Protease
A protease (also called a peptidase or proteinase) is any enzyme that performs proteolysis; protein catabolism by hydrolysis of peptide bonds. Proteases have evolved multiple times, and different classes of protease can perform the same reaction by completely different catalytic mechanisms. Proteases can be found in Animalia, Plantae, Fungi, Bacteria, Archaea and viruses.
Classifications
Catalytic residue
Proteases can be classified into seven broad groups:[1]
- Serine proteases - using a serine alcohol
- Cysteine proteases - using a cysteine thiol
- Threonine proteases - using a threonine secondary alcohol
- Aspartic proteases - using an aspartate carboxylic acid
- Glutamic proteases - using a glutamate carboxylic acid
- Metalloproteases - using a metal, usually zinc
- Asparagine peptide lyases - using an asparagine to perform an elimination reaction (not requiring water)
Proteases were first grouped into 84 families according to their evolutionary relationship in 1993, and classified under four catalytic types: serine, cysteine, aspartic, and metallo proteases.[2] The threonine and glutamic-acid proteases were not described until 1995 and 2004 respectively. The mechanism used to cleave a peptide bond involves making an amino acid residue that has the cysteine and threonine (proteases) or a water molecule (aspartic acid, metallo- and acid proteases) nucleophilic so that it can attack the peptide carboxyl group. One way to make a nucleophile is by a catalytic triad, where a histidine residue is used to activate serine, cysteine, or threonine as a nucleophile. This is not an evolutionary grouping, however, as the nucleophile types have evolved convergently in different superfamilies, and some superfamilies show divergent evolution to multiple different nucleophiles.
Peptide lyases
A seventh catalytic type of proteolytic enzymes, asparagine peptide lyase, was described in 2011. Its proteolytic mechanism is unusual since, rather than hydrolysis, it performs an elimination reaction.[3] During this reaction, the catalytic asparagine forms a cyclic chemical structure that cleaves itself at asparagine residues in proteins under the right conditions. Given its fundamentally different mechanism, its inclusion as a peptidase may be debatable.[3]
Evolutionary relatedness
An up-to-date classification of protease evolutionary superfamilies is found in the MEROPS database.[4] In this database, proteases are classified firstly by 'clan' (superfamily) based on structure, mechanism and catalytic residue order (e.g. the PA clan where P indicates a mixture of nucleophile families). Within each 'clan', proteases are classified into families based on sequence similarity (e.g. the S1 and C3 families within the PA clan). Each family may contain many hundreds of related proteases (e.g. trypsin, elastase, thrombin and streptogrisin within the S1 family).
Currently more than 50 clans are known, each indicating an independent evolutionary origin of proteolysis.[4]
Optimal pH
Alternatively, proteases may be classified by the optimal pH in which they are active:
- Acid proteases
- Neutral proteases involved in type 1 hypersensitivity. Here, it is released by mast cells and causes activation of complement and kinins.[5] This group includes the calpains.
- Basic proteases (or alkaline proteases)
Function and mechanism
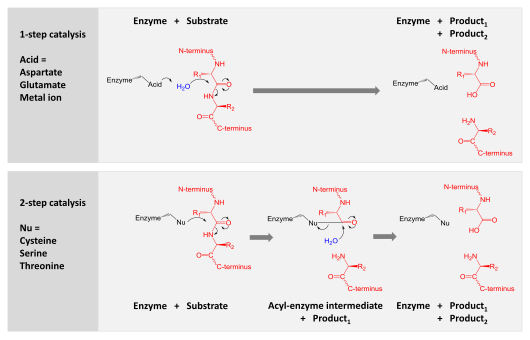
Proteases are involved in digesting long protein chains into shorter fragments by splitting the peptide bonds that link amino acid residues. Some detach the terminal amino acids from the protein chain (exopeptidases, such as aminopeptidases, carboxypeptidase A); others attack internal peptide bonds of a protein (endopeptidases, such as trypsin, chymotrypsin, pepsin, papain, elastase).
Catalysis
Catalysis is achieved by one of two mechanisms:
- Aspartic, glutamic and metallo- proteases activate a water molecule which performs a nucleophilic attack on the peptide bond to hydrolyse it.
- Serine, threonine and cysteine proteases use a nucleophilic residue in a (usually in a catalytic triad). That residue performs a nucleophilic attack to covalently link the protease to the substrate protein, releasing the first half of the product. This covalent acyl-enzyme intermediate is then hydrolysed by activated water to complete catalysis by releasing the second half of the product and regenerating the free enzyme.
Specificity
Proteolysis can be highly promiscuous such that a wide range of protein substrates are hydrolysed. This is the case for digestive enzymes such as trypsin which have to be able to cleave the array of proteins ingested into smaller peptide fragments. Promiscuous proteases typically bind to a single amino acid on the substrate and so only have specificity for that residue. For example, trypsin is specific for the sequences ...K\... or ...R\... ('\'=cleavage site).
Conversely some proteases are highly specific and only cleave substrates with a certain sequence. Blood clotting (such as thrombin) and viral polyprotein processing (such as TEV protease) requires this level of specificity in order to achieve precise cleavage events. This is achieved by proteases having a long binding cleft or tunnel with several pockets along it which bind the specified residues. For example, TEV protease is specific for the sequence ...ENLYFQ\S... ('\'=cleavage site).
Degradation and autolysis
Proteases, being themselves proteins, are cleaved by other protease molecules, sometimes of the same variety. This acts as a method of regulation of protease activity. Some proteases are less active after autolysis (e.g. TEV protease) whilst others are more active (e.g. trypsinogen).
Occurrence
Proteases occur in all organisms, from prokaryotes to eukaryotes to viruses. These enzymes are involved in a multitude of physiological reactions from simple digestion of food proteins to highly regulated cascades (e.g., the blood-clotting cascade, the complement system, apoptosis pathways, and the invertebrate prophenoloxidase-activating cascade). Proteases can either break specific peptide bonds (limited proteolysis), depending on the amino acid sequence of a protein, or break down a complete peptide to amino acids (unlimited proteolysis). The activity can be a destructive change (abolishing a protein's function or digesting it to its principal components), it can be an activation of a function, or it can be a signal in a signalling pathway.
Plants
Plant genomes encode hundreds of proteases, largely of unknown function. Those with known function are largely involved in developmental regulation.[6] Plant proteases also play a role in regulation of photosynthesis.[7]
Animals
Proteases are used throughout an organism for various metabolic processes. Acid proteases secreted into the stomach (such as pepsin) and serine proteases present in duodenum (trypsin and chymotrypsin) enable us to digest the protein in food. Proteases present in blood serum (thrombin, plasmin, Hageman factor, etc.) play important role in blood-clotting, as well as lysis of the clots, and the correct action of the immune system. Other proteases are present in leukocytes (elastase, cathepsin G) and play several different roles in metabolic control. Some snake venoms are also proteases, such as pit viper haemotoxin and interfere with the victim's blood clotting cascade. Proteases determine the lifetime of other proteins playing important physiological role like hormones, antibodies, or other enzymes. This is one of the fastest "switching on" and "switching off" regulatory mechanisms in the physiology of an organism.
By complex cooperative action the proteases may proceed as cascade reactions, which result in rapid and efficient amplification of an organism's response to a physiological signal.
Bacteria
Bacteria secrete proteases to hydrolyse (digest) the peptide bonds in proteins and therefore break the proteins down into their constituent monomers (amino acids). Bacterial and fungal proteases are particularly important to the global carbon and nitrogen cycles in the recycling of proteins, and such activity tends to be regulated by nutritional signals in these organisms.[8] The net impact of nutritional regulation of protease activity among the thousands of species present in soil can be observed at the overall microbial community level as proteins are broken down in response to carbon, nitrogen, or sulfur limitation.[9]
Bacteria contain proteases responsible for general protein quality control (e.g. the AAA+ proteasome) by degrading unfolded or misfolded proteins.
A secreted bacterial protease may also act as an exotoxin, and be an example of a virulence factor in bacterial pathogenesis (for example, exfoliative toxin). Bacterial exotoxic proteases destroy extracellular structures.
Viruses
Some viruses express their entire genome as one massive polyprotein and use a protease to cleave this into functional units (e.g. polio, norovirus, and TEV proteases). These proteases (e.g. TEV protease) have high specificity and only cleave very restricted set of substrate sequences.
Uses
The field of protease research is enormous. Since 2004, approximately 8000 papers related to this field were published each year.[10] Proteases are used in industry, medicine and as a basic biological research tool.[11][12]
Digestive proteases are part of many laundry detergents and are also used extensively in the bread industry in bread improver. A variety of proteases are used medically both for their native function (e.g. controlling blood clotting) or for completely artificial functions (e.g. for the targeted degradation of pathogenic proteins). Highly specific proteases such as TEV protease and thrombin are commonly used to cleave fusion proteins and affinity tags in a controlled fashion.
Inhibitors
The activity of proteases is inhibited by protease inhibitors.[13] One example of protease inhibitors is the serpin superfamily, which includes alpha 1-antitrypsin, C1-inhibitor, antithrombin, alpha 1-antichymotrypsin, plasminogen activator inhibitor-1, and neuroserpin.[14]
Natural protease inhibitors include the family of lipocalin proteins, which play a role in cell regulation and differentiation. Lipophilic ligands, attached to lipocalin proteins, have been found to possess tumor protease inhibiting properties. The natural protease inhibitors are not to be confused with the protease inhibitors used in antiretroviral therapy. Some viruses, with HIV/AIDS among them, depend on proteases in their reproductive cycle. Thus, protease inhibitors are developed as antiviral means.
Other natural protease inhibitors are used as defense mechanisms. Common examples are the trypsin inhibitors found in the seeds of some plants, most notable for humans being soybeans, a major food crop, where they act to discourage predators. Raw soybeans are toxic to many animals, including humans, until the protease inhibitors they contain have been denatured.
See also
- Protease
- PA clan
- Convergent evolution
- Proteolysis
- Catalytic triad
- The Proteolysis Map
- Proteases in angiogenesis
- Intramembrane proteases
- Protease inhibitor (pharmacology)
- Protease inhibitor (biology)
- TopFIND - database of protease specificity, substrates, products and inhibitors
- MEROPS - Database of protease evolutionary groups
References
- ^ Kohei Oda (2012). "New families of carboxyl peptidases: serine-carboxyl peptidases and glutamic peptidases". Journal of Biochemistry. 151 (1): 13–25. doi:10.1093/jb/mvr129. PMID 22016395.
- ^ "Evolutionary families of peptidases". Biochemical Journal. 290 (Pt 1): 205–218. 1993. doi:10.1042/bj2900205. PMC 1132403. PMID 8439290.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ a b "Asparagine peptide lyases: a seventh catalytic type of proteolytic enzymes". Journal of Biological Chemistry. 286 (44): 38321–8. 2011. doi:10.1074/jbc.M111.260026. PMID 21832066.
{{cite journal}}: Unknown parameter|authors=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b Rawlings ND, Barrett AJ, Bateman A (January 2010). "MEROPS: the peptidase database". Nucleic Acids Res. 38 (Database issue): D227–33. doi:10.1093/nar/gkp971. PMC 2808883. PMID 19892822.
- ^ Mitchell, Richard Sheppard; Kumar, Vinay; Abbas, Abul K.; Fausto, Nelson (2007). Robbins Basic Pathology. Philadelphia: Saunders. p. 122. ISBN 1-4160-2973-7. 8th edition.
- ^ van der Hoorn, RA (2008). "Plant proteases: from phenotypes to molecular mechanisms". Annual Review of Plant Biology. 59: 191–223. doi:10.1146/annurev.arplant.59.032607.092835. PMID 18257708.
- ^ Zelisko, A; Jackowski, G (October 2004). "Senescence-dependent degradation of Lhcb3 is mediated by a thylakoid membrane-bound protease". Journal of plant physiology. 161 (10): 1157–70. doi:10.1016/j.jplph.2004.01.006. PMID 15535125.
- ^ Sims, G.K. (2006). "Nitrogen Starvation Promotes Biodegradation of N-Heterocyclic Compounds in Soil". Soil Biology & Biochemistry. 38: 2478–2480. doi:10.1016/j.soilbio.2006.01.006.
- ^ Sims, G. K.; Wander, M. M. (2002). "Proteolytic activity under nitrogen or sulfur limitation". Appl. Soil Ecol. 568: 1–5.
- ^ Woessner, edited by Alan J. Barrett, Neil D. Rawlings, J. Fred (2004). Handbook of proteolytic enzymes (2nd ed.). London, UK: Elsevier Academic Press. ISBN 0-12-079610-4.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ Hooper, ed. by N. M. (2002). Proteases in biology and medicine. London: Portland Press. ISBN 1-85578-147-6.
{{cite book}}:|first=has generic name (help) - ^ Feijoo-Siota, Lucía; Villa, Tomás G. (28 September 2010). "Native and Biotechnologically Engineered Plant Proteases with Industrial Applications". Food and Bioprocess Technology. 4 (6): 1066–1088. doi:10.1007/s11947-010-0431-4.
- ^ Southan, C (Jul 1, 2001). "A genomic perspective on human proteases as drug targets". Drug Discovery Today. 6 (13): 681–688. doi:10.1016/s1359-6446(01)01793-7. PMID 11427378.
- ^ Puente, XS; López-Otín, C (April 2004). "A genomic analysis of rat proteases and protease inhibitors". Genome Research. 14 (4): 609–22. doi:10.1101/gr.1946304. PMC 383305. PMID 15060002.
External links
- International Proteolysis Society
- MEROPS - the peptidase database
- List of protease inhibitors
- Protease cutting predictor
- List of proteases and their specificities (see also [1])
- Proteolysis MAP from Center for Proteolytic Pathways
- Proteolysis Cut Site database - curated expert annotation from users
- Protease cut sites graphical interface
- TopFIND protease database covering cut sites, substrates and protein termini
- Proteases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
