Coronavirus
| Coronavirus | |
|---|---|
| |
| Virus classification | |
| Group: | Group IV ((+)ssRNA)
|
| Order: | |
| Family: | |
| Subfamily: | |
| Type species | |
| Coronavirus | |
| Species | |
|
Human coronavirus HKU1 | |
Coronaviruses are species in the genera of virus belonging to one of two subfamilies Coronavirinae and Torovirinae in the family Coronaviridae, in the order Nidovirales.[1][2] Coronaviruses are enveloped viruses with a positive-sense RNA genome and with a nucleocapsid of helical symmetry. The genomic size of coronaviruses ranges from approximately 26 to 32 kilobases, extraordinarily large for an RNA virus.
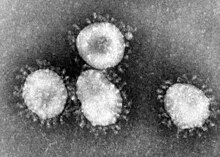
The name "coronavirus" is derived from the Latin corona, meaning crown or halo, and refers to the characteristic appearance of virions under electron microscopy (E.M.) with a fringe of large, bulbous surface projections creating an image reminiscent of the solar corona. This morphology is created by the viral spike (S) peplomers, which are proteins that populate the surface of the virus and determine host tropism.
Proteins that contribute to the overall structure of all coronaviruses are the spike (S), envelope (E), membrane (M) and nucleocapsid (N). In the specific case of the SARS coronavirus (see below), a defined receptor-binding domain on S mediates the attachment of the virus to its cellular receptor, angiotensin-converting enzyme 2 (ACE2).[3] Some coronaviruses (specifically the members of Betacoronavirus subgroup A) also have a shorter spike-like protein called hemagglutinin esterase (HE).[1]
History
Coronaviruses were first described in the 1960s from the nasal cavities of patients with the common cold. These viruses were subsequently named human coronavirus 229E and human coronavirus OC43.
Coronaviruses primarily infect the upper respiratory and gastrointestinal tract of mammals and birds. Six different currently known strains of coronaviruses infect humans. The most recently publicized human coronavirus, SARS-CoV which causes SARS, has a unique pathogenesis because it causes both upper and lower respiratory tract infections and can also cause gastroenteritis.
Coronaviruses are believed to cause a significant percentage of all common colds in human adults. Coronaviruses cause colds in humans primarily in the winter and early spring seasons. The significance and economic impact of coronaviruses as causative agents of the common cold are hard to assess because, unlike rhinoviruses (another common cold virus), human coronaviruses are difficult to grow in the laboratory. Coronaviruses can even cause pneumonia, either direct viral pneumonia or a secondary bacterial pneumonia.
In chickens, the infectious bronchitis virus (IBV), a coronavirus, targets not only the respiratory tract but also the uro-genital tract. The virus can spread to different organs throughout the chicken.
Coronaviruses also cause a range of diseases in farm animals and domesticated pets, some of which can be serious and are a threat to the farming industry. Economically significant coronaviruses of farm animals include porcine coronavirus (transmissible gastroenteritis coronavirus, TGE) and bovine coronavirus, which both result in diarrhea in young animals. Feline Coronavirus: two forms, Feline enteric coronavirus is a pathogen of minor clinical significance, but spontaneous mutation of this virus can result in feline infectious peritonitis (FIP), a disease associated with high mortality. Similarly, there are two types of coronavirus that infect ferrets: Ferret enteric coronavirus causes a gastrointestinal syndrome known as epizootic catarrhal enteritis (ECE), and a more lethal systemic version of the virus (like FIP in cats) known in ferrets as ferret systemic coronavirus (FSC).[4] There are two types of canine coronavirus (CCoV), one that causes mild gastrointestinal disease and one that has been found to cause respiratory disease. Mouse hepatitis virus (MHV) is a coronavirus that causes an epidemic murine illness with high mortality, especially among colonies of laboratory mice.
Prior to the discovery of SARS-CoV, MHV had been the best-studied coronavirus both in vivo and in vitro as well as at the molecular level. Some strains of MHV cause a progressive demyelinating encephalitis in mice which has been used as a murine model for multiple sclerosis. Significant research efforts have been focused on elucidating the viral pathogenesis of these animal coronaviruses, especially by virologists interested in veterinary and zoonotic diseases.
Replication

Replication of Coronavirus begins with entry to the cell which takes place in the cytoplasm in a membrane-protected microenvironment. Upon entry to the cell the virus particle is uncoated and the RNA genome is deposited into the cytoplasm.
The Coronavirus genome has a 5’ methylated cap and a 3’polyadenylated tail. This allows the RNA to attach to ribosomes for translation.
Coronaviruses also have a protein known as a replicase encoded in its genome which allows the RNA viral genome to be transcribed into new RNA copies using the host cell's machinery. The replicase is the first protein to be made; once the gene encoding the replicase is translated, the translation is stopped by a stop codon. This is known as a nested transcript. When the mRNA transcript only encodes one gene, it is monocistronic.
The RNA genome is replicated and a long polyprotein is formed, where all of the proteins are attached. Coronaviruses have a non-structural protein - a protease - which is able to separate the proteins in the chain. This is a form of genetic economy for the virus, allowing it to encode the greatest number of genes in a small number of nucleotides.
Severe acute respiratory syndrome
In 2003, following the outbreak of Severe acute respiratory syndrome (SARS) which had begun the prior year in Asia, and secondary cases elsewhere in the world, the World Health Organization (WHO) issued a press release stating that a novel coronavirus identified by a number of laboratories was the causative agent for SARS. The virus was officially named the SARS coronavirus (SARS-CoV).
The epidemic resulted in over 8,000 infections, about 10% of which resulted in death.[3] X-ray crystallography studies performed at the Advanced Light Source of Lawrence Berkeley National Laboratory have begun to give hope of a vaccine against the disease "since [the spike protein] appears to be recognized by the immune system of the host."[5]
Following the high-profile publicity of SARS outbreaks, there has been a renewed interest in coronaviruses among virologists. For many years, scientists knew about only two human coronaviruses (HCoV-229E and HCoV-OC43). The discovery of SARS-CoV added a third human coronavirus.
By the end of 2004, three independent research labs reported the discovery of a fourth human coronavirus. It has been named NL63, NL, and the New Haven coronavirus by different research groups.[6] The three labs are still arguing over which one discovered the virus first and has the right to name it.
Early in 2005, a research team at the University of Hong Kong reported finding a fifth human coronavirus in two patients with pneumonia. They named it Human coronavirus HKU1.
In September 2012, a sixth new type of coronavirus was identified, initially called Novel Coronavirus 2012, and now officially Middle East respiratory syndrome coronavirus (MERS-CoV) [7][8]
The World Health Organisation accordingly issued a global alert[9] The WHO update on 28 September 2012 said that the virus did not seem to pass easily from person to person.[10] However, on May 12, 2013, a case of contamination from human to human in France was confirmed by the French Ministry of Social Affairs and Health.[11] In addition, cases of person-to-person transmission have been reported by the Ministry of Health in Tunisia. Two confirmed cases seem to have caught the disease from their late father, who became ill after a visit to Qatar and Saudi Arabia.
By Oct. 30 2013, there were 124 cases and 52 deaths in Saudi Arabia.[12] After the Dutch Erasmus Medical Centre sequenced the virus, the virus was given a new name, Human Corona Virus-Erasmus Medical Centre (HCoV-EMC). The final name for the virus is Middle East respiratory syndrome coronavirus (MERS-CoV).
- Human coronavirus 229E
- Human coronavirus OC43
- SARS-CoV
- Human Coronavirus NL63 (HCoV-NL63, New Haven coronavirus)
- Human coronavirus HKU1
- Middle East respiratory syndrome coronavirus (MERS-CoV), previously known as Novel coronavirus 2012 and HCoV-EMC. see also here.
Coronaviruses have been recognized as causing pathological conditions in veterinary medicine since the early 1970s. Except for avian infectious bronchitis, the major related diseases have mainly an intestinal location.
(listed following their estimated economical importance)
- Infectious bronchitis virus (IBV) causes avian infectious bronchitis.
- Porcine coronavirus (transmissible gastroenteritis coronavirus of pigs, TGEV).
- Bovine coronavirus (BCV), responsible for severe profuse enteritis in of young calves.
- Feline coronavirus (FCoV) causes mild enteritis in cats as well as severe Feline infectious peritonitis (other variants of the same virus).
- the two types of canine coronavirus (CCoV) (one causing enteritis, the other found in respiratory diseases).
- Turkey coronavirus (TCV) causes enteritis in turkeys.
- Ferret enteric coronavirus causes epizootic catarrhal enteritis in ferrets.
- Ferret systemic coronavirus causes FIP-like systemic syndrome in ferrets.[13]
- Pantropic canine coronavirus
Another new veterinary disease, Porcine epidemic diarrhea virus, has emerged around the world. Its economic importance is as yet unclear, but shows high mortality in piglets.
Taxonomy
- Genus: Alphacoronavirus; type species: Alphacoronavirus 1
- Genus Betacoronavirus; type species: Murine coronavirus
- Genus Gammacoronavirus; type species: Avian coronavirus
- Genus Deltacoronavirus; type species: Bulbul coronavirus HKU11
Evolution
The most recent common ancestor of the coronovirus has been placed at 10,000 years before the present.[14] They may be considerably older than this.
Another estimate places the most recent common ancestor (MRCA) of all coronaviruses ~8100 BC.[15] The MRCA of Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus have been placed at ~2400 BC, ~3300 BC, ~2800 BC and ~3000 BC, respectively. It appears that bats and birds, the warm blooded flying vertebrates, are ideal hosts for the coronavirus gene source with bats for Alphacoronavirus and Betacoronavirus and birds for Gammacoronavirus and Deltacoronavirus, to fuel coronavirus evolution and dissemination.
Bovine coronavirus and canine respiratory coronavirus diverged from a common ancestor in 1951.[16] Bovine coronavirus and human coronavirus OC43 diverged in 1899. Bovine coronavirus diverged from the equine coronavirus species at the end of the 18th century.
Another estimate suggests that human coronavirus OC43 diverged from bovine coronavirus in 1890.[17]
The MRCA of human coronavirus OC43 has been dated to the 1950s.[18]
Middle East respiratory syndrome coronavirus although related to several bat species appears to have diverged from these several centuries ago.[18]
The most closely related bat coronovirus and the SARS coronavirus diverged in 1986.[19]
A path of evolution of the SARS virus and keen relationship with bats have been proposed.[20][21] The authors suggest that the coronaviruses have been coevolved with bats for a long time and the ancestors of SARS virus first infected the species of the genus Hipposideridae, subsequently spread to species of the Rhinolophidae and then to civets and finally to humans.
Alpaca coronavirus and human coronavirus 229E diverged before 1960.[22]
The human coronavirus NL63 and a bat coronovirus shared an MRCA 563 to 822 years ago.[23]
See also
References
- ^ a b de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebuhr J (2011). "Family Coronaviridae". Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier, Oxford. pp. 806–828. ISBN 978-0-12-384684-6.
{{cite book}}: Unknown parameter|editors=ignored (|editor=suggested) (help)CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link) - ^ International Committee on Taxonomy of Viruses (24 August 2010). "ICTV Master Species List 2009 – v10" (xls).
- ^ a b Li F, Li W, Farzan M, Harrison SC (September 2005). "Structure of SARS coronavirus spike receptor-binding domain complexed with receptor". Science. 309 (5742): 1864–8. doi:10.1126/science.1116480. PMID 16166518.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jerry Murray, DVM (16 April 2014). "What's New With Ferret FIP-like Disease?" (xls).
- ^ "Learning How SARS Spikes Its Quarry". Press Release PR-HHMI-05-4. Chevy Chase, MD: Howard Hughes Medical Institute. Retrieved September 16, 2005.
- ^ van der Hoek L; Pyrc; Jebbink; et al. (April 2004). "Identification of a new human coronavirus". Nature Medicine. 10 (4): 368–73. doi:10.1038/nm1024. PMID 15034574.
- ^ Doucleef, Michaeleen (26 September 2012). "Scientists Go Deep On Genes Of SARS-Like Virus". Associated Press. Retrieved 27 September 2012.
- ^ Falco, Miriam (24 September 2012). "New SARS-like virus poses medical mystery". CNN Health. Retrieved 16 March 2013.
- ^ "New SARS-like virus found in Middle East". Al-Jazeera. 24 September 2012. Retrieved 16 March 2013.
- ^ Kelland, Kate (28 September 2012). "New virus not spreading easily between people: WHO". Reuters. Retrieved 16 March 2013.
- ^ Nouveau coronavirus - Point de situation : Un nouveau cas d’infection confirmé (Novel coronavirus - Status report: A new case of confirmed infection) May 12, 2013 social-sante.gouv.fr
- ^ "Novel coronavirus infection - update". World Health Association. 22 May 2013. Retrieved 23 May 2013.
- ^ http://www.merckmanuals.com/vet/ the Merck Veterinary Manual
- ^ Wertheim, JO; Chu, DK; Peiris, JS; Kosakovsky Pond, SL; Poon, LL (2013). "A case for the ancient origin of coronaviruses". J Virol. 87 (12): 7039–7045. doi:10.1128/JVI.03273-12. PMID 23596293.
- ^ Woo, PC; Lau, SK; Lam, CS; Lau, CC; Tsang, AK; Lau, JH; Bai, R; Teng, JL; Tsang, CC; et al. (2012). "Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus". J Virol. 86 (7): 3995–4008. doi:10.1128/JVI.06540-11. PMC 3302495. PMID 22278237.
{{cite journal}}: Explicit use of et al. in:|author10=(help); Explicit use of et al. in:|first9=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Bidokhti, MR; Tråvén, M; Krishna, NK; Munir, M; Belák, S; Alenius, S; Cortey, M (2013). "Evolutionary dynamics of bovine coronaviruses: natural selection pattern of the spike gene implies adaptive evolution of the strains". J Gen Virol. 94 (9): 2036–2049. doi:10.1099/vir.0.054940-0.
- ^ Vijgen, L; Keyaerts, E; Moës, E; Thoelen, I; Wollants, E; Lemey, P; Vandamme, AM; Van Ranst, M (2005). "Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event". J Virol. 79 (3): 1595–1604. doi:10.1128/jvi.79.3.1595-1604.2005. PMID 15650185.
- ^ a b Lau, SK; Lee, P; Tsang, AK; Yip, CC; Tse, H; Lee, RA; So, LY; Lau, YL; Chan, KH; et al. (2011). "Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination". J Virol. 85 (21): 11325–37. doi:10.1128/JVI.05512-11. PMC 3194943. PMID 21849456.
{{cite journal}}: Explicit use of et al. in:|author10=(help); Explicit use of et al. in:|first9=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) Cite error: The named reference "Lau2013" was defined multiple times with different content (see the help page). - ^ Vijaykrishna, D; Smith, GJ; Zhang, JX; Peiris, JS; Chen, H; Guan, Y (2007). "Evolutionary insights into the ecology of coronaviruses". J Virol. 81 (8): 4012–4020. doi:10.1128/jvi.02605-06. PMID 17267506.
- ^ Gouilh, MA; Puechmaille, SJ; Gonzalez, JP; Teeling, E; Kittayapong, P; Manuguerra, JC (2011). "SARS-Coronavirus ancestor's foot-prints in South-East Asian bat colonies and the refuge theory". Infect Genet Evol. 11 (7): 1690–702. doi:10.1016/j.meegid.2011.06.021. PMID 21763784.
- ^ Cui J, Han N, Streicker D, Li G, Tang X, Shi Z, Hu Z, Zhao G, Fontanet A, Guan Y, Wang L, Jones G, Field HE, Daszak P, Zhang S; Han; Streicker; Li; Tang; Shi; Hu; Zhao; Fontanet; Guan; Wang; Jones; Field; Daszak; Zhang (Oct 2007). "Evolutionary relationships between bat coronaviruses and their hosts". Emerg. Infect. Dis. 13 (10): 1526–32. doi:10.3201/eid1310.070448. PMC 2851503. PMID 18258002.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Crossley BM, Mock RE, Callison SA, Hietala SK (2012) Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses 4(12):3689-3700. doi:10.3390/v4123689
- ^ Huynh, J; Li, S; Yount, B; Smith, A; Sturges, L; Olsen, JC; Nagel, J; Johnson, JB; Agnihothram, S; et al. (2012). "Evidence supporting a zoonotic origin of human coronavirus strain NL63". J Virol. 86 (23): 12816–25. doi:10.1128/JVI.00906-12. PMID 22993147.
{{cite journal}}: Explicit use of et al. in:|author10=(help); Explicit use of et al. in:|first9=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help)
External links
- [1] (World Health Organization, Eastern Mediterranean Health Journal, supplement on coronavirus)
- Laude H, Rasschaert D, Delmas B, Godet M, Gelfi J, Charley B; Rasschaert; Delmas; Godet; Gelfi; Charley (June 1990). "Molecular biology of transmissible gastroenteritis virus". Veterinary Microbiology. 23 (1–4): 147–54. doi:10.1016/0378-1135(90)90144-K. PMID 2169670.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Sola I, Alonso S, Zúñiga S, Balasch M, Plana-Durán J, Enjuanes L; Alonso; Zúñiga; Balasch; Plana-Durán; Enjuanes (April 2003). "Engineering the transmissible gastroenteritis virus genome as an expression vector inducing lactogenic immunity". Journal of Virology. 77 (7): 4357–69. doi:10.1128/JVI.77.7.4357-4369.2003. PMC 150661. PMID 12634392.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Tajima M (1970). "Morphology of transmissible gastroenteritis virus of pigs. A possible member of coronaviruses. Brief report". Archiv Für Die Gesamte Virusforschung. 29 (1): 105–8. doi:10.1007/BF01253886. PMID 4195092.
- Virus Pathogen Database and Analysis Resource (ViPR): Coronaviridae
- German Research Foundation (Coronavirus Consortium)
