Pharmacogenomics: Difference between revisions
m tweak cite |
→Web-based resources: moved in-line external links to cites and templated bare URLs |
||
| Line 179: | Line 179: | ||
|- |
|- |
||
|- |
|- |
||
! Data Source !! Main Use !! |
! Data Source !! Main Use !! Citation |
||
|- |
|- |
||
| ''PharmVar'' || A central repository for pharmacogene variation that focuses on haplotype structure and allelic variation || |
| ''PharmVar'' || A central repository for pharmacogene variation that focuses on haplotype structure and allelic variation || <ref name="pmid29134625">{{cite journal | vauthors = Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE | title = The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database | journal = Clinical Pharmacology and Therapeutics | volume = 103 | issue = 3 | pages = 399–401 | date = March 2018 | pmid = 29134625 | pmc = 5836850 | doi = 10.1002/cpt.910 }}</ref> |
||
|- |
|- |
||
| ''SuperCYP Bioinformatics Tool'' || Containing 1170 drugs with more than 3800 interactions, and approximately 2000 known SNPs. These SNPs are listed and ordered according to their effect on expression and/or activity || |
| ''SuperCYP Bioinformatics Tool'' || Containing 1170 drugs with more than 3800 interactions, and approximately 2000 known SNPs. These SNPs are listed and ordered according to their effect on expression and/or activity || <ref name="pmid19934256">{{cite journal | vauthors = Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, Guenther S, Winnenburg R, Schroeder M, Preissner R | display-authors = 6 | title = SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions | journal = Nucleic Acids Research | volume = 38 | issue = Database issue | pages = D237-43 | date = January 2010 | pmid = 19934256 | pmc = 2808967 | doi = 10.1093/nar/gkp970 }}</ref> |
||
|- |
|- |
||
| ''PharmGKB'' || The Pharmacogenomics Knowledge Base (PharmGKB) is an interactive tool for researchers investigating how genetic variation affects drug response || |
| ''PharmGKB'' || The Pharmacogenomics Knowledge Base (PharmGKB) is an interactive tool for researchers investigating how genetic variation affects drug response || <ref name="pmid23824865">{{cite journal | vauthors = Thorn CF, Klein TE, Altman RB | title = PharmGKB: the Pharmacogenomics Knowledge Base | journal = Methods in Molecular Biology | location = Clifton, N.J. | volume = 1015 | issue = | pages = 311–20 | date = 2013 | pmid = 23824865 | pmc = 4084821 | doi = 10.1007/978-1-62703-435-7_20 }}</ref> |
||
|- |
|- |
||
| ''dbSNP database'' || A repository of SNPs and other variants that have been reported after discovery, compiled and officially named. These are SNPs across the board || https://www.ncbi.nlm.nih.gov/SNP/ |
| ''dbSNP database'' || A repository of SNPs and other variants that have been reported after discovery, compiled and officially named. These are SNPs across the board || <ref>{{Cite web|url=https://www.ncbi.nlm.nih.gov/SNP/|title=DBSNP Home Page | work = National Center for Biotechnology Information, U.S. National Library of Medicine }}</ref><ref name="pmid10592272">{{cite journal | vauthors = Smigielski EM, Sirotkin K, Ward M, Sherry ST | title = dbSNP: a database of single nucleotide polymorphisms | journal = Nucleic Acids Research | volume = 28 | issue = 1 | pages = 352–5 | date = January 2000 | pmid = 10592272 | pmc = 102496 | doi = 10.1093/nar/28.1.352 }}</ref> |
||
|- |
|- |
||
| ''FINDbase'' || Repository of allele frequencies of pharmacogenetic markers in different populations || <ref name="pmid24234438">{{cite journal | vauthors = Papadopoulos P, Viennas E, Gkantouna V, Pavlidis C, Bartsakoulia M, Ioannou ZM, Ratbi I, Sefiani A, Tsaknakis J, Poulas K, Tzimas G, Patrinos GP | display-authors = 6 | title = Developments in FINDbase worldwide database for clinically relevant genomic variation allele frequencies | journal = Nucleic Acids Research | volume = 42 | issue = Database issue | pages = D1020-6 | date = January 2014 | pmid = 24234438 | pmc = 3964978 | doi = 10.1093/nar/gkt1125 }}</ref> |
|||
| ''FINDbase'' || Repository of allele frequencies of pharmacogenetic markers in different populations || http://www.findbase.org/ |
|||
|- |
|- |
||
| ''Pharmacogenomics Biomarkers in Drug Labelling'' || A table that identifies which FDA-approved drugs have pharmacogenomics-related warning labels || https://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm |
| ''Pharmacogenomics Biomarkers in Drug Labelling'' || A table that identifies which FDA-approved drugs have pharmacogenomics-related warning labels || <ref>{{Cite journal|url=https://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm|title = Table of Pharmacogenomic Biomarkers in Drug Labeling|journal = FDA|date = 18 August 2021}}</ref> |
||
|- |
|- |
||
| ''SNPedia'' || A wiki-based bioinformatics database of SNPs || http://www.snpedia.com/index.php/SNPedia |
| ''SNPedia'' || A wiki-based bioinformatics database of SNPs || <ref>{{Cite web|url=http://www.snpedia.com/index.php/SNPedia|title = SNPedia}}</ref><ref name="pmid22140107">{{cite journal | vauthors = Cariaso M, Lennon G | title = SNPedia: a wiki supporting personal genome annotation, interpretation and analysis | journal = Nucleic Acids Research | volume = 40 | issue = Database issue | pages = D1308–12 | date = January 2012 | pmid = 22140107 | pmc = 3245045 | doi = 10.1093/nar/gkr798 }}</ref> |
||
|- |
|- |
||
|''Pharmacogenomics Research Network (PGRN)'' |
|''Pharmacogenomics Research Network (PGRN)'' |
||
|The PGRN hosts resources and information to stimulate collaborative research in pharmacogenomics and precision medicine. |
|The PGRN hosts resources and information to stimulate collaborative research in pharmacogenomics and precision medicine. |
||
|<ref>{{cite web | title = Pharmacogenomics Research Network PGRN | url = https://www.pgrn.org}}</ref> |
|||
|http://www.pgrn.org/ |
|||
|} |
|} |
||
Revision as of 18:33, 9 October 2021
| Part of a series on |
| Genetics |
|---|
 |
Pharmacogenomics is the study of the role of the genome in drug response. Its name (pharmaco- + genomics) reflects its combining of pharmacology and genomics. Pharmacogenomics analyzes how the genetic makeup of an individual affects their response to drugs.[1] It deals with the influence of acquired and inherited genetic variation on drug response in patients by correlating gene expression or single-nucleotide polymorphisms with pharmacokinetics (drug absorption, distribution, metabolism, and elimination) and pharmacodynamics (effects mediated through a drug's biological targets).[2][3][4] The term pharmacogenomics is often used interchangeably with pharmacogenetics. Although both terms relate to drug response based on genetic influences, pharmacogenetics focuses on single drug-gene interactions, while pharmacogenomics encompasses a more genome-wide association approach, incorporating genomics and epigenetics while dealing with the effects of multiple genes on drug response.[5][6][7]
Pharmacogenomics aims to develop rational means to optimize drug therapy, with respect to the patients' genotype, to ensure maximum efficiency with minimal adverse effects.[8] Through the utilization of pharmacogenomics, it is hoped that pharmaceutical drug treatments can deviate from what is dubbed as the "one-dose-fits-all" approach. Pharmacogenomics also attempts to eliminate the trial-and-error method of prescribing, allowing physicians to take into consideration their patient's genes, the functionality of these genes, and how this may affect the efficacy of the patient's current or future treatments (and where applicable, provide an explanation for the failure of past treatments).[5][9] Such approaches promise the advent of precision medicine and even personalized medicine, in which drugs and drug combinations are optimized for narrow subsets of patients or even for each individual's unique genetic makeup.[10][11] Whether used to explain a patient's response or lack thereof to a treatment, or act as a predictive tool, it hopes to achieve better treatment outcomes, greater efficacy, minimization of the occurrence of drug toxicities and adverse drug reactions (ADRs). For patients who have lack of therapeutic response to a treatment, alternative therapies can be prescribed that would best suit their requirements. In order to provide pharmacogenomic recommendations for a given drug, two possible types of input can be used: genotyping or exome or whole genome sequencing.[12] Sequencing provides many more data points, including detection of mutations that prematurely terminate the synthesized protein (early stop codon).[12]
History
Pharmacogenomics was first recognized by Pythagoras around 510 BC when he made a connection between the dangers of fava bean ingestion with hemolytic anemia and oxidative stress. This identification was later validated and attributed to deficiency of G6PD in the 1950s and called favism.[13][14] Although the first official publication dates back to 1961,[15] circa 1950s marked the unofficial beginnings of this science. Reports of prolonged paralysis and fatal reactions linked to genetic variants in patients who lacked butyryl-cholinesterase (‘pseudocholinesterase’) following administration of succinylcholine injection during anesthesia were first reported in 1956.[2][16] The term pharmacogenetic was first coined in 1959 by Friedrich Vogel of Heidelberg, Germany (although some papers suggest it was 1957 or 1958).[17] In the late 1960s, twin studies supported the inference of genetic involvement in drug metabolism, with identical twins sharing remarkable similarities to drug response compared to fraternal twins.[18] The term pharmacogenomics first began appearing around the 1990s.[13]
The first FDA approval of a pharmacogenetic test was in 2005[19] (for alleles in CYP2D6 and CYP2C19).
Drug-metabolizing enzymes
There are several known genes which are largely responsible for variances in drug metabolism and response. The focus of this article will remain on the genes that are more widely accepted and utilized clinically for brevity.
- Cytochrome P450s
- VKORC1
- TPMT
Cytochrome P450
The most prevalent drug-metabolizing enzymes (DME) are the Cytochrome P450 (CYP) enzymes. These enzymes introduce reactive or polar groups into xenobiotics such as drugs. The term Cytochrome P450 was coined by Omura and Sato in 1962 to describe the membrane-bound, heme-containing protein characterized by 450 nm spectral peak when complexed with carbon monoxide.[20] The human CYP family consists of 57 genes, with 18 families and 44 subfamilies. CYP proteins are conveniently arranged into these families and subfamilies on the basis of similarities identified between the amino acid sequences. Enzymes that share 35-40% identity are assigned to the same family by an Arabic numeral, and those that share 55-70% make up a particular subfamily with a designated letter.[21] For example, CYP2D6 refers to family 2, subfamily D, and gene number 6.
From a clinical perspective, the most commonly tested CYPs include: CYP2D6, CYP2C19, CYP2C9, CYP3A4 and CYP3A5. These genes account for the metabolism of approximately 70-90% of currently available prescription drugs.[22][23] The table below provides a summary for some of the medications that take these pathways.
| Drug Metabolism of Major CYPs[24][25] | |||||
|---|---|---|---|---|---|
| Enzyme | Fraction of drug metabolism (%) | Example Drugs | |||
| CYP2C9 | 10 | Tolbutamide, ibuprofen, mefenamic acid, tetrahydrocannabinol, losartan, diclofenac | |||
| CYP2C19 | 5 | S-mephenytoin, amitriptyline, diazepam, omeprazole, proguanil, hexobarbital, propranolol, imipramine | |||
| CYP2D6 | 20-30 | Debrisoquine, metoprolol, sparteine, propranolol, encainide, codeine, dextromethorphan, clozapine, desipramine, haloperidol, amitriptyline, imipramine | |||
| CYP3A4 | 40-45 | Erythromycin, ethinylestradiol, nifedipine, triazolam, cyclosporine, amitriptyline, imipramine | |||
| CYP3A5 | <1 | Erythromycin, ethinylestradiol, nifedipine, triazolam, cyclosporine, amitriptyline, aldosterone | |||
CYP2B6
CYP2B6 plays an important role in the metabolism of drugs including the anti-HIV drug efavirenz, the anti-malarial artemisinin, the antidepressants bupropion and ketamine, the anticancer drug cyclophosphamide, and the opioid methadone.[26] This is a highly polymorphic enzyme with the variant CYP2B6*6 having special importance, as it leads to errors in RNA processing and reduced enzyme levels. A second important variant CYP2B6*18 also fails to produce functional protein. The CYP2B6*6 variant occurs with prevalences of 15% to 60% in various populations worldwide, while the CYP2B68*18 is found predominantly in Africans. The higher prevalence of central nervous system side effects in African as compared to American and European patients treated with efavirenz has been attributed to the higher frequency of the CYP2B6 slow metabolizer phenotype in sub-Saharan African populations.[27]
CYP2D6
Also known as debrisoquine hydroxylase (named after the drug that led to its discovery), CYP2D6 is the most well-known and extensively studied CYP gene.[28] It is a gene of great interest also due to its highly polymorphic nature, and involvement in a high number of medication metabolisms (both as a major and minor pathway). More than 100 CYP2D6 genetic variants have been identified.[25] Both polymorphisms in the CYP2D6 gene (leading to versions of the enzyme having differing levels of metabolic activity) and copy number variants are known. For certain drugs predominantly metabolized by CYP2D6, these variations can lead to unusually high or low drug concentrations in serum (Referred to as poor metabolizer and ultra metabolizer phenotypes, respectively), thus leading to increased side effects or reduced efficacy. Commonly affected drugs include tramadol, venlafaxine, morphine, mirtazapine, and metoprolol.[29] The frequency of CYP2D6 varies geographically, with the highest prevalence of slow metabolizers found in east Asia and the lowest prevalence in the Americas.[30]
CYP2C19
Discovered in the early 1980s, CYP2C19 is the second most extensively studied and well understood gene in pharmacogenomics.[24] Over 28 genetic variants have been identified for CYP2C19,[31] of which affects the metabolism of several classes of drugs, such as antidepressants and proton pump inhibitors.[32]
CYP2C9
CYP2C9 constitutes the majority of the CYP2C subfamily, representing approximately 20% of the liver content. It is involved in the metabolism of approximately 10% of all drugs, which include medications with narrow therapeutic windows such as warfarin and tolbutamide.[32][33] There are approximately 57 genetic variants associated with CYP2C9.[31]
CYP3A4 and CYP3A5
The CYP3A family is the most abundantly found in the liver, with CYP3A4 accounting for 29% of the liver content.[24] These enzymes also cover between 40-50% of the current prescription drugs, with the CYP3A4 accounting for 40-45% of these medications.[14] CYP3A5 has over 11 genetic variants identified at the time of this publication.[31]
VKORC1
The vitamin K epoxide reductase complex subunit 1 (VKORC1) is responsible for the pharmacodynamics of warfarin.[34] VKORC1 along with CYP2C9 are useful for identifying the risk of bleeding during warfarin administration. Warfarin works by inhibiting VKOR, which is encoded by the VKORC1 gene. Individuals with polymorphism in this have an affected response to warfarin treatment.[35]
TPMT
Thiopurine methyltransferase (TPMT) catalyzes the S-methylation of thiopurines, thereby regulating the balance between cytotoxic thioguanine nucleotide and inactive metabolites in hematopoietic cells.[36] TPMT is highly involved in 6-MP metabolism and TMPT activity and TPMT genotype is known to affect the risk of toxicity. Excessive levels of 6-MP can cause myelosuppression and myelotoxicity.[37] Related patent litigation arose in Mayo Collaborative Services v. Prometheus Laboratories, Inc., in which the Supreme Court of the United States found that patent around measuring doses of the drug was patent-eligible.
Codeine, clopidogrel, tamoxifen, and warfarin a few examples of medications that follow the above metabolic pathways.
Predictive prescribing
Patient genotypes are usually categorized into the following predicted phenotypes:
- UM: Ultra-rapid metabolizer: patients with substantially increased metabolic activity;
- EM: Extensive metabolizer: normal metabolic activity;
- IM: Intermediate metabolizer: patients with reduced metabolic activity; and
- PM: Poor metabolizer: patients with little to no functional metabolic activity.
The two extremes of this spectrum are the poor metabolizers and ultra-rapid metabolizers. Efficacy of a medication is not only based on the above metabolic statuses, but also the type of drug consumed. Drugs can be classified into two main groups: active drugs and prodrugs. Active drugs refer to drugs that are inactivated during metabolism, and prodrugs are inactive until they are metabolized.

For example, we have two patients who are taking codeine for pain relief. Codeine is a prodrug, so it requires conversion from its inactive form to its active form. The active form of codeine is morphine, which provides the therapeutic effect of pain relief. If person A receives one *1 allele each from mother and father to code for the CYP2D6 gene, then that person is considered to have an extensive metabolizer (EM) phenotype, as allele *1 is considered to have a normal-function (this would be represented as CYP2D6 *1/*1). If person B on the other hand had received one *1 allele from the mother and a *4 allele from the father, that individual would be an Intermediate Metabolizer (IM) (the genotype would be CYP2D6 *1/*4). Although both individuals are taking the same dose of codeine, person B could potentially lack the therapeutic benefits of codeine due to the decreased conversion rate of codeine to its active counterpart morphine.
Each phenotype is based upon the allelic variation within the individual genotype. However, several genetic events can influence a same phenotypic trait, and establishing genotype-to-phenotype relationships can thus be far from consensual with many enzymatic patterns. For instance, the influence of the CYP2D6*1/*4 allelic variant on the clinical outcome in patients treated with Tamoxifen remains debated today. In oncology, genes coding for DPD, UGT1A1, TPMT, CDA involved in the pharmacokinetics of 5-FU/capecitabine, irinotecan, 6-mercaptopurine and gemcitabine/cytarabine, respectively, have all been described as being highly polymorphic. A strong body of evidence suggests that patients affected by these genetic polymorphisms will experience severe/lethal toxicities upon drug intake, and that pre-therapeutic screening does help to reduce the risk of treatment-related toxicities through adaptive dosing strategies.[38]
Applications
The list below provides a few more commonly known applications of pharmacogenomics:[39]
- Improve drug safety, and reduce ADRs;
- Tailor treatments to meet patients' unique genetic pre-disposition, identifying optimal dosing;
- Improve drug discovery targeted to human disease; and
- Improve proof of principle for efficacy trials.
Pharmacogenomics may be applied to several areas of medicine, including pain management, cardiology, oncology, and psychiatry. A place may also exist in forensic pathology, in which pharmacogenomics can be used to determine the cause of death in drug-related deaths where no findings emerge using autopsy.[citation needed]
In cancer treatment, pharmacogenomics tests are used to identify which patients are most likely to respond to certain cancer drugs. In behavioral health, pharmacogenomic tests provide tools for physicians and care givers to better manage medication selection and side effect amelioration. Pharmacogenomics is also known as companion diagnostics, meaning tests being bundled with drugs. Examples include KRAS test with cetuximab and EGFR test with gefitinib. Beside efficacy, germline pharmacogenetics can help to identify patients likely to undergo severe toxicities when given cytotoxics showing impaired detoxification in relation with genetic polymorphism, such as canonical 5-FU.[40] In particular, genetic deregulations affecting genes coding for DPD, UGT1A1, TPMT, CDA and CYP2D6 are now considered as critical issues for patients treated with 5-FU/capecitabine, irinotecan, mercaptopurine/azathioprine, gemcitabine/capecitabine/AraC and tamoxifen, respectively.[41]
In cardiovascular disorders, the main concern is response to drugs including warfarin, clopidogrel, beta blockers, and statins.[12] In patients with CYP2C19, who take clopidogrel, cardiovascular risk is elevated, leading to medication package insert updates by regulators.[42] In patients with type 2 diabetes, haptoglobin (Hp) genotyping shows an effect on cardiovascular disease, with Hp2-2 at higher risk and supplemental vitamin E reducing risk by affecting HDL.[43]
In psychiatry, as of 2010, research has focused particularly on 5-HTTLPR and DRD2.[44]
Clinical implementation
Initiatives to spur adoption by clinicians include the Ubiquitous Pharmacogenomics program in Europe and the Clinical Pharmacogenetics Implementation Consortium (CPIC) in the United States.[45] In a 2017 survey of European clinicians, in the prior year two-thirds had not ordered a pharmacogenetic test.[46]
In 2010, Valderbilt University Medical Center launched Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT);[47] in 2015 survey, two-thirds of the clinicians had ordered a pharmacogenetic test.[48]
In the United States, the FDA has updated medication package inserts based on genomic evidence.[49]
In 2019, the largest private health insurer, UnitedHealthcare, announced that it would pay for genetic testing to predict response to psychiatric drugs; as of 2019, it is the only private insurer to offer such coverage.[50]
In 2020, Canada's 4th largest health and dental insurer, Green Shield Canada, announced that it would pay for pharmacogenetic testing and its associated clinical decision support software to optimize and personalize mental health prescriptions.[51]
Example case studies
Case A – Antipsychotic adverse reaction[52]
Patient A suffers from schizophrenia. Their treatment included a combination of ziprasidone, olanzapine, trazodone and benztropine. The patient experienced dizziness and sedation, so they were tapered off ziprasidone and olanzapine, and transitioned to quetiapine. Trazodone was discontinued. The patient then experienced excessive sweating, tachycardia and neck pain, gained considerable weight and had hallucinations. Five months later, quetiapine was tapered and discontinued, with ziprasidone re-introduction into their treatment due to the excessive weight gain. Although the patient lost the excessive weight they gained, they then developed muscle stiffness, cogwheeling, tremors and night sweats. When benztropine was added they experienced blurry vision. After an additional five months, the patient was switched from ziprasidone to aripiprazole. Over the course of 8 months, patient A gradually experienced more weight gain, sedation, developed difficulty with their gait, stiffness, cogwheeling and dyskinetic ocular movements. A pharmacogenomics test later proved the patient had a CYP2D6 *1/*41, which has a predicted phenotype of IM and CYP2C19 *1/*2 with a predicted phenotype of IM as well.
Case B – Pain Management [53]
Patient B is a woman who gave birth by caesarian section. Her physician prescribed codeine for post-caesarian pain. She took the standard prescribed dose, however experienced nausea and dizziness while she was taking codeine. She also noticed that her breastfed infant was lethargic and feeding poorly. When the patient mentioned these symptoms to her physician, they recommended that she discontinue codeine use. Within a few days, both the patient and her infant's symptoms were no longer present. It is assumed that if the patient underwent a pharmacogenomic test, it would have revealed she may have had a duplication of the gene CYP2D6 placing her in the Ultra-rapid metabolizer (UM) category, explaining her ADRs to codeine use.
Case C – FDA Warning on Codeine Overdose for Infants[54]
On February 20, 2013, the FDA released a statement addressing a serious concern regarding the connection between children who are known as CYP2D6 UM and fatal reactions to codeine following tonsillectomy and/or adenoidectomy (surgery to remove the tonsils and/or adenoids). They released their strongest Boxed Warning to elucidate the dangers of CYP2D6 UMs consuming codeine. Codeine is converted to morphine by CYP2D6, and those who have UM phenotypes are at danger of producing large amounts of morphine due to the increased function of the gene. The morphine can elevate to life-threatening or fatal amounts, as became evident with the death of three children in August 2012.
Polypharmacy
A potential role pharmacogenomics may play would be to reduce the occurrence of polypharmacy. It is theorized that with tailored drug treatments, patients will not have the need to take several medications that are intended to treat the same condition. In doing so, they could potentially minimize the occurrence of ADRs, have improved treatment outcomes, and can save costs by avoiding purchasing extraneous medications. An example of this can be found in psychiatry, where patients tend to be receiving more medications than even age-matched non-psychiatric patients. This has been associated with an increased risk of inappropriate prescribing.[55]
The need for pharmacogenomics tailored drug therapies may be most evident in a survey conducted by the Slone Epidemiology Center at Boston University from February 1998 to April 2007. The study elucidated that an average of 82% of adults in the United States are taking at least one medication (prescription or nonprescription drug, vitamin/mineral, herbal/natural supplement), and 29% are taking five or more. The study suggested that those aged 65 years or older continue to be the biggest consumers of medications, with 17-19 % in this age group taking at least ten medications in a given week. Polypharmacy has also shown to have increased since 2000 from 23% to 29%.[56]
Drug labeling
The U.S. Food and Drug Administration (FDA) appears to be very invested in the science of pharmacogenomics[57] as is demonstrated through the 120 and more FDA-approved drugs that include pharmacogenomic biomarkers in their labels.[58] This number increased varies over the years.[59] A study of the labels of FDA-approved drugs as of 20 June 2014 found that there were 140 different drugs with a pharmacogenomic biomarker in their label.[60] Because a drug can have different biomarkers, this corresponded to 158 drug–biomarker pairs.[60] Only 29% stated a requirement or recommendation for genetic biomarker testing but this was higher for oncology drugs (62%).[60] On May 22, 2005, the FDA issued its first Guidance for Industry: Pharmacogenomic Data Submissions, which clarified the type of pharmacogenomic data required to be submitted to the FDA and when.[61] Experts recognized the importance of the FDA's acknowledgement that pharmacogenomics experiments will not bring negative regulatory consequences.[62] The FDA had released its latest guide Clinical Pharmacogenomics (PGx): Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling in January, 2013. The guide is intended to address the use of genomic information during drug development and regulatory review processes.
Challenges
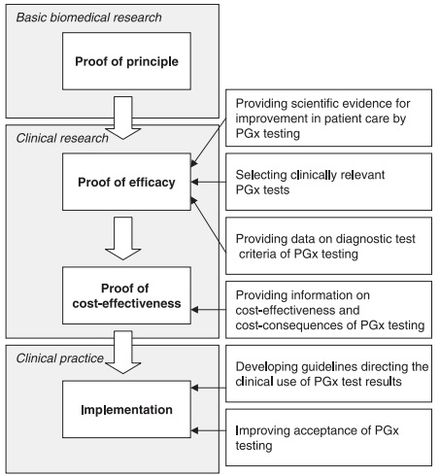
Although there appears to be a general acceptance of the basic tenet of pharmacogenomics amongst physicians and healthcare professionals,[64] several challenges exist that slow the uptake, implementation, and standardization of pharmacogenomics. Some of the concerns raised by physicians include:[7][64][65]
- Limitation on how to apply the test into clinical practices and treatment;
- A general feeling of lack of availability of the test;
- The understanding and interpretation of evidence-based research;
- Combining test results with other patient data for prescription optimization; and
- Ethical, legal and social issues.
Issues surrounding the availability of the test include:[63]
- The lack of availability of scientific data: Although there are considerable number of DME involved in the metabolic pathways of drugs, only a fraction have sufficient scientific data to validate their use within a clinical setting; and
- Demonstrating the cost-effectiveness of pharmacogenomics: Publications for the pharmacoeconomics of pharmacogenomics are scarce, therefore sufficient evidence does not at this time exist to validate the cost-effectiveness and cost-consequences of the test.
Although other factors contribute to the slow progression of pharmacogenomics (such as developing guidelines for clinical use), the above factors appear to be the most prevalent. Increasingly substantial evidence and industry body guidelines for clinical use of pharmacogenetics have made it a population wide approach to precision medicine. Cost, reimbursement, education, and easy use at the point of care remain significant barriers to widescale adoption.
Controversies
Some alleles that vary in frequency between specific populations have been shown to be associated with differential responses to specific drugs. The beta blocker atenolol is an anti-hypertensive medication that is shown to more significantly lower the blood pressure of Caucasian patients than African American patients in the United States.[citation needed] This observation suggests that Caucasian and African American populations have different alleles governing oleic acid biochemistry, which react differentially with atenolol.[66] Similarly, hypersensitivity to the antiretroviral drug abacavir is strongly associated with a single-nucleotide polymorphism that varies in frequency between populations.[67]
The FDA approval of the drug BiDil (isosorbide dinitrate/hydralazine) with a label specifying African-Americans with congestive heart failure, produced a storm of controversy over race-based medicine and fears of genetic stereotyping,[68] even though the label for BiDil did not specify any genetic variants but was based on racial self-identification.[69][70]
Future
Computational advances have enabled cheaper and faster sequencing.[71] Research has focused on combinatorial chemistry,[72] genomic mining, omic technologies and high throughput screening.
As the cost per genetic test decreases, the development of personalized drug therapies will increase.[73] Technology now allows for genetic analysis of hundreds of target genes involved in medication metabolism and response in less than 24 hours for under $1,000. This a huge step towards bringing pharmacogenetic technology into everyday medical decisions. Likewise, companies like deCODE genetics, MD Labs Pharmacogenetics, Navigenics and 23andMe offer genome scans. The companies use the same genotyping chips that are used in GWAS studies and provide customers with a write-up of individual risk for various traits and diseases and testing for 500,000 known SNPs. Costs range from $995 to $2500 and include updates with new data from studies as they become available. The more expensive packages even included a telephone session with a genetics counselor to discuss the results.[74]
Ethics
Pharmacogenetics has become a controversial issue in the area of bioethics. Privacy and confidentiality are major concerns.[75] The evidence of benefit or risk from a genetic test may only be suggestive, which could cause dilemmas for providers.[75]: 145 Drug development may be affected, with rare genetic variants possibly receiving less research.[75] Access and patient autonomy are also open to discussion.[76]: 680
Web-based resources
| Web Resources for Pharmacogenomics[77][78] | |||||
|---|---|---|---|---|---|
| Data Source | Main Use | Citation | |||
| PharmVar | A central repository for pharmacogene variation that focuses on haplotype structure and allelic variation | [79] | |||
| SuperCYP Bioinformatics Tool | Containing 1170 drugs with more than 3800 interactions, and approximately 2000 known SNPs. These SNPs are listed and ordered according to their effect on expression and/or activity | [80] | |||
| PharmGKB | The Pharmacogenomics Knowledge Base (PharmGKB) is an interactive tool for researchers investigating how genetic variation affects drug response | [81] | |||
| dbSNP database | A repository of SNPs and other variants that have been reported after discovery, compiled and officially named. These are SNPs across the board | [82][83] | |||
| FINDbase | Repository of allele frequencies of pharmacogenetic markers in different populations | [84] | |||
| Pharmacogenomics Biomarkers in Drug Labelling | A table that identifies which FDA-approved drugs have pharmacogenomics-related warning labels | [85] | |||
| SNPedia | A wiki-based bioinformatics database of SNPs | [86][87] | |||
| Pharmacogenomics Research Network (PGRN) | The PGRN hosts resources and information to stimulate collaborative research in pharmacogenomics and precision medicine. | [88] | |||
See also
- Genomics
- Metabolomics
- Pharmacovigilance
- Population groups in biomedicine
- Toxgnostics
- Medical terminology
- HL7
References
- ^ Ermak G (2015). Emerging Medical Technologies. World Scientific. ISBN 978-981-4675-80-2.
- ^ a b Johnson JA (November 2003). "Pharmacogenetics: potential for individualized drug therapy through genetics". Trends in Genetics. 19 (11): 660–6. doi:10.1016/j.tig.2003.09.008. PMID 14585618.
- ^ "Center for Pharmacogenomics and Individualized Therapy". Retrieved 2014-06-25.
- ^ "overview of pharmacogenomics". Up-to-Date. May 16, 2014. Retrieved 2014-06-25.
- ^ a b Sheffield LJ, Phillimore HE (May 2009). "Clinical use of pharmacogenomic tests in 2009". The Clinical Biochemist. Reviews. 30 (2): 55–65. PMC 2702214. PMID 19565025.
- ^ Shin J, Kayser SR, Langaee TY (April 2009). "Pharmacogenetics: from discovery to patient care". American Journal of Health-System Pharmacy. 66 (7): 625–37. doi:10.2146/ajhp080170. PMID 19299369.
- ^ a b "Center for Genetics Education".[permanent dead link]
- ^ Becquemont L (June 2009). "Pharmacogenomics of adverse drug reactions: practical applications and perspectives". Pharmacogenomics. 10 (6): 961–9. doi:10.2217/pgs.09.37. PMID 19530963.
- ^ Hauser AS, Chavali S, Masuho I, Jahn LJ, Martemyanov KA, Gloriam DE, Babu MM (January 2018). "Pharmacogenomics of GPCR Drug Targets". Cell. 172 (1–2): 41–54.e19. doi:10.1016/j.cell.2017.11.033. PMC 5766829. PMID 29249361.
- ^ "Guidance for Industry Pharmacogenomic Data Submissions" (PDF). U.S. Food and Drug Administration. March 2005. Retrieved 2008-08-27.
- ^ Squassina A, Manchia M, Manolopoulos VG, Artac M, Lappa-Manakou C, Karkabouna S, et al. (August 2010). "Realities and expectations of pharmacogenomics and personalized medicine: impact of translating genetic knowledge into clinical practice". Pharmacogenomics. 11 (8): 1149–67. doi:10.2217/pgs.10.97. PMID 20712531.
- ^ a b c Huser V, Cimino JJ (2013). "Providing pharmacogenomics clinical decision support using whole genome sequencing data as input". AMIA Joint Summits on Translational Science Proceedings. AMIA Joint Summits on Translational Science. 2013: 81. PMID 24303303.
- ^ a b Pirmohamed M (October 2001). "Pharmacogenetics and pharmacogenomics". British Journal of Clinical Pharmacology. 52 (4): 345–7. doi:10.1046/j.0306-5251.2001.01498.x. PMC 2014592. PMID 11678777.
- ^ a b Prasad K (January 2009). "Role of regulatory agencies in translating pharmacogenetics to the clinics". Clinical Cases in Mineral and Bone Metabolism. 6 (1): 29–34. PMC 2781218. PMID 22461095.
- ^ Evans DA, Clarke CA (September 1961). "Pharmacogenetics". British Medical Bulletin. 17 (3): 234–40. doi:10.1093/oxfordjournals.bmb.a069915. PMID 13697554.
- ^ Kalow W (2006). "Pharmacogenetics and pharmacogenomics: origin, status, and the hope for personalized medicine". The Pharmacogenomics Journal. 6 (3): 162–5. doi:10.1038/sj.tpj.6500361. PMID 16415920.
- ^ Vogel F (1959). "Moderne probleme der humangenetik" [Modern human genetics problems]. Ergebnisse der inneren medizin und kinderheilkunde [Results of internal medicine and pediatrics] (in German). Berlin, Heidelberg: Springer: 52–125.
- ^ Motulsky AG, Qi M (February 2006). "Pharmacogenetics, pharmacogenomics and ecogenetics". Journal of Zhejiang University. Science. B. 7 (2): 169–70. doi:10.1631/jzus.2006.B0169. PMC 1363768. PMID 16421980.
- ^ Squassina A, Manchia M, Manolopoulos VG, Artac M, Lappa-Manakou C, Karkabouna S, et al. (August 2010). "Realities and expectations of pharmacogenomics and personalized medicine: impact of translating genetic knowledge into clinical practice". Pharmacogenomics. 11 (8): 1149–67. doi:10.2217/pgs.10.97. PMID 20712531.
- ^ Debose-Boyd RA (February 2007). "A helping hand for cytochrome p450 enzymes". Cell Metabolism. 5 (2): 81–3. doi:10.1016/j.cmet.2007.01.007. PMID 17276348.
- ^ Nebert DW, Russell DW (October 2002). "Clinical importance of the cytochromes P450". Lancet. 360 (9340): 1155–62. doi:10.1016/s0140-6736(02)11203-7. PMID 12387968. S2CID 13577054.
- ^ Hart SN, Wang S, Nakamoto K, Wesselman C, Li Y, Zhong XB (January 2008). "Genetic polymorphisms in cytochrome P450 oxidoreductase influence microsomal P450-catalyzed drug metabolism". Pharmacogenetics and Genomics. 18 (1): 11–24. doi:10.1097/FPC.0b013e3282f2f121. PMID 18216718. S2CID 38880360.
- ^ Gomes AM, Winter S, Klein K, Turpeinen M, Schaeffeler E, Schwab M, Zanger UM (April 2009). "Pharmacogenomics of human liver cytochrome P450 oxidoreductase: multifactorial analysis and impact on microsomal drug oxidation". Pharmacogenomics. 10 (4): 579–99. doi:10.2217/pgs.09.7. PMID 19374516.
- ^ a b c Hasler JA (February 1999). "Pharmacogenetics of cytochromes P450". Molecular Aspects of Medicine. 20 (1–2): 12–24, 25–137. doi:10.1016/s0098-2997(99)00005-9. PMID 10575648.
- ^ a b Ingelman-Sundberg M (April 2004). "Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future". Trends in Pharmacological Sciences. 25 (4): 193–200. doi:10.1016/j.tips.2004.02.007. PMID 15063083.
- ^ Zanger UM, Klein K (2013). "Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance". Frontiers in Genetics. 4: 24. doi:10.3389/fgene.2013.00024. PMC 3588594. PMID 23467454.
- ^ Masimirembwa C, Dandara C, Leutscher PD (October 2016). "Rolling out Efavirenz for HIV Precision Medicine in Africa: Are We Ready for Pharmacovigilance and Tackling Neuropsychiatric Adverse Effects?". Omics. 20 (10): 575–580. doi:10.1089/omi.2016.0120. PMID 27627692.
- ^ Badyal DK, Dadhich AP (October 2001). "Cytochrome P450 and drug interactions" (PDF). Indian Journal of Pharmacology. 33: 248–259.
- ^ Zhou SF (2009). "Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I". Clinical Pharmacokinetics. 48 (11): 689–723. doi:10.2165/11318030-000000000-00000. PMID 19817501. S2CID 5936561.
- ^ Zhou Y, Ingelman-Sundberg M, Lauschke VM (October 2017). "Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects". Clinical Pharmacology and Therapeutics. 102 (4): 688–700. doi:10.1002/cpt.690. PMC 5600063. PMID 28378927.
- ^ a b c Ingelman-Sundberg M, Nebert DW, Sim SC. "The Human Cytochrome P450 (CYP) Allele Nomenclature Database". Retrieved 2014-09-03.
- ^ a b Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (December 2007). "Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects". Pharmacology & Therapeutics. 116 (3): 496–526. doi:10.1016/j.pharmthera.2007.09.004. PMID 18001838.
- ^ Sikka R, Magauran B, Ulrich A, Shannon M (December 2005). "Bench to bedside: Pharmacogenomics, adverse drug interactions, and the cytochrome P450 system". Academic Emergency Medicine. 12 (12): 1227–35. doi:10.1111/j.1553-2712.2005.tb01503.x. PMID 16282513.
- ^ Teh LK, Langmia IM, Fazleen Haslinda MH, Ngow HA, Roziah MJ, Harun R, et al. (April 2012). "Clinical relevance of VKORC1 (G-1639A and C1173T) and CYP2C9*3 among patients on warfarin". Journal of Clinical Pharmacy and Therapeutics. 37 (2): 232–6. doi:10.1111/j.1365-2710.2011.01262.x. PMID 21507031. S2CID 22671298.
- ^ U.S. Food and Drug Administration (FDA). "Table of Pharmacogenomic Biomarkers in Drug Labels". Retrieved 2014-09-03.
- ^ a b Crews KR, Hicks JK, Pui CH, Relling MV, Evans WE (October 2012). "Pharmacogenomics and individualized medicine: translating science into practice". Clinical Pharmacology and Therapeutics. 92 (4): 467–75. doi:10.1038/clpt.2012.120. PMC 3589526. PMID 22948889.
- ^ Sim SC, Kacevska M, Ingelman-Sundberg M (February 2013). "Pharmacogenomics of drug-metabolizing enzymes: a recent update on clinical implications and endogenous effects". The Pharmacogenomics Journal. 13 (1): 1–11. doi:10.1038/tpj.2012.45. PMID 23089672.
- ^ Lee SY, McLeod HL (January 2011). "Pharmacogenetic tests in cancer chemotherapy: what physicians should know for clinical application". The Journal of Pathology. 223 (1): 15–27. doi:10.1002/path.2766. PMID 20818641. S2CID 6330198.
- ^ Cohen N (November 2008). Pharmacogenomics and Personalized Medicine (Methods in Pharmacology and Toxicology). Totowa, NJ: Humana Press. p. 6. ISBN 978-1934115046.
- ^ Ciccolini J, Gross E, Dahan L, Lacarelle B, Mercier C (October 2010). "Routine dihydropyrimidine dehydrogenase testing for anticipating 5-fluorouracil-related severe toxicities: hype or hope?". Clinical Colorectal Cancer. 9 (4): 224–8. doi:10.3816/CCC.2010.n.033. PMID 20920994.
- ^ Yang CG, Ciccolini J, Blesius A, Dahan L, Bagarry-Liegey D, Brunet C, et al. (January 2011). "DPD-based adaptive dosing of 5-FU in patients with head and neck cancer: impact on treatment efficacy and toxicity". Cancer Chemotherapy and Pharmacology. 67 (1): 49–56. doi:10.1007/s00280-010-1282-4. PMID 20204365. S2CID 25362813.
- ^ Dean L (2012). "Clopidogrel Therapy and CYP2C19 Genotype". In Pratt VM, McLeod HL, Rubinstein WS, et al. (eds.). Medical Genetics Summaries. National Center for Biotechnology Information (NCBI). PMID 28520346. Bookshelf ID: NBK84114.
- ^ Bale BF, Doneen AL, Vigerust DJ (2018). "Precision Healthcare of Type 2 Diabetic Patients Through Implementation of Haptoglobin Genotyping". Frontiers in Cardiovascular Medicine. 5: 141. doi:10.3389/fcvm.2018.00141. PMC 6198642. PMID 30386783.
- ^ Malhotra AK (2010). "The state of pharmacogenetics". Psychiatr Times. 27 (4): 38–41, 62.
- ^ Williams MS (August 2019). "Early Lessons from the Implementation of Genomic Medicine Programs". Annual Review of Genomics and Human Genetics. 20 (1): 389–411. doi:10.1146/annurev-genom-083118-014924. PMID 30811224.
- ^ Just KS, Steffens M, Swen JJ, Patrinos GP, Guchelaar HJ, Stingl JC (October 2017). "Medical education in pharmacogenomics-results from a survey on pharmacogenetic knowledge in healthcare professionals within the European pharmacogenomics clinical implementation project Ubiquitous Pharmacogenomics (U-PGx)". European Journal of Clinical Pharmacology. 73 (10): 1247–1252. doi:10.1007/s00228-017-2292-5. PMC 5599468. PMID 28669097.
- ^ Carlson B (2012). "Vanderbilt pioneers bedside genetics". Biotechnology Healthcare. 9 (2): 31–2. PMC 3411230. PMID 22876213.
- ^ Peterson JF, Field JR, Shi Y, Schildcrout JS, Denny JC, McGregor TL, et al. (August 2016). "Attitudes of clinicians following large-scale pharmacogenomics implementation". The Pharmacogenomics Journal. 16 (4): 393–8. doi:10.1038/tpj.2015.57. PMC 4751074. PMID 26261062.
- ^ Drozda K, Pacanowski MA, Grimstein C, Zineh I (August 2018). "Pharmacogenetic Labeling of FDA-Approved Drugs: A Regulatory Retrospective". JACC. Basic to Translational Science. 3 (4): 545–549. doi:10.1016/j.jacbts.2018.06.001. PMC 6115648. PMID 30175278.
- ^ "Pharmacogenetic test makers cheer UnitedHealth coverage. Other payers aren't there yet". MedTech Dive. Retrieved 2019-12-29.
- ^ https://www.greenshield.ca/en-ca/blog/post/personalized-drug-treatment-through-pharmacogenetics
- ^ Foster A, Wang Z, Usman M, Stirewalt E, Buckley P (December 2007). "Pharmacogenetics of antipsychotic adverse effects: Case studies and a literature review for clinicians". Neuropsychiatric Disease and Treatment. 3 (6): 965–73. doi:10.2147/ndt.s1752. PMC 2656342. PMID 19300635.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Pharmacogenetics: increasing the safety and effectiveness of drug therapy [Brochure]" (PDF). American Medical Association. 2011.
- ^ "FDA Drug Safety Communication: Safety review update of codeine use in children; new Boxed Warning and Contraindication on use after tonsillectomy and/or adenoidectomy". United States Food and Drug Administration. 2013-02-20.
- ^ Ritsner M (2013). Polypharmacy in Psychiatry Practice, Volume I. Multiple Medication Strategies. Dordrecht: Springer Science and Business Media. ISBN 978-94-007-5804-9.
- ^ "Patterns of Medication Use in the United States". Boston University, Slone Epidemiology Center. 2006.
- ^ "Pharmacogenetics and Pharmacogenomics: State-of-the-art and potential socio-economic impacts in the EU". European Commission, Joint Research Centre, Institute for Prospective Technological Studies. 2006-04-01.
- ^ "Table of Pharmacogenomic Biomarkers in Drug Labels". United States Food and Drug Administration. 2013-06-19.
- ^ Abrahams E, Silver M (July 2009). "The case for personalized medicine". Journal of Diabetes Science and Technology. 3 (4): 680–4. doi:10.1177/193229680900300411. PMC 2769975. PMID 20144313.
- ^ a b c Vivot A, Boutron I, Ravaud P, Porcher R (September 2015). "Guidance for pharmacogenomic biomarker testing in labels of FDA-approved drugs". Genetics in Medicine. 17 (9): 733–8. doi:10.1038/gim.2014.181. PMID 25521333.
- ^ Xie HG, Frueh FW (November 2005). "Pharmacogenomics steps toward personalized medicine". Personalized Medicine. 2 (4): 325–337. doi:10.2217/17410541.2.4.325. PMID 29788578.
- ^ Katsnelson A (May 2005). "Cautious welcome for FDA pharmacogenomics guidance". Nature Biotechnology. 23 (5): 510. doi:10.1038/nbt0505-510. PMID 15877053. S2CID 28473989.
- ^ a b Swen JJ, Huizinga TW, Gelderblom H, de Vries EG, Assendelft WJ, Kirchheiner J, Guchelaar HJ (August 2007). "Translating pharmacogenomics: challenges on the road to the clinic". PLoS Medicine. 4 (8): e209. doi:10.1371/journal.pmed.0040209. PMC 1945038. PMID 17696640.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, Verbrugge RR, et al. (March 2012). "Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey". Clinical Pharmacology and Therapeutics. 91 (3): 450–8. doi:10.1038/clpt.2011.306. PMID 22278335. S2CID 21366195.
- ^ Ma JD, Lee KC, Kuo GM (August 2012). "Clinical application of pharmacogenomics". Journal of Pharmacy Practice. 25 (4): 417–27. doi:10.1177/0897190012448309. PMID 22689709. S2CID 1212666.
- ^ Wikoff WR, Frye RF, Zhu H, Gong Y, Boyle S, Churchill E, et al. (2013). "Pharmacometabolomics reveals racial differences in response to atenolol treatment". PloS One. 8 (3): e57639. doi:10.1371/journal.pone.0057639. PMC 3594230. PMID 23536766.
- ^ Rotimi CN, Jorde LB (October 2010). "Ancestry and disease in the age of genomic medicine". The New England Journal of Medicine. 363 (16): 1551–8. doi:10.1056/NEJMra0911564. PMID 20942671. S2CID 21293807.
- ^ Bloche MG (November 2004). "Race-based therapeutics". The New England Journal of Medicine. 351 (20): 2035–7. doi:10.1056/NEJMp048271. PMID 15533852. S2CID 1467851.
- ^ Frank R (March 30 – April 1, 2006). "Back with a Vengeance: the Reemergence of a Biological Conceptualization of Race in Research on Race/Ethnic Disparities in Health". Annual Meeting of the Population Association of America. Los Angeles, California. Archived from the original on December 1, 2008. Retrieved 2008-11-20.
- ^ Crawley L (July 2007). "The paradox of race in the Bidil debate". Journal of the National Medical Association. 99 (7): 821–2. PMC 2574363. PMID 17668653.
- ^ Kalow W (2005). Pharmacogenomics. New York: Taylor & Francis. pp. 552–3. ISBN 978-1-57444-878-8.
- ^ Thorpe DS (2001). "Combinatorial chemistry: starting the second decade". The Pharmacogenomics Journal. 1 (4): 229–32. doi:10.1038/sj.tpj.6500045. PMID 11908762.
- ^ Paul NW, Fangerau H (December 2006). "Why should we bother? Ethical and social issues in individualized medicine". Current Drug Targets. 7 (12): 1721–7. doi:10.2174/138945006779025428. PMID 17168846.
- ^ Topol E (2012). The Creative Destruction of Medicine: How the Digital Revolution Will Create Better Health Care. New York: Basic Books. ISBN 978-0-465-02550-3.
- ^ a b c Corrigan OP (March 2005). "Pharmacogenetics, ethical issues: review of the Nuffield Council on Bioethics Report". Journal of Medical Ethics. 31 (3): 144–8. doi:10.1136/jme.2004.007229. PMC 1734105. PMID 15738433.
- ^ Breckenridge A, Lindpaintner K, Lipton P, McLeod H, Rothstein M, Wallace H (September 2004). "Pharmacogenetics: ethical problems and solutions". Nature Reviews. Genetics. 5 (9): 676–80. doi:10.1038/nrg1431. PMID 15372090. S2CID 6149591.
- ^ Barh D, Dhawan D, Ganguly NK (2013). Omics for Personalized Medicine. India: Springer Media. doi:10.1007/978-81-322-1184-6. ISBN 978-81-322-1183-9. S2CID 46120003.
- ^ Stram D (2014). "Post-GWAS Analyses". Design, Analysis, and Interpretation of Genome-Wide Association Scans. Statistics for Biology and Health. Los Angeles: Springer Science and Business Media. pp. 285–327. doi:10.1007/978-1-4614-9443-0_8. ISBN 978-1-4614-9442-3.
- ^ Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE (March 2018). "The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database". Clinical Pharmacology and Therapeutics. 103 (3): 399–401. doi:10.1002/cpt.910. PMC 5836850. PMID 29134625.
- ^ Preissner S, Kroll K, Dunkel M, Senger C, Goldsobel G, Kuzman D, et al. (January 2010). "SuperCYP: a comprehensive database on Cytochrome P450 enzymes including a tool for analysis of CYP-drug interactions". Nucleic Acids Research. 38 (Database issue): D237-43. doi:10.1093/nar/gkp970. PMC 2808967. PMID 19934256.
- ^ Thorn CF, Klein TE, Altman RB (2013). "PharmGKB: the Pharmacogenomics Knowledge Base". Methods in Molecular Biology. 1015. Clifton, N.J.: 311–20. doi:10.1007/978-1-62703-435-7_20. PMC 4084821. PMID 23824865.
- ^ "DBSNP Home Page". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Smigielski EM, Sirotkin K, Ward M, Sherry ST (January 2000). "dbSNP: a database of single nucleotide polymorphisms". Nucleic Acids Research. 28 (1): 352–5. doi:10.1093/nar/28.1.352. PMC 102496. PMID 10592272.
- ^ Papadopoulos P, Viennas E, Gkantouna V, Pavlidis C, Bartsakoulia M, Ioannou ZM, et al. (January 2014). "Developments in FINDbase worldwide database for clinically relevant genomic variation allele frequencies". Nucleic Acids Research. 42 (Database issue): D1020-6. doi:10.1093/nar/gkt1125. PMC 3964978. PMID 24234438.
- ^ "Table of Pharmacogenomic Biomarkers in Drug Labeling". FDA. 18 August 2021.
- ^ "SNPedia".
- ^ Cariaso M, Lennon G (January 2012). "SNPedia: a wiki supporting personal genome annotation, interpretation and analysis". Nucleic Acids Research. 40 (Database issue): D1308–12. doi:10.1093/nar/gkr798. PMC 3245045. PMID 22140107.
- ^ "Pharmacogenomics Research Network PGRN".
Further reading
- Katsnelson A (August 2005). "A Drug to Call One's Own: Will medicine finally get personal?". Scientific American.
- Karczewski KJ, Daneshjou R, Altman RB (2012). "Chapter 7: Pharmacogenomics". PLoS Computational Biology. 8 (12): e1002817. doi:10.1371/journal.pcbi.1002817. PMC 3531317. PMID 23300409.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
External links
- "Pharmacogenomics Factsheet". National Center for Biotechnology Information (NCBI), U.S. National Library of Medicine. Retrieved 2011-07-11.
a quick introduction to customised drugs
- "Pharmacogenomics Education Initiatives". U.S. Food and Drug Administration. 2010-09-24. Retrieved 2011-07-11.
- "Personalized Medicine (Pharmacogenetics)". University of Utah's Genetic Science Learning Center. Archived from the original on 2011-05-19. Retrieved 2011-07-11.
- "Center for Pharmacogenomics and Individualized Therapy". University of North Carolina at Chapel Hill Center for Pharmacogenomics and Individualized Therapy. Archived from the original on 2014-08-06. Retrieved 2014-06-25.
Journals:
- "Pharmacogenomics". Future Medicine Ltd.
- "Pharmacogenetics and Genomics". Lippincott Williams & Wilkins. ISSN 1744-6872.
- "The Pharmacogenomics Journal". Nature Publishing Group. ISSN 1470-269X.
