Holozoa
| Holozoans Temporal range:
| |
|---|---|
 | |
| Holozoan diversity | |
| Scientific classification | |
| Domain: | Eukaryota |
| Clade: | Amorphea |
| Clade: | Obazoa |
| (unranked): | Opisthokonta |
| (unranked): | Holozoa Lang et al., 2002[1] |
| Clades[4] | |
Incertae sedis | |
| Synonyms | |
| |
Holozoa (from Ancient Greek ὅλος (holos) 'whole' and ζῷον (zoion) 'animal') is a clade of organisms that includes animals and their closest single-celled relatives, but excludes fungi and all other organisms. Together they amount to more than 1.5 million species of purely heterotrophic organisms, including around 300 unicellular species. It consists of various subgroups, namely Metazoa (or animals) and the protists Choanoflagellata, Filasterea, Pluriformea and Ichthyosporea. Along with fungi and some other groups, Holozoa is part of the Opisthokonta, a supergroup of eukaryotes. Choanofila was previously used as the name for a group similar in composition to Holozoa, but its usage is discouraged now because it excludes animals and is therefore paraphyletic.
The holozoan protists play a crucial role in understanding the evolutionary steps leading to the emergence of multicellular animals from single-celled ancestors. Recent genomic studies have shed light on the evolutionary relationships between the various holozoan lineages, revealing insights into the origins of multicellularity. Some fossils of possible metazoans have been reinterpreted as holozoan protists.
Characteristics
Composition
Holozoa is a clade that includes animals and their closest relatives, as well as their common ancestor, but excludes fungi. It is defined on a branch-based approach as the clade encompassing all relatives of Homo sapiens (an animal), but not Neurospora crassa (a fungus).[4] Holozoa, besides animals, primarily comprises unicellular protist lineages of varied morphologies such as choanoflagellates, filastereans, ichthyosporeans, and the distinct genera Corallochytrium, Syssomonas, and Tunicaraptor.[6][2]
- Choanoflagellata, with around 250 species,[7] are the closest living relatives of animals. They are free-living unicellular or colonial flagellates that feed on bacteria using a characteristic "collar" of microvilli. The collar of choanoflagellates closely resembles sponge collar cells,[8] leading to theories since the 19th century about their relatedness to sponges.[9] The mysterious Proterospongia is an example of a colonial choanoflagellate that was thought to be related to the origin of sponges.[10] The affinities of the other single-celled holozoans only began to be recognized in the 1990s.[11]
- Ichthyosporea, also known as Mesomycetozoea and comprising around 40 species, largely consist of parasites or commensals. They interact with a diverse range of animals, from humans and fish to marine invertebrates. Most reproduce through multinucleated colonies and disperse as flagellates or amoebae.[7]
- Filasterea is a group of 6 amoeboid species belonging to the genera Ministeria, Pigoraptor,[6] Capsaspora and Txikispora,[12] united by the structure of their thread-like pseudopods.[13]
- Pluriformea is a provisional name for the clade composed by the two species Corallochytrium limacisporium and Syssomonas multiformis. These organisms have varied shapes, including cellular aggregations, amoebae, flagellates and amoeboflagellates.[6]
- Tunicaraptor unikontum is the newest discovered clade, whose position within Holozoa has yet to be resolved. It is a flagellate with a specialized "mouth" structure absent in other holozoans.[2]
- Metazoa, known as animals, are multicellular organisms that sum more than 1.5 million living species.[14] They are characterized by a blastula phase during their embryonic development and, except for the amorphous sponges, the formation of germ layers and differentiated tissues.[4]
Genetics
The first sequenced unicellular holozoan genome was that of Monosiga brevicollis, a choanoflagellate. It measures around 41.6 mega–base-pairs (Mbp) and contains around 9200 coding genes, making it comparable in size to the genome of filamentous fungi. Animal genomes are usually larger (e.g. human genome, 2900 Mbp; fruit fly, 180 Mbp), with some exceptions.[15]
Evolution
Phylogeny
Holozoa, along with a clade that contains fungi and their protist relatives (Holomycota), are part of the larger supergroup of eukaryotes known as Opisthokonta. Holozoa diverged from their opisthokont ancestor around 1070 million years ago (Mya).[16] The choanoflagellates, animals and filastereans group together as the clade Filozoa. Within Filozoa, the choanoflagellates and animals group together as the clade Choanozoa.[13] Based on phylogenetic and phylogenomic analyses, the cladogram of Holozoa is shown below:[17][18][6][2]
| Opisthokonta |
|
||||||||||||||||||||||||||||||||||
| 1250 Mya |
Uncertainty remains around the relationship of the two most basal groups, Ichthyosporea and Pluriformea.[4] They may be sister to each other, forming the putative clade Teretosporea.[19] Alternatively, Ichthyosporea may be the earliest-branching of the two, while Pluriformea is sister to the Filozoa clade comprising filastereans, choanoflagellates and animals. This second outcome is more strongly supported after the discovery of Syssomonas.[2][6]
The position of Tunicaraptor, the newest holozoan member, is still unresolved. Three different phylogenetic positions of Tunicaraptor have been obtained from analyses: as the sister group to Filasterea, as sister to Filozoa, or as the most basal group of all Holozoa.[2][20]
Environmental DNA surveys of oceans have revealed new diverse lineages of Holozoa. Most of them nest within known groups, mainly Ichthyosporea and Choanoflagellata. However, one environmental clade does not nest within any known group and is a potential new holozoan lineage. It has been tentatively named MASHOL (for 'marine small Holozoa').[21]
Unicellular ancestry of animals

The quest to elucidate the evolutionary origins of animals from a unicellular ancestor requires an examination of the transition to multicellularity. In the absence of a fossil record documenting this evolution, insights into the unicellular ancestor of animals are obtained from the analysis of shared genes and genetic pathways between animals and their closest living unicellular relatives. The genetic content of these single-celled holozoans has revealed a significant discovery: many genetic characteristics previously thought as unique to animals can also be found in these unicellular relatives. This suggests that the origin of multicellular animals did not happen solely because of the appearance of new genes (i.e. innovation), but because of pre-existing genes that were adapted or utilized in new ways (i.e. co-option).[7][6] For example:
- Adhesion proteins are necessary in allowing cells to stick to each other and to the extracellular matrix, forming layers and tissues in animals. Some unicellular holozoans, like choanoflagellates and filastereans, possess genes that encode proteins involved in cell-cell adhesion and cell-matrix adhesion (e.g. cadherin and integrin, respectively). Other genes, however, seem to be exclusively found in animals (e.g. β-catenin).[7]
- ECM-related proteins, involved in the formation of the extracellular matrix, are present in other holozoans (e.g. laminins, collagens and fibronectins).[22]
- Signal transduction proteins are another requirement for metazoan multicellularity. Some animal cytoplasmic tyrosine kinases (such as focal adhesion kinase) and the Hippo signaling pathway are present in unicellular holozoans. Other signaling pathways highly conserved in animals (e.g. Hedgehog, WNT, TGFβ, JAK-STAT and Notch) are absent in other holozoans, but similar signaling receptors evolved independently in choanoflagellates, filastereans and ichthyosporeans (e.g. receptor tyrosine kinases).[7]
- A considerable portion of animal transcription factors (TF) is already present in unicellular holozoans, including some TF classes previously thought to be animal-specific (e.g. p53 and T-box).[7]
Additionally, many biological processes seen in animals are already present in their unicellular relatives, such as sexual reproduction and gametogenesis in the choanoflagellate Salpingoeca rosetta and several types of multicellular differentiation.[7]
Fossil record
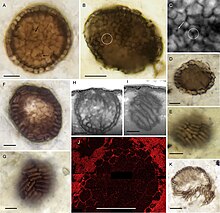
A billion-year-old freshwater microscopic fossil named Bicellum brasieri is possibly the earliest known holozoan. It shows two differentiated cell types or life cycle stages. It consists of a spherical ball of tightly packed cells (stereoblasts) enclosed in a single layer of elongated cells. There are also two populations of stereoblasts with mixed shapes, which have been interpreted as cellular migration to the periphery, a movement that could be explained by differential cell-cell adhesion. These occurrences are consistent with extant unicellular holozoans, which are known to form multicellular stages in complex life cycles.[3]
Proposed Ediacaran fossil "embryos" of early metazoans, discovered in the Doushantuo Formation, have been reinterpreted as non-animal protists within Holozoa. According to some authors, although they present possible embryonic cleavage, they lack metazoan synapomorphies such as tissue differentiation and nearby juveniles or adults. Instead, its development is comparable to the germination stage of non-animal holozoans. They possibly represent an evolutionary grade in which palintomic cleavage (i.e. rapid cell divisions without cytoplasmic growth in between, a characteristic of animal embryonic cleavage)[23] was the method of dispersal and propagation.[24]
Taxonomy
History
Prior to 2002, a relationship between Choanoflagellata, Ichthyosporea and the animal-fungi divergence was considered on the basis of morphology and ultrastructure. Early phylogenetic analyses gave contradicting results, because the amount of available DNA sequences was insufficient to yield unambiguous results. The taxonomic uncertainty was such that, for example, some Ichthyosporea were traditionally treated as trichomycete fungi.[1]
Holozoa was first recognized as a clade in 2002 through a phylogenomic analysis by Franz Bernd Lang, Charles J. O'Kelly and other collaborators, as part of a paper published in the journal Current Biology. The study used complete mitochondrial genomes of a choanoflagellate (Monosiga brevicollis) and an ichthyosporean (Amoebidium parasiticum) to firmly resolve the position of Ichthyosporea as the sister group to Choanoflagellata+Metazoa. This clade was named Holozoa (from Ancient Greek ὅλος (holos) 'whole' and ζῷον (zoion) 'animal'), meaning 'whole animal', referencing the wider animal ancestry that it contains.[1]
Holozoa has since been supported as a robust clade by every posterior analysis,[20] even after the discovery of more taxa nested within it (namely Filasterea since 2008,[13] and the pluriformean species Corallochytrium and Syssomonas since 2014[25] and 2017[6] respectively). As of 2019, the clade is accepted by the International Society of Protistologists, which revises the classification of eukaryotes.[4]
Classification
In classifications that use traditional taxonomic ranks (e.g. kingdom, phylum, class), all holozoan protists are classified as subphylum Choanofila (phylum Choanozoa,[a] kingdom Protozoa) while the animals are classified as a separate kingdom Metazoa or Animalia.[26] This classification excludes animals, even though they descend from the same common ancestor as choanofilan protists, making it a paraphyletic group rather than a true clade. Modern cladistic approaches to eukaryotic classification prioritise monophyletic groupings over traditional ranks, which are increasingly perceived as redundant and superfluous. Because Holozoa is a clade, its use is preferred over the paraphyletic taxon Choanofila.[4]
- Holozoa Lang et al. 2002
- Incertae sedis: †Bicellum brasieri Strother & Wellman 2021[3]
- Tunicaraptor Tikhonenkov, Mikhailov, Hehenberger, Karpov, Prokina, Esaulov, Belyakova, Mazei, Mylnikov, Aleoshin & Keeling 2020[2]
- Ichthyosporea Cavalier-Smith 1998 [Mesomycetozoea Mendoza et al. 2002]
- Dermocystida Cavalier-Smith 1998
- Ichthyophonida Cavalier-Smith 1998
- Pluriformea Hehenberger et al. 2017
- Corallochytrium Raghu-Kumar 1987
- Syssomonas Tikhonenkov, Hehenberger, Mylnikov & Keeling 2017
- Filozoa Shalchian-Tabrizi et al. 2008
- Filasterea Shalchian-Tabrizi et al. 2008
- Capsaspora Hertel, Bayne & Loker, 2002
- Ministeria Patterson et al. 1993
- Pigoraptor Tikhonenkov et al. 2017
- Txikispora Urrutia, Feist & Bass 2022[12]
- Choanozoa Brunet & King 2017 [Choanozoa Cavalier-Smith et al. 1991 (P)][a]
- Choanoflagellata Kent 1880–1882 [Choanoflagellatea Cavalier-Smith 1997 emend. Cavalier-Smith 1998]
- Craspedida Cavalier-Smith 1997, emend. Nitsche et al. 2011
- Acanthoecida Cavalier-Smith 1997, emend. Nitsche et al. 2011
- Metazoa Haeckel 1874, emend. Adl et al. 2005 [Animalia Linnaeus 1758]
- Porifera Grant 1836
- Placozoa Grell 1971
- Ctenophora Eschscholtz 1829
- Cnidaria Hatschek 1888
- Bilateria Hatschek 1888
- Choanoflagellata Kent 1880–1882 [Choanoflagellatea Cavalier-Smith 1997 emend. Cavalier-Smith 1998]
- Filasterea Shalchian-Tabrizi et al. 2008
Notes
- ^ a b The term "Choanozoa" has been used since 1991 by Cavalier-Smith as a paraphyletic phylum of opisthokont protists,[27] and the terms "Apoikozoa" and "choanimal" were proposed as names for the clade Metazoa+Choanoflagellata. However, these terms have not been formally described or adopted, and were rejected in favor of a renamed Choanozoa to fit the clade Metazoa+Choanoflagellata.[4]
References
- ^ a b c Lang BF, O'Kelly C, Nerad T, Gray MW, Burger G (2002). "The Closest Unicellular Relatives of Animals". Current Biology. 12 (20): 1773–1778. doi:10.1016/S0960-9822(02)01187-9. PMID 12401173.
- ^ a b c d e f g Tikhonenkov DV, Mikhailov KV, Hehenberger E, Mylnikov AP, Aleoshin VV, Keeling PJ, et al. (2020). "New Lineage of Microbial Predators Adds Complexity to Reconstructing the Evolutionary Origin of Animals". Current Biology. 30 (22): 4500–4509. doi:10.1016/j.cub.2020.08.061. PMID 32976804.
- ^ a b c Strother, Paul K.; Brasier, Martin D.; Wacey, David; Timpe, Leslie; Saunders, Martin; Wellman, Charles H. (April 2021). "A possible billion-year-old holozoan with differentiated multicellularity". Current Biology. 31 (12): 2658–2665.e2. doi:10.1016/j.cub.2021.03.051. PMID 33852871.
- ^ a b c d e f g Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. PMC 6492006. PMID 30257078.
- ^ Cavalier-Smith, Thomas (2009). "Megaphylogeny, Cell Body Plans, Adaptive Zones: Causes and Timing of Eukaryote Basal Radiations". Journal of Eukaryotic Microbiology. 56: 26–33. doi:10.1111/j.1550-7408.2008.00373.x.
- ^ a b c d e f g Hehenberger, Elisabeth; Tikhonenkov, Denis V.; Kolisko, Martin; Campo, Javier del; Esaulov, Anton S.; Mylnikov, Alexander P.; Keeling, Patrick J. (2017). "Novel Predators Reshape Holozoan Phylogeny and Reveal the Presence of a Two-Component Signaling System in the Ancestor of Animals". Current Biology. 27 (13): 2043–2050.e6. doi:10.1016/j.cub.2017.06.006. PMID 28648822.
- ^ a b c d e f g Sebé-Pedrós A, Degnan B, Ruiz-Trillo I (2017). "The origin of Metazoa: a unicellular perspective". Nature Reviews Genetics. 18 (8): 498–512. doi:10.1038/nrg.2017.21. PMID 28479598. S2CID 30709486.
- ^ Steenkamp, Emma T.; Wright, Jane; Baldauf, Sandra L. (January 2006). "The Protistan Origins of Animals and Fungi". Molecular Biology and Evolution. 23 (1): 93–106. doi:10.1093/molbev/msj011. PMID 16151185.
- ^ Simpson AGB, Slamovits CH, Archibald JM (2017). "Chapter 1. Protist Diversity and Eukaryote Phylogeny". In Archibald JM, Simpson AGB, Slamovits CH (eds.). Handbook of the Protists. Vol. 1 (2 ed.). Springer International Publishing. pp. 1–22. ISBN 978-3-319-28147-6.
- ^ Brunet T, King N (2022). "The Single-Celled Ancestors of Animals: A History of Hypotheses". In Herron MD, Conlin PL, Ratcliff WC (eds.). The Evolution of Multicellularity. Evolutionary Cell Biology. CRC Press. pp. 251–278. doi:10.1201/9780429351907-17. ISBN 9780429351907.
- ^ Ragan, Mark A.; Goggin, C. Louise; Cawthorn, Richard J.; Cerenius, Lage; Jamieson, Angela V.C.; Plourde, Susan M.; Rand, Thomas G.; Söoderhäll, Kenneth; Gutell, Robin R. (15 October 1996). "A novel clade of protistan parasites near the animal-fungal divergence". PNAS. 93 (21): 11907–11912. Bibcode:1996PNAS...9311907R. doi:10.1073/pnas.93.21.11907. PMC 38157. PMID 8876236.
- ^ a b Urrutia A, Mitsi K, Foster R, Ross S, Carr M, Ward GM, et al. (2022). "Txikispora philomaios n. sp., n. g., a micro-eukaryotic pathogen of amphipods, reveals parasitism and hidden diversity in Class Filasterea". Journal of Eukaryotic Microbiology. 69 (2): e12875. doi:10.1111/jeu.12875. PMID 34726818. S2CID 240422937.
- ^ a b c Shalchian-Tabrizi, Kamran; Minge, Marianne A.; Espelund, Mari; Orr, Russell; Ruden, Torgeir; Jakobsen, Kjetill S.; Cavalier-Smith, Thomas; Aramayo, Rodolfo (7 May 2008). Aramayo, Rodolfo (ed.). "Multigene phylogeny of choanozoa and the origin of animals". PLOS ONE. 3 (5): e2098. Bibcode:2008PLoSO...3.2098S. doi:10.1371/journal.pone.0002098. PMC 2346548. PMID 18461162.
- ^ Zhang, Zhi-Qiang (2013). "Animal biodiversity: an update of classification and diversity in 2013+". Zootaxa. 3703 (1): 5–11. doi:10.11646/zootaxa.3703.1.3.
- ^ King N, Westbrook M, Young S, et al. (2008). "The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans". Nature. 451: 783–788. doi:10.1038/nature06617.
- ^ Lawal HM, Schilde C, Kin K, et al. (2020). "Cold climate adaptation is a plausible cause for evolution of multicellular sporulation in Dictyostelia". Scientific Reports. 10: 8797. doi:10.1038/s41598-020-65709-3.
- ^ Parfrey, Laura Wegener; Lahr, Daniel J. G.; Knoll, Andrew H.; Katz, Laura A. (August 16, 2011). "Estimating the timing of early eukaryotic diversification with multigene molecular clocks". Proceedings of the National Academy of Sciences of the United States of America. 108 (33): 13624–13629. Bibcode:2011PNAS..10813624P. doi:10.1073/pnas.1110633108. PMC 3158185. PMID 21810989.
- ^ Torruella, Guifré; de Mendoza, Alex; Grau-Bové, Xavier; Antó, Meritxell; Chaplin, Mark A.; del Campo, Javier; Eme, Laura; Pérez-Cordón, Gregorio; Whipps, Christopher M. (21 September 2015). "Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi". Current Biology. 25 (18): 2404–2410. doi:10.1016/j.cub.2015.07.053. ISSN 0960-9822. PMID 26365255.
- ^ Grau-Bové, Xavier; Torruella, Guifré; Donachie, Stuart; Suga, Hiroshi; Leonard, Guy; Richards, Thomas A; Ruiz-Trillo, Iñaki (2017). "Dynamics of genomic innovation in the unicellular ancestry of animals". eLife. 6: e26036. doi:10.7554/eLife.26036.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Ros-Rocher N, Pérez-Posada A, Michelle LM, Ruiz-Trillo I (February 2021). "The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition". Open Biol. 11 (2): 200359. doi:10.1098/rsob.200359. PMID 33622103.
- ^ Arroyo, Alicia S; Lannes, Romain; Bapteste, Eric; Ruiz-Trillo, Iñaki (September 2020). "Gene Similarity Networks Unveil a Potential Novel Unicellular Group Closely Related to Animals from the Tara Oceans Expedition". Genome Biology and Evolution. 12 (9): 1664–1678. doi:10.1093/gbe/evaa117.
- ^ Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoot M, Sánchez-Pons N, Torruella G, Derelle R, Manning G, Lang BF, Russ C, Haas BJ, Roger AJ, Nusbaum C, Ruiz-Trillo I (2013). "The Capsaspora genome reveals a complex unicellular prehistory of animals". Nature Communications. 4 (2325): 2325. Bibcode:2013NatCo...4.2325S. doi:10.1038/ncomms3325. PMC 3753549. PMID 23942320.
- ^ Chen L, Xiao S, Pang K, Zhou C, Yuan X (September 2014). "Cell differentiation and germ–soma separation in Ediacaran animal embryo-like fossils". Nature. 516 (7530): 238–241. Bibcode:2014Natur.516..238C. doi:10.1038/nature13766. PMID 25252979. S2CID 4448316.
- ^ Huldtgren T, Cunningham JA, Yin C, Stampanoni M, Marone F, Donoghue PCJ, Bengtson S (2011). "Fossilized Nuclei and Germination Structures Identify Ediacaran "Animal Embryos" as Encysting Protists". Science. 334 (6063): 1696–1699. Bibcode:2011Sci...334Q1696H. doi:10.1126/science.1209537. PMID 22194575. S2CID 39813961.
- ^ Torruella G, de Mendoza A, Grau-Bové X, Donachie S, Pérez-Cordón G, Sitjà-Bobadilla A, Paley R, Manohar CS, Nichols K, Eme L, del Campo J (2014). "Phylotranscriptomics reveals ancient and convergent features in Corallochytrium and Ministeria (Holozoa, Opisthokonta)". Phylogeny and evolutionary perspective of Opisthokonta protists (PDF) (PhD thesis). Vol. 75. Universitat de Barcelona. pp. 1–9.
- ^ Cavalier-Smith T (May 2022). "Ciliary transition zone evolution and the root of the eukaryote tree: implications for opisthokont origin and classification of kingdoms Protozoa, Plantae, and Fungi". Protoplasma. 259 (3): 487–593. doi:10.1007/s00709-021-01665-7. PMC 9010356. PMID 34940909.
- ^ Cavalier-Smith T (May 2013). "Early evolution of eukaryote feeding modes, cell structural diversity, and classification of the protozoan phyla Loukozoa, Sulcozoa, and Choanozoa". European Journal of Protistology. 49 (2): 115–178. doi:10.1016/j.ejop.2012.06.001. PMID 23085100.

