Gracilicutes
| Gracilicutes | |
|---|---|

| |
| Escherichia coli cells magnified 25,000 times | |
| Scientific classification | |
| Domain: | |
| (unranked): | Gracilicutes Gibbons and Murray 1978[1]
|
| Superphyla/Phyla | |
Gracilicutes (Latin: gracilis, slender, and cutis, skin, referring to the cell wall) is a clade in bacterial phylogeny.[2]
Traditionally gram staining results were most commonly used as a classification tool, consequently until the advent of molecular phylogeny, the Kingdom Monera (as the domains Bacteria and Archaea were known then) was divided into four phyla,[3][4]
- Gracilicutes (gram-negative, it is split in many groups, but some authors still use it in a narrower sense)
- Firmacutes [sic] (gram-positive, subsequently corrected to Firmicutes,[5] today it excludes the Actinobacteria)
- Mollicutes (gram variable, posteriorly renamed as Tenericutes, e.g. Mycoplasma)
- Mendosicutes (uneven gram stain, "methanogenic bacteria" now known as methanogens and classed as Archaea)
This classification system was abandoned in favour of the three-domain system based on molecular phylogeny started by C. Woese.[6][7]
This taxon was revived in 2006 by Cavalier-Smith as an infrakindgom containing the phyla Spirochaetae, Sphingobacteria (FCB), Planctobacteria (PVC), and Proteobacteria.[8] It is a gram-negative clade that branched off from other bacteria just before the evolutionary loss of the outer membrane or capsule, and just after the evolution of flagella.[8] This taxon is not generally accepted and the three-domain system is followed.[9]
An almost identical taxon called Hydrobacteria was defined in 2009 using bioinfomatic methods. It is in contrast to the other major group of eubacteria called Terrabacteria. The analysis also altered the internal phylogeny of this group, with PVC-FCB-Spirochaetae placed forming a clade next to Proteobacteria. What was PVC is described as polyphyletic.[10] However, in the other subsequent phylogenetic analyzes, the PVC group was recovered as monophyletic.
Relationships
The phylogenetic tree according to the phylogenetic analyzes of Battistuzzi (2009) is the following and with a molecular clock calibration.[10]
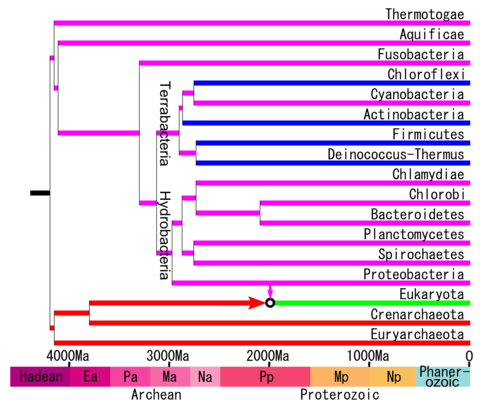
Recent phylogenetic analyzes have found that proteobacteria are a paraphyletic phylum that could encompass several recently discovered candidate phyla and other phyla such as Acidobacteria, Chrysiogenetes, Deferribacteres and possibly Aquificae. This suggests that Gracilicutes or Hydrobacteria as a clade may comprise several candidates more closely related to Proteobacteria, Spirochaetes, PVC group, and FCB group than to bacteria from the clade Terrabacteria. Some of these phyla were classified as part of the proteobacteria. For example, Cavalier-Smith in his proposal of the 6 kingdoms included Acidobacteria, Aquificae, Chrysiogenetes and Deferribacteres as part of the proteobacteria.[11]
Phylogenetic analyzes have found roughly the following phylogeny between the major and some more closely related phyla.[12][13][14][15]
| Gracilicutes |
| ||||||||||||||||||||||||||||||||||||||||||||||||
According to the phylogenetic analysis of Hug (2016), the relationships could be the following.[16]

According to the phylogenetic analysis of Zhu (2019), the relationships could be the following:[15]
The following graph shows Cavalier-Smith's version of the tree of life, indicating the status of Gracilicutes. However, this tree is not supported by any molecular analysis so it should not be considered phylogenetic.
Cavalier-Smith's Tree of Life, 2006[cstol 1]
[A]
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Legend:
[A] Gram-negative with a peptidoglycan cell wall like Chlorosome.
[B] Oxygenic Photosynthesis, Omp85 and four new catalases.
[C] Glycobacterial revolution: outer membrane with insertion of lipopolysaccharides, hopanoids, diaminopimelic acid, ToIC and TonB.
[D] Phycobilin chromophores.
[E] Flagella.
[F] Four sections: an amino acid in HSP60 and FtsZ and a domain in RNA polymerases β and σ.
[G] Endospores.
[H] Gram-positive Bacteria: hypertrophy of the wall peptidoglycan, sortase enzyme and a loss of the outer membrane.
[I] Glycerol 1-P dehydrogenase.
[J] Proteasome and phosphatidylinositol.
[K] Neomura revolution: Replacement of peptidoglycan by glycoproteins and lipoproteins.
[L] Reverse DNA gyrase and ether lipid isoprenoids.
[M] Phagocytosis.
- ^ Cavalier-Smith T (2006). "Cell evolution and Earth history: stasis and revolution". Philos Trans R Soc Lond B Biol Sci. 361 (1470): 969–1006. doi:10.1098/rstb.2006.1842. PMC 1578732. PMID 16754610.
References
- ^ Gibbons, N. E. & Murray, R. G. E. 1978. Proposals concerning the higher taxa of bacteria. Int J Syst Bacteriol 28:1–6, (PDF)
- ^ Boussau, Bastien; et al. (2008). "Phylogenetic tree obtained from 56 genes of Bacteria". BMC Evolutionary Biology. 8: 272. doi:10.1186/1471-2148-8-272.
Accounting for horizontal gene transfers explains conflicting hypotheses regarding the position of aquificales in the phylogeny of Bacteria
- ^ Gibbons, N.E.; Murray, R.G.E. (1978). "Proposals concerning the higher taxa of bacteria" (PDF). International Journal of Systematic Bacteriology. 28: 1–6. doi:10.1099/00207713-28-1-1.
- ^ Krieg, N.R. & Holt, J.C. (eds., 1984). Bergey’s Manual of Systematic Bacteriology, 1st ed., vol. 1, Williams and Wilkins, Baltimore, [1].
- ^ Murray, R.G.E. (1984). The higher taxa, or, a place for everything...?. In: N.R. Krieg & J.G. Holt (ed.) Bergey's Manual of Systematic Bacteriology, vol. 1, The Williams & Wilkins Co., Baltimore, p. 31-34
- ^ Woese, C. R. (1987). "Bacterial evolution". Microbiological Reviews. 51 (2): 221–271. doi:10.1128/MMBR.51.2.221-271.1987. PMC 373105. PMID 2439888.
- ^ Don J. Brenner; Noel R. Krieg; James T. Staley (July 26, 2005) [1984(Williams & Wilkins)]. George M. Garrity (ed.). Introductory Essays. Bergey's Manual of Systematic Bacteriology. Vol. 2A (2nd ed.). New York: Springer. p. 304. ISBN 978-0-387-24143-2. British Library no. GBA561951.
- ^ a b Cavalier-Smith T (2006). "Rooting the tree of life by transition analyses". Biol. Direct. 1: 19. doi:10.1186/1745-6150-1-19. PMC 1586193. PMID 16834776.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Krieg, N.R.; Ludwig, W.; Whitman, W.B.; Hedlund, B.P.; Paster, B.J.; Staley, J.T.; Ward, N.; Brown, D.; Parte, A. (November 24, 2010) [1984(Williams & Wilkins)]. George M. Garrity (ed.). The Bacteroidetes, Spirochaetes, Tenericutes (Mollicutes), Acidobacteria, Fibrobacteres, Fusobacteria, Dictyoglomi, Gemmatimonadetes, Lentisphaerae, Verrucomicrobia, Chlamydiae, and Planctomycetes. Bergey's Manual of Systematic Bacteriology. Vol. 4 (2nd ed.). New York: Springer. p. 908. ISBN 978-0-387-95042-6. British Library no. GBA561951.
- ^ a b Battistuzzi, F. U.; Hedges, S. B. (6 November 2008). "A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land". Molecular Biology and Evolution. 26 (2): 335–343. doi:10.1093/molbev/msn247. PMID 18988685.
- ^ Cavalier-Smith 2006. The 10 phyla of the kingdom Bacteria Biology Direct 2006 1:19 doi:10.1186/1745-6150-1-19
- ^ Karthik Anantharaman et al. 2016, Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system Nature Communications DOI: 10.1038/ncomms13219
- ^ Gareth A. Coleman, Adrián A. Davín, Tara Mahendrarajah, Anja Spang, Philip Hugenholtz, Gergely J. Szöllősi, Tom A. Williams. (2020). A rooted phylogeny resolves early bacterial evolution. Biorxiv.
- ^ Christian Rinke et al 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature Volume: 499, Pages: 431–437 doi:10.1038/nature12352
- ^ a b Zhu, Q., Mai, U., Pfeiffer, W. et al. (2019) «Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea». Nature Communications, 10: 5477
- ^ Hug, L. A. et al. 2016, A new view of the tree of life. Nature Microbiology, 1, 16048.
