Intron
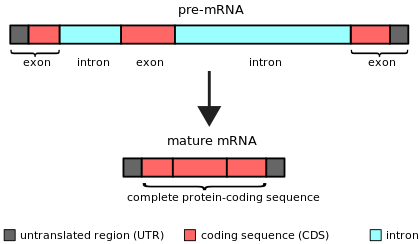
An intron (for intragenic region) is any nucleotide sequence within a gene that is removed by RNA splicing during maturation of the final RNA product.[1][2] In other words, introns are non-coding regions of an RNA transcript, or the DNA encoding it, that are eliminated by splicing before translation.[3][4] The word intron is derived from the term intragenic region, i.e. a region inside a gene.[5] The term intron refers to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts.[6] Sequences that are joined together in the final mature RNA after RNA splicing are exons.
Introns are found in the genes of most organisms and many viruses and can be located in a wide range of genes, including those that generate proteins, ribosomal RNA (rRNA) and transfer RNA (tRNA). When proteins are generated from intron-containing genes, RNA splicing takes place as part of the RNA processing pathway that follows transcription and precedes translation.
Discovery and etymology
Introns were first discovered in protein-coding genes of adenovirus,[7][8] and were subsequently identified in genes encoding transfer RNA and ribosomal RNA genes. Introns are now known to occur within a wide variety of genes throughout organisms and viruses within all of the biological kingdoms.
The fact that genes were split or interrupted by introns was discovered independently in 1977 by Phillip Allen Sharp and Richard J. Roberts, for which they shared the Nobel Prize in Physiology or Medicine in 1993.[9] The term intron was introduced by American biochemist Walter Gilbert:[5]
"The notion of the cistron [i.e., gene] ... must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions which will be expressed – exons." (Gilbert 1978)
The term intron also refers to intracistron, i.e., an additional piece of DNA that arises within a cistron.[10]
Although introns are sometimes called intervening sequences,[11] the term "intervening sequence" can refer to any of several families of internal nucleic acid sequences that are not present in the final gene product, including inteins, untranslated sequences (UTR), and nucleotides removed by RNA editing, in addition to introns.
Distribution
The frequency of introns within different genomes is observed to vary widely across the spectrum of biological organisms. For example, introns are extremely common within the nuclear genome of jawed vertebrates (e.g. humans and mice), where protein-coding genes almost always contain multiple introns, while introns are rare within the nuclear genes of some eukaryotic microorganisms,[12] for example baker's/brewer's yeast (Saccharomyces cerevisiae). In contrast, the mitochondrial genomes of vertebrates are entirely devoid of introns, while those of eukaryotic microorganisms may contain many introns.[13]
A particularly extreme case is the Drosophila dhc7 gene containing a ≥3.6 megabase (Mb) intron, which takes roughly three days to transcribe.[14][15] On the other extreme, a recent study suggests that the shortest known eukaryotic intron length is 30 base pairs (bp) belonging to the human MST1L gene.[16]
Classification
Splicing of all intron-containing RNA molecules is superficially similar, as described above. However, different types of introns were identified through the examination of intron structure by DNA sequence analysis, together with genetic and biochemical analysis of RNA splicing reactions.
At least four distinct classes of introns have been identified:[1]
- Introns in nuclear protein-coding genes that are removed by spliceosomes (spliceosomal introns)
- Introns in nuclear and archaeal transfer RNA genes that are removed by proteins (tRNA introns)
- Self-splicing group I introns that are removed by RNA catalysis
- Self-splicing group II introns that are removed by RNA catalysis
Group III introns are proposed to be a fifth family, but little is known about the biochemical apparatus that mediates their splicing. They appear to be related to group II introns, and possibly to spliceosomal introns.[17]
Spliceosomal introns
Nuclear pre-mRNA introns (spliceosomal introns) are characterized by specific intron sequences located at the boundaries between introns and exons.[18] These sequences are recognized by spliceosomal RNA molecules when the splicing reactions are initiated.[19] In addition, they contain a branch point, a particular nucleotide sequence near the 3' end of the intron that becomes covalently linked to the 5' end of the intron during the splicing process, generating a branched (lariat) intron. Apart from these three short conserved elements, nuclear pre-mRNA intron sequences are highly variable. Nuclear pre-mRNA introns are often much longer than their surrounding exons.
tRNA introns
Transfer RNA introns that depend upon proteins for removal occur at a specific location within the anticodon loop of unspliced tRNA precursors, and are removed by a tRNA splicing endonuclease. The exons are then linked together by a second protein, the tRNA splicing ligase.[20] Note that self-splicing introns are also sometimes found within tRNA genes.[21]
Group I and group II introns
Group I and group II introns are found in genes encoding proteins (messenger RNA), transfer RNA and ribosomal RNA in a very wide range of living organisms.,[22][23] Following transcription into RNA, group I and group II introns also make extensive internal interactions that allow them to fold into a specific, complex three-dimensional architecture. These complex architectures allow some group I and group II introns to be self-splicing, that is, the intron-containing RNA molecule can rearrange its own covalent structure so as to precisely remove the intron and link the exons together in the correct order. In some cases, particular intron-binding proteins are involved in splicing, acting in such a way that they assist the intron in folding into the three-dimensional structure that is necessary for self-splicing activity. Group I and group II introns are distinguished by different sets of internal conserved sequences and folded structures, and by the fact that splicing of RNA molecules containing group II introns generates branched introns (like those of spliceosomal RNAs), while group I introns use a non-encoded guanosine nucleotide (typically GTP) to initiate splicing, adding it on to the 5'-end of the excised intron.
Biological functions and evolution
While introns do not encode protein products, they are integral to gene expression regulation. Some introns themselves encode functional RNAs through further processing after splicing to generate noncoding RNA molecules.[24] Alternative splicing is widely used to generate multiple proteins from a single gene. Furthermore, some introns play essential roles in a wide range of gene expression regulatory functions such as Nonsense-mediated decay[25] and mRNA export.[26]
The biological origins of introns are obscure. After the initial discovery of introns in protein-coding genes of the eukaryotic nucleus, there was significant debate as to whether introns in modern-day organisms were inherited from a common ancient ancestor (termed the introns-early hypothesis), or whether they appeared in genes rather recently in the evolutionary process (termed the introns-late hypothesis). Another theory is that the spliceosome and the intron-exon structure of genes is a relic of the RNA world (the introns-first hypothesis).[27] There is still considerable debate about the extent to which of these hypotheses is most correct. The popular consensus at the moment is that introns arose within the eukaryote lineage as selfish elements.[28]
Early studies of genomic DNA sequences from a wide range of organisms show that the intron-exon structure of homologous genes in different organisms can vary widely.[29] More recent studies of entire eukaryotic genomes have now shown that the lengths and density (introns/gene) of introns varies considerably between related species. For example, while the human genome contains an average of 8.4 introns/gene (139,418 in the genome), the unicellular fungus Encephalitozoon cuniculi contains only 0.0075 introns/gene (15 introns in the genome).[30] Since eukaryotes arose from a common ancestor (common descent), there must have been extensive gain or loss of introns during evolutionary time.[31][32] This process is thought to be subject to selection, with a tendency towards intron gain in larger species due to their smaller population sizes, and the converse in smaller (particularly unicellular) species.[33] Biological factors also influence which genes in a genome lose or accumulate introns.[34][35][36]
Alternative splicing of exons within a gene after intron excision acts to introduce greater variability of protein sequences translated from a single gene, allowing multiple related proteins to be generated from a single gene and a single precursor mRNA transcript. The control of alternative RNA splicing is performed by a complex network of signaling molecules that respond to a wide range of intracellular and extracellular signals.
Introns contain several short sequences that are important for efficient splicing, such as acceptor and donor sites at either end of the intron as well as a branch point site, which are required for proper splicing by the spliceosome. Some introns are known to enhance the expression of the gene that they are contained in by a process known as intron-mediated enhancement (IME).
Actively transcribed regions of DNA frequently form R-loops that are vulnerable to DNA damage. In highly expressed yeast genes, introns inhibit R-loop formation and the occurrence of DNA damage.[37] Genome-wide analysis in both yeast and humans revealed that intron-containing genes have decreased R-loop levels and decreased DNA damage compared to intronless genes of similar expression.[37] Insertion of an intron within an R-loop prone gene can also suppress R-loop formation and recombination. Bonnet et al. (2017)[37] speculated that the function of introns in maintaining genetic stability may explain their evolutionary maintenance at certain locations, particularly in highly expressed genes.
Starvation adaptation
The physical presence of introns promotes cellular resistance to starvation via intron enhanced repression of ribosomal protein genes of nutrient-sensing pathways.[38]
As mobile genetic elements
Introns may be lost or gained over evolutionary time, as shown by many comparative studies of orthologous genes. Subsequent analyses have identified thousands of examples of intron loss and gain events, and it has been proposed that the emergence of eukaryotes, or the initial stages of eukaryotic evolution, involved an intron invasion.[39] Two definitive mechanisms of intron loss, Reverse Transcriptase-Mediated Intron Loss (RTMIL) and genomic deletions, have been identified, and are known to occur.[40] The definitive mechanisms of intron gain, however, remain elusive and controversial. At least seven mechanisms of intron gain have been reported thus far: Intron Transposition, Transposon Insertion, Tandem Genomic Duplication, Intron Transfer, Intron Gain during Double-Strand Break Repair (DSBR), Insertion of a Group II Intron, and Intronization. In theory it should be easiest to deduce the origin of recently gained introns due to the lack of host-induced mutations, yet even introns gained recently did not arise from any of the aforementioned mechanisms. These findings thus raise the question of whether or not the proposed mechanisms of intron gain fail to describe the mechanistic origin of many novel introns because they are not accurate mechanisms of intron gain, or if there are other, yet to be discovered, processes generating novel introns.[41]
In intron transposition, the most commonly purported intron gain mechanism, a spliced intron is thought to reverse splice into either its own mRNA or another mRNA at a previously intron-less position. This intron-containing mRNA is then reverse transcribed and the resulting intron-containing cDNA may then cause intron gain via complete or partial recombination with its original genomic locus. Transposon insertions can also result in intron creation. Such an insertion could intronize the transposon without disrupting the coding sequence when a transposon inserts into the sequence AGGT, resulting in the duplication of this sequence on each side of the transposon. It is not yet understood why these elements are spliced, whether by chance, or by some preferential action by the transposon. In tandem genomic duplication, due to the similarity between consensus donor and acceptor splice sites, which both closely resemble AGGT, the tandem genomic duplication of an exonic segment harboring an AGGT sequence generates two potential splice sites. When recognized by the spliceosome, the sequence between the original and duplicated AGGT will be spliced, resulting in the creation of an intron without alteration of the coding sequence of the gene. Double-stranded break repair via non-homologous end joining was recently identified as a source of intron gain when researchers identified short direct repeats flanking 43% of gained introns in Daphnia.[41] These numbers must be compared to the number of conserved introns flanked by repeats in other organisms, though, for statistical relevance. For group II intron insertion, the retrohoming of a group II intron into a nuclear gene was proposed to cause recent spliceosomal intron gain.
Intron transfer has been hypothesized to result in intron gain when a paralog or pseudogene gains an intron and then transfers this intron via recombination to an intron-absent location in its sister paralog. Intronization is the process by which mutations create novel introns from formerly exonic sequence. Thus, unlike other proposed mechanisms of intron gain, this mechanism does not require the insertion or generation of DNA to create a novel intron.[41]
The only hypothesized mechanism of recent intron gain lacking any direct evidence is that of group II intron insertion, which when demonstrated in vivo, abolishes gene expression.[42] Group II introns are therefore likely the presumed ancestors of spliceosomal introns, acting as site-specific retroelements, and are no longer responsible for intron gain.[43][44] Tandem genomic duplication is the only proposed mechanism with supporting in vivo experimental evidence: a short intragenic tandem duplication can insert a novel intron into a protein-coding gene, leaving the corresponding peptide sequence unchanged.[45] This mechanism also has extensive indirect evidence lending support to the idea that tandem genomic duplication is a prevalent mechanism for intron gain. The testing of other proposed mechanisms in vivo, particularly intron gain during DSBR, intron transfer, and intronization, is possible, although these mechanisms must be demonstrated in vivo to solidify them as actual mechanisms of intron gain. Further genomic analyses, especially when executed at the population level, may then quantify the relative contribution of each mechanism, possibly identifying species-specific biases that may shed light on varied rates of intron gain amongst different species.[41]
See also
Structure:
Splicing:
Function
Others:
References
- ^ a b Alberts, Bruce (2008). Molecular biology of the cell. New York: Garland Science. ISBN 978-0-8153-4105-5.
- ^ Stryer, Lubert; Berg, Jeremy Mark; Tymoczko, John L. (2007). Biochemistry. San Francisco: W.H. Freeman. ISBN 978-0-7167-6766-4.
- ^ Ghosh, Shampa; Sinha, Jitendra Kumar (2017), "Intron", in Vonk, Jennifer; Shackelford, Todd (eds.), Encyclopedia of Animal Cognition and Behavior, Springer International Publishing, pp. 1–5, doi:10.1007/978-3-319-47829-6_70-1, ISBN 978-3-319-47829-6
- ^ Editors, B. D. (6 August 2017). "Intron". Biology Dictionary. Retrieved 1 December 2019.
{{cite web}}:|last=has generic name (help) - ^ a b Gilbert, Walter (1978). "Why genes in pieces". Nature. 271 (5645): 501. doi:10.1038/271501a0. PMID 622185.
- ^ Kinniburgh, Alan; mertz, j; Ross, J. (July 1978). "The precursor of mouse β-globin messenger RNA contains two intervening RNA sequences". Cell. 14 (3): 681–693. doi:10.1016/0092-8674(78)90251-9. PMID 688388.
- ^ Chow LT, Gelinas RE, Broker TR, Roberts RJ (September 1977). "An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA". Cell. 12 (1): 1–8. doi:10.1016/0092-8674(77)90180-5. PMID 902310.
- ^ Berget SM, Moore C, Sharp PA (August 1977). "Spliced segments at the 5' terminus of adenovirus 2 late mRNA". Proc. Natl. Acad. Sci. U.S.A. 74 (8): 3171–5. doi:10.1073/pnas.74.8.3171. PMC 431482. PMID 269380.
- ^ https://www.nobelprize.org/nobel_prizes/medicine/laureates/1993/press.html
- ^ Tonegawa, S.; Maxam, A. M.; Tizard, R.; Bernard, O.; Gilbert, W. (1 March 1978). "Sequence of a mouse germ-line gene for a variable region of an immunoglobulin light chain". Proceedings of the National Academy of Sciences. 75 (3): 1485–1489. doi:10.1073/pnas.75.3.1485. ISSN 0027-8424. PMC 411497. PMID 418414.
- ^ Tilghman, S. M.; Tiemeier, D. C.; Seidman, J. G.; Peterlin, B. M.; Sullivan, M.; Maizel, J. V.; Leder, P. (1 February 1978). "Intervening sequence of DNA identified in the structural portion of a mouse beta-globin gene". Proceedings of the National Academy of Sciences. 75 (2): 725–729. doi:10.1073/pnas.75.2.725. ISSN 0027-8424. PMC 411329. PMID 273235.
- ^ Stajich JE, Dietrich FS, Roy SW (2007). "Comparative genomic analysis of fungal genomes reveals intron-rich ancestors". Genome Biol. 8 (10): R223. doi:10.1186/gb-2007-8-10-r223. PMC 2246297. PMID 17949488.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Taanman, Jan-Willem (1999). "The mitochondrial genome: structure, transcription, translation and replication". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1410 (2): 103–123. doi:10.1016/s0005-2728(98)00161-3. PMID 10076021 – via Elsevier Science Direct.
- ^ Tollervey, David; Caceres, Javier F (November 2000). "RNA Processing Marches on". Cell. 103 (5): 703–709. doi:10.1016/S0092-8674(00)00174-4. PMID 11114327.
- ^ Reugels, AM; Kurek, R; Lammermann, U; Bünemann, H (February 2000). "Mega-introns in the dynein gene DhDhc7(Y) on the heterochromatic Y chromosome give rise to the giant threads loops in primary spermatocytes of Drosophila hydei". Genetics. 154 (2): 759–69. PMC 1460963. PMID 10655227. Retrieved 12 December 2014.
- ^ Piovesan, Allison; Caracausi, Maria; Ricci, Marco; Strippoli, Pierluigi; Vitale, Lorenza; Pelleri, Maria Chiara (1 December 2015). "Identification of minimal eukaryotic introns through GeneBase, a user-friendly tool for parsing the NCBI Gene databank". DNA Research. 22 (6): 495–503. doi:10.1093/dnares/dsv028. PMC 4675715. PMID 26581719.
- ^ Copertino DW, Hallick RB (December 1993). "Group II and group III introns of twintrons: potential relationships with nuclear pre-mRNA introns". Trends Biochem. Sci. 18 (12): 467–71. doi:10.1016/0968-0004(93)90008-b. PMID 8108859.
- ^ Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA (1986). "Splicing of messenger RNA precursors". Annu. Rev. Biochem. 55: 1119–50. doi:10.1146/annurev.bi.55.070186.005351. PMID 2943217.
- ^ Guthrie C, Patterson B (1988). "Spliceosomal snRNAs". Annu. Rev. Genet. 22: 387–419. doi:10.1146/annurev.ge.22.120188.002131. PMID 2977088.
- ^ Greer CL, Peebles CL, Gegenheimer P, Abelson J (February 1983). "Mechanism of action of a yeast RNA ligase in tRNA splicing". Cell. 32 (2): 537–46. doi:10.1016/0092-8674(83)90473-7. PMID 6297798.
- ^ Reinhold-Hurek B, Shub DA (May 1992). "Self-splicing introns in tRNA genes of widely divergent bacteria". Nature. 357 (6374): 173–6. doi:10.1038/357173a0. PMID 1579169.
- ^ Cech TR (1990). "Self-splicing of group I introns". Annu. Rev. Biochem. 59: 543–68. doi:10.1146/annurev.bi.59.070190.002551. PMID 2197983.
- ^ Michel F, Ferat JL (1995). "Structure and activities of group II introns". Annu. Rev. Biochem. 64: 435–61. doi:10.1146/annurev.bi.64.070195.002251. PMID 7574489.
- ^ Rearick D, Prakash A, McSweeny A, Shepard SS, Fedorova L, Fedorov A (March 2011). "Critical association of ncRNA with introns". Nucleic Acids Res. 39 (6): 2357–66. doi:10.1093/nar/gkq1080. PMC 3064772. PMID 21071396.
- ^ Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ (December 2012). "Introns in UTRs: why we should stop ignoring them". BioEssays. 34 (12): 1025–34. doi:10.1002/bies.201200073. PMID 23108796.
- ^ Cenik, Can; Chua, Hon Nian; Zhang, Hui; Tarnawsky, Stefan P.; Akef, Abdalla; Derti, Adnan; Tasan, Murat; Moore, Melissa J.; Palazzo, Alexander F.; Roth, Frederick P. (2011). Snyder, Michael (ed.). "Genome Analysis Reveals Interplay between 5′UTR Introns and Nuclear mRNA Export for Secretory and Mitochondrial Genes". PLOS Genetics. 7 (4): e1001366. doi:10.1371/journal.pgen.1001366. ISSN 1553-7404. PMC 3077370. PMID 21533221.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Penny D, Hoeppner MP, Poole AM, Jeffares DC (November 2009). "An overview of the introns-first theory". Journal of Molecular Evolution. 69 (5): 527–40. doi:10.1007/s00239-009-9279-5. PMID 19777149.
- ^ Cavalier-Smith, T (1985). "Selfish DNA and the origin of introns". Nature. 315 (6017): 283–4. doi:10.1038/315283b0. PMID 2987701.
- ^ Rodríguez-Trelles F, Tarrío R, Ayala FJ (2006). "Origins and evolution of spliceosomal introns". Annu. Rev. Genet. 40: 47–76. doi:10.1146/annurev.genet.40.110405.090625. PMID 17094737.
- ^ Mourier T, Jeffares DC (May 2003). "Eukaryotic intron loss". Science. 300 (5624): 1393. doi:10.1126/science.1080559. PMID 12775832.
- ^ Roy SW, Gilbert W (March 2006). "The evolution of spliceosomal introns: patterns, puzzles and progress". Nature Reviews Genetics. 7 (3): 211–21. doi:10.1038/nrg1807. PMID 16485020.
- ^ de Souza SJ (July 2003). "The emergence of a synthetic theory of intron evolution". Genetica. 118 (2–3): 117–21. doi:10.1023/A:1024193323397. PMID 12868602.
- ^ Lynch M (April 2002). "Intron evolution as a population-genetic process". Proceedings of the National Academy of Sciences. 99 (9): 6118–23. doi:10.1073/pnas.092595699. PMC 122912. PMID 11983904.
- ^ Jeffares DC, Mourier T, Penny D (January 2006). "The biology of intron gain and loss". Trends in Genetics. 22 (1): 16–22. doi:10.1016/j.tig.2005.10.006. PMID 16290250.
- ^ Jeffares DC, Penkett CJ, Bähler J (August 2008). "Rapidly regulated genes are intron poor". Trends in Genetics. 24 (8): 375–8. doi:10.1016/j.tig.2008.05.006. PMID 18586348.
- ^ Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA (August 2002). "Selection for short introns in highly expressed genes". Nature Genetics. 31 (4): 415–8. doi:10.1038/ng940. PMID 12134150.
- ^ a b c Bonnet A, Grosso AR, Elkaoutari A, Coleno E, Presle A, Sridhara SC, Janbon G, Géli V, de Almeida SF, Palancade B (2017). "Introns Protect Eukaryotic Genomes from Transcription-Associated Genetic Instability". Mol. Cell. 67 (4): 608–621.e6. doi:10.1016/j.molcel.2017.07.002. PMID 28757210.
- ^ Parenteau, Julie; Maignon, Laurine; Berthoumieux, Mélodie; Catala, Mathieu; Gagnon, Vanessa; Abou Elela, Sherif (16 January 2019). "Introns are mediators of cell response to starvation". Nature. 565 (7741): 612–617. doi:10.1038/s41586-018-0859-7. ISSN 1476-4687. PMID 30651641.
- ^ Rogozin, I. B.; Carmel, L.; Csuros, M.; Koonin, E. V. (2012). "Origin and evolution of spliceosomal introns". Biology Direct. 7: 11. doi:10.1186/1745-6150-7-11. PMC 3488318. PMID 22507701.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Derr, L. K.; Strathern, J. N. (1993). "A role for reverse transcripts in gene conversion". Nature. 361 (6408): 170–173. doi:10.1038/361170a0. PMID 8380627.
- ^ a b c d Yenerall, P.; Zhou, L. (2012). "Identifying the mechanisms of intron gain: Progress and trends". Biology Direct. 7: 29. doi:10.1186/1745-6150-7-29. PMC 3443670. PMID 22963364.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Chalamcharla, V. R.; Curcio, M. J.; Belfort, M. (2010). "Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry". Genes & Development. 24 (8): 827–836. doi:10.1101/gad.1905010. PMC 2854396. PMID 20351053.
- ^ Cech, T. R. (1986). "The generality of self-splicing RNA: Relationship to nuclear mRNA splicing". Cell. 44 (2): 207–210. doi:10.1016/0092-8674(86)90751-8. PMID 2417724.
- ^ Dickson, L.; Huang, H. -R.; Liu, L.; Matsuura, M.; Lambowitz, A. M.; Perlman, P. S. (2001). "Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites". Proceedings of the National Academy of Sciences. 98 (23): 13207–13212. doi:10.1073/pnas.231494498. PMC 60849. PMID 11687644.
- ^ Hellsten, U.; Aspden, J. L.; Rio, D. C.; Rokhsar, D. S. (2011). "A segmental genomic duplication generates a functional intron". Nature Communications. 2: 454–. doi:10.1038/ncomms1461. PMC 3265369. PMID 21878908.
External links
- A search engine for exon/intron sequences defined by NCBI
- Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter Molecular Biology of the Cell, 2007, ISBN 978-0-8153-4105-5. Fourth edition is available online through the NCBI Bookshelf: link
- Jeremy M Berg, John L Tymoczko, and Lubert Stryer, Biochemistry 5th edition, 2002, W H Freeman. Available online through the NCBI Bookshelf: link
- Intron finding tool for plant genomic sequences
- Exon-intron graphic maker
