last common ancestor as an earliest ancestor. Examples include the book The Link: Uncovering Our Earliest Ancestor by Colin Tudge,[7] and a documentary of the same name screened on the History Channel (US) and BBC One (UK) [8], both referring to the primate fossil dubbed Ida.
MRCA of a population identified by a single genetic marker
It is also possible to consider the ancestry of individual genes (coalescent theory), instead of a person (an organism) as a whole. Unlike organisms, a gene is passed down from a generation of organisms to the next generation either as perfect replicas of itself or as slightly mutated descendant genes. While organisms have ancestry graphs and progeny graphs via sexual reproduction, a gene has a single chain of ancestors and a tree of descendants. An organism produced by sexual, allogamous reproduction has at least two ancestors (immediate parents), but a gene always has one single ancestor.
Given any gene in the body of a person, we can trace a single chain of human ancestors back in time, following the lineage of this one gene. Because a typical organism is built from tens of thousands of genes, there are numerous ways to trace the ancestry of organisms using this mechanism. But all these inheritance pathways share one common feature. If we start with all humans alive in 1995 and trace their ancestry by one particular gene (actually a locus), we find that the farther we move back in time, the smaller the number of ancestors becomes. The pool of ancestors continues to shrink until we find the most recent common ancestor (MRCA) of all humans who were alive in 1995, via this particular gene pathway.[1]
Patrilineal and matrilineal MRCA
It is not possible to trace human ancestry via autosomal chromosomes. Although an autosomal chromosome contains genes that are passed down from parents to children via independent assortment from only one of the two parents, genetic recombination (chromosomal crossover) mixes genes from non-sister chromatids from both parents during meiosis, thus changing the genetic composition of the chromosome.
However, the mitochondrial DNA (mtDNA) is nearly immune to sexual mixing, unlike the nuclear DNA whose chromosomes are shuffled and recombined in Mendelian inheritance. Mitochondrial DNA, therefore, can be used to trace matrilineal inheritance and to find the Mitochondrial Eve (also known as the African Eve), the most recent common ancestor of all humans via the mitochondrial DNA pathway.
Similarly Y chromosome is present as a single sex chromosome in the male individual and is passed on to sons and grandsons without recombination. It can be used to trace patrilineal inheritance and to find the Y-chromosomal Adam, the most recent common ancestor of all humans via the Y-DNA pathway.
Mitochondrial Eve and Y-chromosomal Adam have been established by researchers using genealogical DNA tests. Mitochondrial Eve is estimated to have lived about 140,000 years ago. Y-chromosomal Adam is estimated to have lived around 60,000 years ago. The MRCA of humans alive today would therefore need to have lived more recently than either.[3] [9]
Time to MRCA estimates
Different types of MRCAs are estimated to have lived at different times in the past. These time to MRCA (TMRCA) estimates are also computed differently depending on the type of MRCA being considered. Patrilineal and matrilineal MRCAs (Mitochondrial Eve and Y-chromosomal Adam) are traced by single gene markers, thus their TMRCA are computed based on DNA test results and established mutation rates as practiced in genetic genealogy. Time to genealogical MRCA of all living humans is computed based on non-genetic, methematical models and computer simulations.
Since Mitochondrial Eve and Y-chromosomal Adam are traced by single genes via a single ancestral parent lines, the time to these genetic MRCAs will necessarily be greater than that for the genealogical MRCA. This is because single genes will coalesce slower than tracing of conventional human genealogy via both parents. The latter considers only individual humans, without taking into account whether any gene from the computed MRCA actually survives in every single person in the current population.[10]
TMRCA via genetic markers
Mitochondrial DNA can be used to trace the ancestry of a set of populations. In this case, populations are defined by the accumulation of mutations on the mtDNA, and special trees are created for the mutations and the order in which they occurred in each population. The tree is formed through the testing of a large number of individuals all over the world for the presence or lack of a certain set of mutations. Once this is done it is possible to determine how many mutations separate one population from another. The number of mutations, together with estimated mutation rate of the mtDNA in the regions tested, allows scientists to determine the approximate time to MRCA (TMRCA) which indicates time passed since the populations last shared the same set of mutations or belonged to the same haplogroup.
In the case of Y-Chrosomal DNA, TMRCA is arrived at in a different way. Y-DNA haplogroups are defined by single-nucleotide polymorphism in various regions of the Y-DNA. The tim to MRCA within a haplogroup is defined by the accumulation of mutations in STR sequences of the Y-Chromosome of that haplogroup only. Y-DNA network analysis of Y-STR haplotypes showing a non-star cluster indicates Y-STR variability due to multiple founding individuals. Analysis yielding a star cluster can be regarded as representing a population descended from a single ancestor. In this case the variability of the Y-STR sequence, also called the microsatellite variation, can be regarded as a measure of the time passed since the ancestor founded this particular population. The descendants of Genghis Khan or one of his ancestors represents a famous star cluster than can be dated back to the time of Genghis Khan.[11]
TMRCA calculations are considered critical evidence when attempting to determine migration dates of various populations as they spread around the world. For example, if a mutation is deemed to have occurred 30,000 years ago, then this mutation should be found amongst all populations that diverged after this date. If archeological evidence indicates cultural spread and formation of regionally isolated populations then this must be reflected in the isolation of subsequent genetic mutations in this region. If genetic divergence and regional divergence coincide it can be concluded that the observed divergence is due to migration as evidenced by the archaeological record. However, if the date of genetic divergence occurs at a different time than the archaeological record, then scientists will have to look at alternate archaeological evidence to explain the genetic divergence. The issue is best illustrated in the debate surrounding the demic diffusion versus cultural diffusion during the European Neolithic.[12]
TMRCA of all living humans
Estimating time to MRCA of all humans based on the common genealogical usage of the term 'ancestor' is much harder and less accurate compared to estimates of Patrilineal and matrilineal MRCAs. Researches must trace ancestry along both female and male parental lines, and rely on historical and archaeological records.
Depending on the survival of isolated lineages without admixture from modern migrations and taking into account long-isolated peoples, such as historical tribes in central Africa, Australia, and remote islands in the South Pacific, the human MRCA was generally assumed to have lived in the Upper Paleolithic period. With the advent of mathematical models and computer simulations, however, researchers now find that the MRCA of all humans lived remarkably recently, between 2,000 and 4,000 years ago.
Rohde, Olson, and Chang (2004) constructed a mathematical model that considered the tendency of individuals to choose mates from the same social group, as well as the relative geographical isolation of such groups[4]. The mathematical model with one particular set of parameters showed that the MRCA lived about the year 300 BC, and yielded an identical ancestor point (IAP) of 3,000 BC.
The same 2004 Rohde paper also presented results from a computer program based on Monte Carlo simulation designed to overcome some of the limitations of the mathematical model. The program took into consideration realistic population substructure and migration patterns, allowing the researchers to simulate historical human demography. A conservative simulation yielded a mean MRCA date of 1,415 BC and a mean IA date of 5,353 BC. A less conservative simulation gave an MRCA date of AD 55 and an IA date of 2,158 BC.[4]
An explanation of this recent MRCA date is that, while humanity's MRCA was indeed a Paleolithic individual up to early modern times, the European explorers of the 16th and 17th centuries would have fathered enough offspring so that some "mainland" ancestry by today pervades even remote habitats. The possibility remains, however, that a single isolated population with no recent "mainland" admixture persists somewhere, which would immediately push back the date of humanity's MRCA by many millennia. While simulations help estimate probabilities, the question can be resolved authoritatively only by genetically testing every living human individual.
The paper suggests, "No matter the languages we speak or the color of our skin, we share ancestors who planted rice on the banks of the Yangtze, who first domesticated horses on the steppes of the Ukraine, who hunted giant sloths in the forests of North and South America, and who labored to build the Great Pyramid of Khufu".[4]
However, the assumption that there are no isolated populations anymore is questionable in view of the continuing existence of various uncontacted peoples, who are suspected to have been isolated completely for potentially many lillenia, including the Sentinelese who have not only been isoltated from the western world, but also from the neighboring Asian mainland.
See also
- Clade - a group consisting of an organism and all its descendants
- Coalescent theory - a retrospective model of population genetics
- Genealogy - the study of families and the tracing of their lineages and history
- Genetic distance - the genetic divergence between species or between populations within a species
- Genetic genealogy - the application of genetics to traditional genealogy
- Genealogical DNA tests - examination of nucleotides at specific locations on a person's DNA for genetic genealogy purposes
- Lowest common ancestor - an analogous concept in graph theory and computer science
- Phylogenetic tree - a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species
- Timeline of evolution - outlines the major events in the development of life on the planet Earth
- Timeline of human evolution - outlines the major events in the development of human species
References
- ^ a b c See the chapter All Africa and her progenies in Dawkins, Richard (1995). River Out of Eden. New York: Basic Books. ISBN 0-465-06990-8.
- ^ Zhaxybayeva, Olga; Lapierre, Pascal; Gogarten, J. Peter (May 2004). "Genome mosaicism and organismal lineages" (PDF). Trends in Genetics. 20 (5). Department of Molecular and Cell Biology, University of Connecticut: Elsevier: 254–260. doi:10.1016/j.tig.2004.03.009. PMID 15109780. Retrieved 2009-02-19.
The Ship of Theseus paradox […] is frequently invoked to illustrate this point […]. Even moderate levels of gene transfer will make it impossible to reconstruct the genomes of early ancestors; …
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Dawkins, Richard (2004). The Ancestor's Tale, A Pilgrimage to the Dawn of Life. Boston: Houghton Mifflin Company. ISBN 0-618-00583-8.
- ^ a b c d e f g Rohde DL, Olson S, Chang JT (2004). "Modelling the recent common ancestry of all living humans". Nature. 431 (7008): 562–6. doi:10.1038/nature02842. PMID 15457259.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Rohde, DLT , On the common ancestors of all living humans. Submitted to American Journal of Physical Anthropology. (2003)
- ^ Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–7. doi:10.1126/science.1123061. PMID 16513982.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tudge, Colin. (2009). The Link: Uncovering Our Earliest Ancestors. Little Brown.
- ^ Elizabeth Cline (May 22, 2009). "Ida-lized! The Branding of a Fossil". Seed Magazine.
- ^ Notions such as Mitochondrial Eve and Y-chromosomal Adam yield common ancestors that are more ancient than for all living humans (Hartwell 2004:539).
- ^ Chang, Joseph T. (1999). "Recent common ancestors of all present-day individuals" (PDF). Advances in Applied Probability. 31 (4): 1002–26, discussion and author's reply, 1027–38. doi:10.1239/aap/1029955256. Retrieved 2008-01-29.
{{cite journal}}: Invalid|ref=harv(help) - ^ Tatiana Zerjal (2003), The Genetic Legacy of the Mongols, http://web.unife.it/progetti/genetica/Giorgio/PDFfiles/ajhg2003.pdf
- ^ Morelli L, Contu D, Santoni F, Whalen MB, Francalacci P; et al. (2010). "A Comparison of Y-Chromosome Variation in Sardinia and Anatolia Is More Consistent with Cultural Rather than Demic Diffusion of Agriculture". PLoS ONE. 5: e10419. doi:10.1371/journal.pone.0010419. PMC 2861676. PMID 20454687.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link)
Further reading
- Hartwell, Leland (2004). Genetics: From Genes to Genomes (2nd ed.). Maidenhead: McGraw-Hill. ISBN 0072919302.
- Walsh B (2001). "Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals" (PDF). Genetics. 158 (2): 897–912. PMC 1461668. PMID 11404350.
{{cite journal}}: Unknown parameter|month=ignored (help)
