Hematopoietic stem cell
This article may be too technical for most readers to understand. (August 2015) |
| Hematopoietic stem cell | |
|---|---|
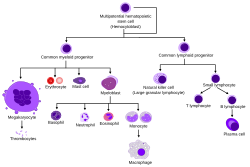 Overview of normal human hematopoiesis | |
| Details | |
| Identifiers | |
| Latin | Cellula haematopoietica praecursoria |
| MeSH | D006412 |
| TH | H2.00.01.0.00006 |
| Anatomical terminology | |
Hematopoietic stem cells (HSCs) or hemocytoblasts are the stem cells that give rise to all the other blood cells through the process of haematopoiesis.[1] They are derived from mesoderm and located in the red bone marrow, which is contained in the core of most bones.
HSCs give rise to both the myeloid and lymphoid lineages of blood cells. (Myeloid cells include monocytes, macrophages, neutrophils, basophils, eosinophils, erythrocytes, dendritic cells, and megakaryocytes or platelets. Lymphoid cells include T cells, B cells, and natural killer cells.) The definition of hematopoietic stem cells has evolved since HSCs were first discovered in 1961.[2] The hematopoietic tissue contains cells with long-term and short-term regeneration capacities and committed multipotent, oligopotent, and unipotent progenitors. HSCs constitute 1:10.000 of cells in myeloid tissue.
HSC studies through much of the past half century have led to a much deeper understanding. More recent advances have resulted in the use of HSC transplants in the treatment of cancers and other immune system disorders.[3]
Sources


HSCs are found in the bone marrow of adults, especially in the pelvis, femur, and sternum. They are also found in umbilical cord blood and, in small numbers, in peripheral blood.[4]
Stem and progenitor cells can be taken from the pelvis, at the iliac crest, using a needle and syringe.[5] The cells can be removed as liquid (to perform a smear to look at the cell morphology) or they can be removed via a core biopsy (to maintain the architecture or relationship of the cells to each other and to the bone).[citation needed]
In order to harvest stem cells from the circulating peripheral blood, blood donors are injected with a cytokine, such as granulocyte-colony stimulating factor (G-CSF), that induces cells to leave the bone marrow and circulate in the blood vessels.[citation needed]
In mammalian embryology, the first definitive HSCs are detected in the AGM (aorta-gonad-mesonephros), and then massively expanded in the fetal liver prior to colonising the bone marrow before birth.[6]
Functional characteristics
Multipotency and self-renewal
HSCs can replenish all blood cell types (i.e., are multipotent) and self-renew. A small number of HSCs can expand to generate a very large number of daughter HSCs. This phenomenon is used in bone marrow transplantation, when a small number of HSCs reconstitute the hematopoietic system. This process indicates that, subsequent to bone marrow transplantation, symmetrical cell divisions into two daughter HSCs must occur.
Stem cell self-renewal is thought to occur in the stem cell niche in the bone marrow, and it is reasonable to assume that key signals present in this niche will be important in self-renewal.[1] There is much interest in the environmental and molecular requirements for HSC self-renewal, as understanding the ability of HSC to replenish themselves will eventually allow the generation of expanded populations of HSC in vitro that can be used therapeutically.
Stem cell heterogeneity
It was originally believed that all HSCs were alike in their self-renewal and differentiation abilities. This view was first challenged by the 2002 discovery by the Muller-Sieburg group in San Diego, who illustrated that different stem cells can show distinct repopulation patterns that are epigenetically predetermined intrinsic properties of clonal Thy-1lo Sca-1+ lin− c-kit+ HSC.[7][8][9] The results of these clonal studies led to the notion of lineage bias. Using the ratio of lymphoid (L) to myeloid (M) cells in blood as a quantitative marker, the stem cell compartment can be split into three categories of HSC. Balanced (Bala) HSCs repopulate peripheral white blood cells in the same ratio of myeloid to lymphoid cells as seen in unmanipulated mice (on average about 15% myeloid and 85% lymphoid cells, or 3 ≤ ρ ≤ 10). Myeloid-biased (My-bi) HSCs give rise to very few lymphocytes resulting in ratios 0 < ρ < 3, while lymphoid-biased (Ly-bi) HSCs generate very few myeloid cells, which results in lymphoid-to-myeloid ratios of ρ > 10. All three types are normal types of HSC, and they do not represent stages of differentiation. Rather, these are three classes of HSC, each with an epigenetically fixed differentiation program. These studies also showed that lineage bias is not stochastically regulated or dependent on differences in environmental influence. My-bi HSC self-renew longer than balanced or Ly-bi HSC. The myeloid bias results from reduced responsiveness to the lymphopoetin interleukin 7 (IL-7).[8]
Subsequently, other groups confirmed and highlighted the original findings.[10] For example, the Eaves group confirmed in 2007 that repopulation kinetics, long-term self-renewal capacity, and My-bi and Ly-bi are stably inherited intrinsic HSC properties.[11] In 2010, the Goodell group provided additional insights about the molecular basis of lineage bias in side population (SP) SCA-1+ lin− c-kit+ HSC.[12] As previously shown for IL-7 signaling, it was found that a member of the transforming growth factor family (TGF-beta) induces and inhibits the proliferation of My-bi and Ly-bi HSC, respectively.
Behavior in culture
A cobblestone area-forming cell (CAFC) assay is a cell culture-based empirical assay. When plated onto a confluent culture of stromal feeder layer, a fraction of HSCs creep between the gaps (even though the stromal cells are touching each other) and eventually settle between the stromal cells and the substratum (here the dish surface) or trapped in the cellular processes between the stromal cells. Emperipolesis is the in vivo phenomenon in which one cell is completely engulfed into another (e.g. thymocytes into thymic nurse cells); on the other hand, when in vitro, lymphoid lineage cells creep beneath nurse-like cells, the process is called pseudoemperipolesis. This similar phenomenon is more commonly known in the HSC field by the cell culture terminology cobble stone area-forming cells (CAFC), which means areas or clusters of cells look dull cobblestone-like under phase contrast microscopy, compared to the other HSCs, which are refractile. This happens because the cells that are floating loosely on top of the stromal cells are spherical and thus refractile. However, the cells that creep beneath the stromal cells are flattened and, thus, not refractile. The mechanism of pseudoemperipolesis is only recently coming to light. It may be mediated by interaction through CXCR4 (CD184) the receptor for CXC Chemokines (e.g., SDF1) and α4β1 integrins.[13]
Mobility
HSCs have a higher potential than other immature blood cells to pass the bone marrow barrier, and, thus, may travel in the blood from the bone marrow in one bone to another bone. If they settle in the thymus, they may develop into T cells. In the case of fetuses and other extramedullary hematopoiesis, HSCs may also settle in the liver or spleen and develop.
This enables HSCs to be harvested directly from the blood.
Physical characteristics
With regard to morphology, hematopoietic stem cells resemble lymphocytes. They are non-adherent, and rounded, with a rounded nucleus and low cytoplasm-to-nucleus ratio. Since HSCs cannot be isolated as a pure population, it is not possible to identify them in a microscope.
Markers
This article may contain an excessive amount of intricate detail that may interest only a particular audience. (August 2015) |
HSCs can be identified or isolated by the use of flow cytometry where the combination of several different cell surface markers are used to separate the rare HSCs from the surrounding blood cells. HSCs lack expression of mature blood cell markers and are thus, called Lin-. Lack of expression of lineage markers is used in combination with detection of several positive cell-surface markers to isolate HSCs. In addition, HSCs are characterised by their small size and low staining with vital dyes such as rhodamine 123 (rhodamine lo) or Hoechst 33342 (side population).
Cluster of differentiation and other markers
The classical marker of human HSC is CD34 first described independently by Civin et al. and Tindle et al.[14][15][16][17] It is used to isolate HSC for reconstitution of patients who are haematologically incompetent as a result of chemotherapy or disease.
Many markers belong to the cluster of differentiation series, like: CD34, CD38, CD90, CD133, CD105, CD45, and also c-kit, - the receptor for stem cell factor.
There are many differences between the human and mice hematopoietic cell markers for the commonly accepted type of hematopoietic stem cells.[18]
- Mouse HSC: CD34lo/−, SCA-1+, Thy1.1+/lo, CD38+, C-kit+, lin−
- Human HSC: CD34+, CD59+, Thy1/CD90+, CD38lo/−, C-kit/CD117+, lin−
However, not all stem cells are covered by these combinations that, nonetheless, have become popular. In fact, even in humans, there are hematopoietic stem cells that are CD34−/CD38−.[19][20] Also some later studies suggested that earliest stem cells may lack c-kit on the cell surface.[21] For human HSCs use of CD133 was one step ahead as both CD34+ and CD34− HSCs were CD133+.
Traditional purification method used to yield a reasonable purity level of mouse hematopoietic stem cells, in general, requires a large(~10-12) battery of markers, most of which were surrogate markers with little functional significance, and thus partial overlap with the stem cell populations and sometimes other closely related cells that are not stem cells. Also, some of these markers (e.g., Thy1) are not conserved across mouse species, and use of markers like CD34− for HSC purification requires mice to be at least 8 weeks old.
SLAM code
Alternative methods that could give rise to a similar or better harvest of stem cells is an active area of research, and are presently[when?] emerging. One such method uses a signature of SLAM family of cell surface molecules. The SLAM (Signaling lymphocyte activation molecule) family is a group of more than 10 molecules whose genes are located mostly tandemly in a single locus on chromosome 1 (mouse), all belonging to a subset of the immunoglobulin gene superfamily, and originally thought to be involved in T-cell stimulation. This family includes CD48, CD150, CD244, etc., CD150 being the founding member, and, thus, also known as slamF1, i.e., SLAM family member 1.
The signature SLAM codes for the hemopoietic hierarchy are:
- Hematopoietic stem cells (HSC): CD150+CD48−CD244−
- Multipotent progenitor cells (MPPs): CD150−CD48−CD244+
- Lineage-restricted progenitor cells (LRPs): CD150−CD48+CD244+
- Common myeloid progenitor (CMP): lin−SCA-1−c-kit+CD34+CD16/32mid
- Granulocyte-macrophage progenitor (GMP): lin−SCA-1−c-kit+CD34+CD16/32hi
- Megakaryocyte-erythroid progenitor (MEP): lin−SCA-1−c-kit+CD34−CD16/32low
For HSCs, CD150+CD48− was sufficient instead of CD150+CD48−CD244− because CD48 is a ligand for CD244, and both would be positive only in the activated lineage-restricted progenitors. It seems that this code was more efficient than the more tedious earlier set of the large number of markers, and are also conserved across the mouse strains; however, recent work has shown that this method excludes a large number of HSCs and includes an equally large number of non-stem cells.[22][23] CD150+CD48− gave stem cell purity comparable to Thy1loSCA-1+lin−c-kit+ in mice.[24]
LT-HSC/ST-HSC/early MPP/late MPP
Irving Weissman's group at Stanford University was the first to isolate mouse hematopoietic stem cells in 1988[citation needed] and was also the first to work out the markers to distinguish the mouse long-term (LT-HSC) and short-term (ST-HSC) hematopoietic stem cells (self-renew-capable), and the Multipotent progenitors (MPP, low or no self-renew capability — the later the developmental stage of MPP, the lesser the self-renewal ability and the more of some of the markers like CD4 and CD135):
- LT-HSC: CD34−, CD38−, SCA-1+, Thy1.1+/lo, C-kit+, lin−, CD135−, Slamf1/CD150+
- ST-HSC: CD34+, CD38+, SCA-1+, Thy1.1+/lo, C-kit+, lin−, CD135−, Slamf1/CD150+, Mac-1 (CD11b)lo
- Early MPP: CD34+, SCA-1+, Thy1.1−, C-kit+, lin−, CD135+, Slamf1/CD150−, Mac-1 (CD11b)lo, CD4lo
- Late MPP: CD34+, SCA-1+, Thy1.1−, C-kit+, lin−, CD135high, Slamf1/CD150−, Mac-1 (CD11b)lo, CD4lo
Nomenclature of hematopoietic colonies and lineages
Between 1948 and 1950, the Committee for Clarification of the Nomenclature of Cells and Diseases of the Blood and Blood-forming Organs issued reports on the nomenclature of blood cells.[25][26] An overview of the terminology is shown below, from earliest to final stage of development:
- [root]blast
- pro[root]cyte
- [root]cyte
- meta[root]cyte
- mature cell name
The root for erythrocyte colony-forming units (CFU-E) is "rubri", for granulocyte-monocyte colony-forming units (CFU-GM) is "granulo" or "myelo" and "mono", for lympocyte colony-forming units (CFU-L) is "lympho" and for megakaryocyte colony-forming units (CFU-Meg) is "megakaryo". According to this terminology, the stages of red blood cell formation would be: rubriblast, prorubricyte, rubricyte, metarubricyte, and erythrocyte. However, the following nomenclature seems to be, at present, the most prevalent:
| Committee | "lympho" | "rubri" | "granulo" or "myelo" | "mono" | "megakaryo" |
|---|---|---|---|---|---|
| Lineage | Lymphoid | Myeloid | Myeloid | Myeloid | Myeloid |
| CFU | CFU-L | CFU-GEMM→CFU-E | CFU-GEMM→CFU-GM→CFU-G | CFU-GEMM→CFU-GM→CFU-M | CFU-GEMM→CFU-Meg |
| Process | lymphocytopoiesis | erythropoiesis | granulocytopoiesis | monocytopoiesis | thrombocytopoiesis |
| [root]blast | Lymphoblast | Proerythroblast | Myeloblast | Monoblast | Megakaryoblast |
| pro[root]cyte | Prolymphocyte | Polychromatophilic erythrocyte | Promyelocyte | Promonocyte | Promegakaryocyte |
| [root]cyte | - | Normoblast | Eosino/neutro/basophilic myelocyte | Megakaryocyte | |
| meta[root]cyte | Large lymphocyte | Reticulocyte | Eosinophilic/neutrophilic/basophilic metamyelocyte, Eosinophilic/neutrophilic/basophilic band cell | Early monocyte | - |
| mature cell name | Small lymphocyte | Erythrocyte | granulocytes (Eosino/neutro/basophil) | Monocyte | thrombocytes (Platelets) |
Osteoclasts also arise from hemopoietic cells of the monocyte/neutrophil lineage, specifically CFU-GM.
Colony-forming units
In the context of hematopoietic stem cells, a colony-forming unit is a subtype of HSC. (This sense of the term is different from colony-forming units of microbes, which is a cell counting unit.) There are various kinds of HSC colony-forming units:
- Colony-forming unit–granulocyte-erythrocyte-monocyte-megakaryocyte (CFU-GEMM)
- Colony-forming unit–lymphocyte (CFU-L)
- Colony-forming unit–erythrocyte (CFU-E)
- Colony-forming unit–granulocyte-macrophage (CFU-GM)
- Colony-forming unit–megakaryocyte (CFU-Meg)
- Colony-forming unit–basophil (CFU-B)
- Colony-forming unit–eosinophil (CFU-Eos)
The above CFUs are based on the lineage. Another CFU, the colony-forming unit–spleen (CFU-S), was the basis of an in vivo clonal colony formation, which depends on the ability of infused bone marrow cells to give rise to clones of maturing hematopoietic cells in the spleens of irradiated mice after 8 to 12 days. It was used extensively in early studies, but is now considered to measure more mature progenitor or transit-amplifying cells rather than stem cells.
HSC repopulation kinetics
Hematopoietic stem cells (HSC) cannot be easily observed directly, and, therefore, their behaviors need to be inferred indirectly. Clonal studies are likely the closest technique for single cell in vivo studies of HSC. Here, sophisticated experimental and statistical methods are used to ascertain that, with a high probability, a single HSC is contained in a transplant administered to a lethally irradiated host. The clonal expansion of this stem cell can then be observed over time by monitoring the percent donor-type cells in blood as the host is reconstituted. The resulting time series is defined as the repopulation kinetic of the HSC.
The reconstitution kinetics are very heterogeneous. However, using symbolic dynamics, one can show that they fall into a limited number of classes.[27] To prove this, several hundred experimental repopulation kinetics from clonal Thy-1lo SCA-1+ lin− c-kit+ HSC were translated into symbolic sequences by assigning the symbols "+", "-", "~" whenever two successive measurements of the percent donor-type cells have a positive, negative, or unchanged slope, respectively. By using the Hamming distance, the repopulation patterns were subjected to cluster analysis yielding 16 distinct groups of kinetics. To finish the empirical proof, the Laplace add-one approach[clarification needed] was used to determine that the probability of finding kinetics not contained in these 16 groups is very small. By corollary, this result shows that the hematopoietic stem cell compartment is also heterogeneous by dynamical criteria.
DNA damage and aging
DNA strand breaks accumulate in long term HSCs during aging.[28] This accumulation is associated with a broad attenuation of DNA repair and response pathways that depends on HSC quiescence.[28] Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template. The NHEJ pathway depends on several proteins including ligase 4, DNA polymerase mu and NHEJ factor 1 (NHEJ1, also known as Cernunnos or XLF).
DNA ligase 4 (Lig4) has a highly specific role in the repair of double-strand breaks by NHEJ. Lig4 deficiency in the mouse causes a progressive loss of HSCs during aging.[29] Deficiency of lig4 in pluripotent stem cells results in accumulation of DNA double-strand breaks and enhanced apoptosis.[30]
In polymerase mu mutant mice, hematopoietic cell development is defective in several peripheral and bone marrow cell populations with about a 40% decrease in bone marrow cell number that includes several hematopoietic lineages.[31] Expansion potential of hematopoietic progenitor cells is also reduced. These characteristics correlate with reduced ability to repair double-strand breaks in hematopoietic tissue.
Deficiency of NHEJ factor 1 in mice leads to premature aging of hematopoietic stem cells as indicated by several lines of evidence including evidence that long-term repopulation is defective and worsens over time.[32] Using a human induced pluripotent stem cell model of NHEJ1 deficiency, it was shown that NHEJ1 has an important role in promoting survival of the primitive hematopoietic progenitors.[33] These NHEJ1 deficient cells possess a weak NHEJ1-mediated repair capacity that is apparently incapable of coping with DNA damages induced by physiological stress, normal metabolism, and ionizing radiation.[33]
The sensitivity of haematopoietic stem cells to Lig4, DNA polymerase mu and NHEJ1 deficiency suggests that NHEJ is a key determinant of the ability of stem cells to maintain themselves against physiological stress over time.[29] Rossi et al.[34] found that endogenous DNA damage accumulates with age even in wild type HSCs, and suggested that DNA damage accrual may be an important physiological mechanism of stem cell aging.
See also
References
- ^ a b Birbrair, Alexander; Frenette, Paul S. (2016-03-01). "Niche heterogeneity in the bone marrow". Annals of the New York Academy of Sciences. 1370: 82–96. doi:10.1111/nyas.13016. ISSN 1749-6632. PMC 4938003. PMID 27015419.
- ^ Till, J. E.; McCULLOCH, E. A. (1961-02-01). "A direct measurement of the radiation sensitivity of normal mouse bone marrow cells". Radiation Research. 14: 213–222. doi:10.2307/3570892. ISSN 0033-7587. PMID 13776896.
- ^ "5. Hematopoietic Stem Cells." Stem Cell Information. National Institutes of Health, U.S. Department of Health and Human Services, 17 Jun 2011. Web. 9 Nov 2013. <http://stemcells.nih.gov/info/scireport/pages/chapter5.aspx >
- ^ http://cordadvantage.com/cord-blood-101/hematopoietic-stem-cell
- ^ "Bone Marrow Transplant Process". Mayo Clinic. Retrieved 18 March 2015.
- ^ Dzierzak & Speck, Of lineage and legacy: the development of mammalian hematopoietic stem cells, Nature Immunology, 2008
- ^ Muller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB, "Deterministc regulation of haematopoietic stem cell self-renewal and differentiation" Blood 2002; 100; 1302-9
- ^ a b Muller-Sieburg CE, Cho RH, Karlson L, Huang JF, Sieburg HB (2004). "Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished progeny with impaired IL-7 responsiveness". Blood. 103 (11): 4111–8. doi:10.1182/blood-2003-10-3448. PMID 14976059.
- ^ Sieburg HB, Cho RH, Dykstra B, Eaves CJ, Muller-Sieburg CE (2006). "The haematopoietic stem cell compartment consists of a limited number of discrete stem cell subsets". Blood. 107 (6): 2311–6. doi:10.1182/blood-2005-07-2970. PMC 1456063. PMID 16291588.
- ^ Schroeder T (2010). "Haematopoietic Stem Cell Heterogeneity: Subtypes, Not Unpredictable Behavior". Cell Stem Cell. 6 (3): 203–207. doi:10.1016/j.stem.2010.02.006. PMID 20207223.
- ^ Dykstra B, et al. (2007). "Long-Term Propagation of Distinct Hematopoietic Differentiation Programs In Vivo". Cell Stem Cell. 1 (2): 218–229. doi:10.1016/j.stem.2007.05.015. PMID 18371352.
- ^ Challen G, Boles NC, Chambers SM, Goodell MA (2010). "Distinct Haematopoietic Stem Cell Subtypes Are Differentially Regulated by TGF-beta1". Cell Stem Cell. 6 (3): 265–278. doi:10.1016/j.stem.2010.02.002. PMC 2837284. PMID 20207229.
- ^ Burger JA, Spoo A, Dwenger A, Burger M, Behringer D. CXCR4 chemokine receptors (CD184) and alpha4beta1 integrins mediate spontaneous migration of human CD34+ progenitors and acute myeloid leukaemia cells beneath marrow stromal cells (pseudoemperipolesis).
- ^ Civin CI. Strauss LC. Brovall C. Fackler MJ. Schwartz jf. Shaper JH. (1984). "Antigenic analysis of haematopoiesis:III. hematopoietic cell surface antigen defined by a monoclonal antibody raised against KG-1a cells". Journal of Immunology. 133 (1): 157–165.
- ^ Tindle RW. Nichols R. Chan L. Campana D. Catovsky D. Birnie GD (1985). "A novel monoclonal antibody BI-3C5 recognises myeloblasts and non-B non-T lymphoblasts in acute leukaemias and CGL blast crises and reacts with immature cells in normal bone marrow". Leukaemia Research. 9: 1–9. doi:10.1016/0145-2126(85)90016-5.
- ^ Tindle RW. Katz F. Martin SH. Watt D. Catovsky D. Janossy G. Greaves M. (1987). "BI-3C5 (CD34) defines multipotential and lineage restricted progenitor cells and their leukaemic counterparts". In: Leucocyte Typing III:White cell differentiation antigens. Oxford University Press 654-655.
- ^ Loken M. Shah V. Civin CI.. (1987). ""Characterization of myeloid antigens on human bone marrow using multicolour immunofluorescence"". In: Leucocyte Typing III:White cell differentiation antigens.Oxford University Press 630-635.
- ^ http://stemcells.nih.gov/info/scireport/chapter5.asp
- ^ Bhatia M.; Bonnet D.; Murdoch B.; Gan O.I.; Dick J.E. (1998). "A newly discovered class of human haematopoietic cells with SCID-repopulating activity". Nature Medicine. 4 (9): 1038–1045. doi:10.1038/2023. PMID 9734397.
- ^ Guo, Yalin; Lübbert, Michael; Engelhardt, Monika (2003). "CD34- Hematopoietic Stem Cells: Current Concepts and Controversies". Stem Cells. 21 (1): 15–20. doi:10.1634/stemcells.21-1-15. PMID 12529547.
- ^ Doi H, et al. (1997). "Pluripotent hemopoietic stem cells are c-kit<low". Proc. Natl. Acad. Sci. USA. 94 (6): 2513–2517. Bibcode:1997PNAS...94.2513D. doi:10.1073/pnas.94.6.2513. PMC 20119. PMID 9122226.
- ^ David C Weksberg, Stuart M Chambers, Nathan C Boles, and Margaret A Goodell CD150 negative Side Population cells represent a functionally distinct population of long-term haematopoietic stem cells" Blood 2007 : blood-2007-09-115006v1
- ^ Van Zant Gary. "Stem cell markers: less is more!". Blood. 107: 855–856. doi:10.1182/blood-2005-11-4400.
- ^ Kiel; et al. (2005). "SLAM Family Receptors Distinguish Hematopoietic Stem and Progenitor Cells and Reveal Endothelial Niches for Stem Cells". Cell. 121 (7): 1109–1121. doi:10.1016/j.cell.2005.05.026. PMID 15989959.
- ^ "First report of the Committee for Clarification of the Nomenclature of Cells and Diseases of the Blood and Blood-forming Organs". Am J Clin Pathol. 18 (5): 443–450. 1948. PMID 18913573.
- ^ "Third, fourth and fifth reports of the committee for clarification of the nomenclature of cells and diseases of the blood and blood-forming organs". Am J Clin Pathol. 20 (6): 562–79. 1950. PMID 15432355.
- ^ Sieburg HB, Muller-Sieburg CE (2004). "Classification of short kinetics by shape". In Silico Biol. 4 (2): 209–17. PMID 15107024.
- ^ a b Beerman I, Seita J, Inlay MA, Weissman IL, Rossi DJ (2014). "Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle". Cell Stem Cell. 15 (1): 37–50. doi:10.1016/j.stem.2014.04.016. PMC 4082747. PMID 24813857.
- ^ a b Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ (2007). "DNA repair is limiting for haematopoietic stem cells during ageing". Nature. 447 (7145): 686–90. Bibcode:2007Natur.447..686N. doi:10.1038/nature05875. PMID 17554302.
- ^ Tilgner K, Neganova I, Moreno-Gimeno I, Al-Aama JY, Burks D, Yung S, Singhapol C, Saretzki G, Evans J, Gorbunova V, Gennery A, Przyborski S, Stojkovic M, Armstrong L, Jeggo P, Lako M (2013). "A human iPSC model of Ligase IV deficiency reveals an important role for NHEJ-mediated-DSB repair in the survival and genomic stability of induced pluripotent stem cells and emerging haematopoietic progenitors". Cell Death Differ. 20 (8): 1089–100. doi:10.1038/cdd.2013.44. PMC 3705601. PMID 23722522.
- ^ Lucas D, Escudero B, Ligos JM, Segovia JC, Estrada JC, Terrados G, Blanco L, Samper E, Bernad A (2009). "Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair". PLoS Genet. 5 (2): e1000389. doi:10.1371/journal.pgen.1000389. PMC 2638008. PMID 19229323.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Avagyan S, Churchill M, Yamamoto K, Crowe JL, Li C, Lee BJ, Zheng T, Mukherjee S, Zha S (2014). "Hematopoietic stem cell dysfunction underlies the progressive lymphocytopenia in XLF/Cernunnos deficiency". Blood. 124 (10): 1622–5. doi:10.1182/blood-2014-05-574863. PMC 4155271. PMID 25075129.
- ^ a b Tilgner K, Neganova I, Singhapol C, Saretzki G, Al-Aama JY, Evans J, Gorbunova V, Gennery A, Przyborski S, Stojkovic M, Armstrong L, Jeggo P, Lako M (2013). "Brief report: a human induced pluripotent stem cell model of cernunnos deficiency reveals an important role for XLF in the survival of the primitive hematopoietic progenitors". Stem Cells. 31 (9): 2015–23. doi:10.1002/stem.1456. PMID 23818183.
- ^ Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL (2007). "Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age". Nature. 447 (7145): 725–9. Bibcode:2007Natur.447..725R. doi:10.1038/nature05862. PMID 17554309.
External links
- Fact sheet about blood stem cells on EuroStemCell
- Hematopoietic+stem+cells at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

