Human genome
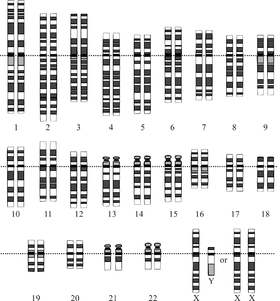
The human genome is the genome of Homo sapiens, which is composed of 24 distinct chromosomes with a total of approximately 3 billion DNA base pairs organized into an estimated 20,000-25,000 genes.[1] The Human Genome Project produced a reference sequence of the euchromatic human genome, which is used worldwide in biomedical sciences. The human genome is much more gene-sparse than was initially predicted at the outset of the Human Genome Project, with only about 1.5% of the total length serving as protein-coding genes.[2]
Features
Chromosomes

There are 24 distinct human chromosomes: 22 autosomal chromosomes, plus the sex-determining X and Y chromosomes. Chromosomes 1-22 are numbered roughly in order of decreasing size. Somatic cells usually have one copy of chromosomes 1-22 from each parent, plus an X chromosome from the mother, and either an X or Y chromosome from the father, for a total of 46.
Genes
There are an estimated 20,000-25,000 human protein-coding genes. The estimate of the number of human genes has been repeatedly revised down from initial predictions of 100,000 or more as genome sequence quality and gene finding methods have improved, and could continue to drop further.
Surprisingly, the number of human genes seems to be less than a factor of two greater that of many much simpler organisms, such as the roundworm and the fruit fly. However, human cells make extensive use of alternative splicing to produce several different proteins from a single gene, and the human proteome is thought to be much larger than those of the aforementioned organisms.
Most human genes have multiple exons, and human introns are frequently much longer than the flanking exons.
Human genes are distributed unevenly across the chromosomes. Each chromosome contains various gene-rich and gene-poor regions, which seem to be correlated with chromosome bands and GC-content. The significance of these nonrandom patterns of gene density is not well understood.
In addition to protein coding genes, the human genome contains several thousand RNA genes, including tRNA, ribosomal RNA, miRNA, and other non-coding RNA genes.
Regulatory sequences
The human genome has many different regulatory sequences which are crucial to controlling gene expression. These are typically short sequences that appear near and within genes. A systematic understanding of these regulatory sequences and how they together act as a gene regulatory network is only beginning to emerge from high-throughput expression and comparative genomics studies.
Identification of regulatory sequences relies in part on evolutionary conservation. The evolutionary branch between the human and mouse, for example, occurred 70-90 million years ago.[3] So computer comparisons of gene sequences that identify conserved non-coding sequences will be an indication of their importance in duties such as gene regulation. [4]
Another comparative genomic approach to locating regulatory sequences in humans is the gene sequencing of the puffer fish. These vertebrates have essentially the same genes and regulatory gene sequences as humans, but with only one-eighth the "junk" DNA. The compact DNA sequence of the puffer fish makes it much easier to locate the regulatory genes.[5]
Other DNA
Protein-coding sequences (specifically exons) comprise less than 1.5% of the human genome.[2] Aside from genes and known regulatory sequences, the human genome contains vast regions of DNA the function of which, if any, remains unknown. These regions in fact comprise the vast majority, by some estimates 97%, of the human genome size. Much of this is comprised of repeat elements, transposons, and pseudogenes, but there is also a large amount of sequence that does not fall under any known classification.
Most of this sequence is possibly an evolutionary artifact that serves no present-day purpose, and these regions are sometimes collectively referred to as "junk" DNA. There are, however, a variety of emerging indications that some sequences within may function in ways that are not currently understood. Recent experiments using microarrays have revealed that a large fraction of non-genic DNA is in fact transcribed into RNA,[6] which leads to the possibility that the resulting transcripts may have some unknown function. Also, the evolutionary conservation across the mammalian genomes of much more sequence than can be explained by protein-coding regions indicates that many, and perhaps most, functional elements in the genome remain unknown.[7] The investigation of the vast quantity of sequence information in the human genome whose function remains unknown is currently a major avenue of scientific inquiry.
Variation
Most studies of human genetic variation have focused on single nucleotide polymorphisms (SNPs), which are substitutions in individual bases along a chromosome. Most analyses estimate that SNPs occur on average somewhere between every 1 in 100 and 1 in 1,000 base pairs in the euchromatic human genome, although they do not occur at a uniform density. Thus follows the popular statement that "all humans are at least 99% genetically identical", although this would be somewhat qualified by most geneticists. A large-scale collaborative effort to catalog SNP variations in the human genome is being undertaken by the International HapMap Project.
The genomic loci and length of certain types of small repetitive sequences are highly variable from person to person, which is the basis of DNA fingerprinting and DNA paternity testing technologies. The heterochromatic portions of the human genome, which total several hundred million base pairs, are also thought to be quite variable within the human population (they are so repetitive and so long that they cannot be accurately sequenced with current technology). These regions contain no genes, and it seems unlikely that any significant phenotypic effect results from typical variation in repeats or heterochromatin.
Most gross genomic mutations in germ cells probably result in inviable embryos; however, a number of human diseases are related to large-scale genomic abnormalities. Down syndrome, Turner Syndrome, and a number of other diseases result from nondisjunction of entire chromosomes. Cancer cells frequently have aneuploidy of chromosomes and chromosome arms, although a cause and effect relationship between aneuploidy and cancer has not been established.
Genetic disorders
These conditions are caused by abnormal expression of one or more genes that matches a clinical phenotype. The disorder may be caused by a gene mutation, an abnormal number of chromosomes, or triplet expansion repeat mutations. Defective genes can be inherited from the parents, in which case it is known as a hereditary disease. The current number of genetic disorders is around 4,000, with the most common being cystic fibrosis.
Studies of genetic disorders is often performed by means of population genetics. Treatment is performed by a geneticist-physician trained in clinical genetics. The results of the Human Genome Project are likely to provide increased availability of genetic testing for gene-related disorders, and eventually improved treatment. Parents can be screened for hereditary conditions and counselled on the consequences, the probability it will be inherited, and how to avoid or ameliorate it in their offspring.
One major gross effect on human phenotypes derives from gene dosage, whose effects play a role in disorders caused by duplication, omission, or disruption of chromosomes. For example, those afflicted with Down syndrome, or trisomy 21, experience high rates of Alzheimer's disease, an effect thought to be related to the overexpression of the Alzheimer's-related amyloid precursor protein whose gene is located on chromosome 21.[8] By contrast, Down's syndrome sufferers experience lower rates of breast cancer, possibly due to the overexpression of a tumor-suppressor gene.[9]
Evolution
Comparative genomics studies of mammalian genomes suggest that approximately 5% of the human genome has been conserved by evolution since the divergence of those species approximately 200 million years ago, containing the vast majority of genes and regulatory sequences. Intriguingly, since genes and known regulatory sequences probably comprise less than 2% of the genome, this suggests that there may be more unknown functional sequence than known functional sequence. A smaller, but large, fraction of human genes seem to be shared among most known vertebrates.
The chimpanzee genome is approximately 95% identical to the human genome. On average, a typical human protein-coding gene differs from its chimpanzee ortholog by only two amino acid substitutions; nearly one third of human genes have exactly the same protein translation as their chimpanzee orthologs. A major difference between the two genomes is human chromosome 2, which is the product of a fusion between chimpanzee chromosomes 12 and 13.[10]
Humans have undergone an extraordinary loss of olfactory receptor genes during our recent evolution, which explains our relatively crude sense of smell compared to most other mammals. Evolutionary evidence suggests that the emergence of color vision in humans and several other primate species has diminished the need for the sense of smell.[11]
Mitochondrial genome
The human mitochondrial genome, while usually not included when referring to the "human genome", is of tremendous interest to geneticists, since it undoubtedly plays a role in mitochondrial disease. It also sheds light on human evolution; for example, analysis of variation in the human mitochondrial genome has led to the postulation of a Mitochondrial Eve from whom all modern humans are descended.
Due to the lack of a system for checking for copying errors, Mitochondrial DNA (mDNA) has a more rapid rate of variation than nuclear DNA. This 20-fold increase in the mutation rate allows mDNA to be used for more accurate tracing of maternal ancestry. Studies of mDNA in populations have allowed ancient migration paths to be traced, such as the migration of Native Americans from Siberia or Polynesians from southeastern Asia. It has also been used to show that there is no trace of Neanderthal DNA in the European gene mixture.[12]
See also
- Dysgenics
- Eukaryotic chromosome fine structure
- Eugenics
- Human Genome Project
- Karyotype
- Mitochondrial Eve
- Y-chromosomal Adam
References
- ^ International Human Genome Sequencing Consortium (2004). "Finishing the euchromatic sequence of the human genome". Nature. 431 (7011): 931–45. PMID 15496913. [1]
- ^ a b International Human Genome Sequencing Consortium (2001). "Initial sequencing and analysis of the human genome". Nature. 409 (6822): 860–921. PMID 11237011. [2]
- ^ Nei M, Xu P, Glazko G (2001). "Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms". Proc Natl Acad Sci U S A. 98 (5): 2497–502. PMID 11226267.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Loots G, Locksley R, Blankespoor C, Wang Z, Miller W, Rubin E, Frazer K (2000). "Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons". Science. 288 (5463): 136–40. PMID 10753117.
{{cite journal}}: CS1 maint: multiple names: authors list (link) Summary - ^ Meunier, Monique. "Genoscope and Whitehead announce a high sequence coverage of the Tetraodon nigroviridis genome". Genoscope. Retrieved 2006-09-12.
- ^ "...a tiling array with 5-nucleotide resolution that mapped transcription activity along 10 human chromosomes revealed that an average of 10% of the genome (compared to the 1 to 2% represented by bona fide exons) corresponds to polyadenylated transcripts, of which more than half do not overlap with known gene locations.Claverie J (2005). "Fewer genes, more noncoding RNA". Science. 309 (5740): 1529–30. PMID 16141064.
- ^ "...the proportion of small (50-100 bp) segments in the mammalian genome that is under (purifying) selection can be estimated to be about 5%. This proportion is much higher than can be explained by protein-coding sequences alone, implying that the genome contains many additional features (such as untranslated regions, regulatory elements, non-protein-coding genes, and chromosomal structural elements) under selection for biological function." Mouse Genome Sequencing Consortium (2002). "Initial sequencing and comparative analysis of the mouse genome". Nature. 420 (6915): 520–62. PMID 12466850.
- ^ Armstrong R, Cairns N, Myers D, Smith C, Lantos P, Rossor M (1996). "A comparison of beta-amyloid deposition in the medial temporal lobe in sporadic Alzheimer's disease, Down's syndrome and normal elderly brains". Neurodegeneration. 5 (1): 35–41. PMID 8731380.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kwak HI, Gustafson T, Metz RP, Laffin B, Schedin P, Porter WW. "Inhibition of breast cancer growth and invasion by single-minded 2s". Carcinogenesis. epub. PMID 16840439.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Human chromosome 2 resulted from a fusion of two ancestral chromosomes that remained separate in the chimpanzee lineage" The Chimpanzee Sequencing and Analysis Consortium (2005). "Initial sequence of the chimpanzee genome and comparison with the human genome". Nature. 437 (7055): 69–87. PMID 16136131.
"Large-scale sequencing of the chimpanzee genome is now imminent."Olson M, Varki A (2003). "Sequencing the chimpanzee genome: insights into human evolution and disease". Nat Rev Genet. 4 (1): 20–8. PMID 12509750. - ^ "Our findings suggest that the deterioration of the olfactory repertoire occurred concomitant with the acquisition of full trichromatic color vision in primates." Gilad Y, Wiebe V, Przeworski M, Lancet D, Pääbo S (2004). "Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates". PLoS Biol. 2 (1): E5. PMID 14737185.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sykes, Bryan (2003-10-09). "Mitochondrial DNA and human history". The Human Genome. Retrieved 2006-09-19.
- Lindblad-Toh K; et al. (2005). "Genome sequence, comparative analysis and haplotype structure of the domestic dog". Nature. 438 (7069): 803–19. PMID 16341006.
{{cite journal}}: Explicit use of et al. in:|author=(help)[3]
External links
- The National Human Genome Research Institute
- National Library of Medicine human genome viewer .
- UCSC Genome Browser.
- Human Genome Project.
- Sabancı University School of Languages Podcasts What makes us different from chimpanzees? by Andrew Berry (MP3 file)
- The National Office of Public Health Genomics
