Log page index: User:ProteinBoxBot/PBB_Log_Index
Protein Status Quick Log - Date: 10:03, 7 November 2007 (UTC)
[edit]Proteins without matches (15)
[edit]Proteins with a High Potential Match (10)
[edit]Manual Inspection (Page not found) (18)
[edit]
Protein Status Grid - Date: 10:03, 7 November 2007 (UTC)
[edit]Vebose Log - Date: 10:03, 7 November 2007 (UTC)
[edit]- INFO: Beginning work on ANXA5... {November 7, 2007 1:36:11 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein ANXA5 image.jpg {November 7, 2007 1:36:42 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:36:56 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_ANXA5_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1a8a.
| PDB = {{PDB2|1a8a}}, {{PDB2|1a8b}}, {{PDB2|1anw}}, {{PDB2|1anx}}, {{PDB2|1avh}}, {{PDB2|1avr}}, {{PDB2|1bc0}}, {{PDB2|1bc1}}, {{PDB2|1bc3}}, {{PDB2|1bcw}}, {{PDB2|1bcy}}, {{PDB2|1bcz}}, {{PDB2|1g5n}}, {{PDB2|1hak}}, {{PDB2|1hvd}}, {{PDB2|1hve}}, {{PDB2|1hvf}}, {{PDB2|1hvg}}, {{PDB2|1n41}}, {{PDB2|1n42}}, {{PDB2|1n44}}, {{PDB2|1sav}}, {{PDB2|2ie6}}, {{PDB2|2ie7}}, {{PDB2|2ran}}
| Name = Annexin A5
| HGNCid = 543
| Symbol = ANXA5
| AltSymbols =; ANX5; ENX2; PP4
| OMIM = 131230
| ECnumber =
| Homologene = 20312
| MGIid = 106008
| GeneAtlas_image1 = PBB_GE_ANXA5_200782_at_tn.png
| Function = {{GNF_GO|id=GO:0004859 |text = phospholipase inhibitor activity}} {{GNF_GO|id=GO:0005509 |text = calcium ion binding}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005544 |text = calcium-dependent phospholipid binding}}
| Component = {{GNF_GO|id=GO:0005622 |text = intracellular}} {{GNF_GO|id=GO:0005737 |text = cytoplasm}}
| Process = {{GNF_GO|id=GO:0006916 |text = anti-apoptosis}} {{GNF_GO|id=GO:0007165 |text = signal transduction}} {{GNF_GO|id=GO:0007596 |text = blood coagulation}} {{GNF_GO|id=GO:0050819 |text = negative regulation of coagulation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 308
| Hs_Ensembl = ENSG00000164111
| Hs_RefseqProtein = NP_001145
| Hs_RefseqmRNA = NM_001154
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 4
| Hs_GenLoc_start = 122808598
| Hs_GenLoc_end = 122837626
| Hs_Uniprot = P08758
| Mm_EntrezGene = 11747
| Mm_Ensembl = ENSMUSG00000027712
| Mm_RefseqmRNA = NM_009673
| Mm_RefseqProtein = NP_033803
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 3
| Mm_GenLoc_start = 36640893
| Mm_GenLoc_end = 36656884
| Mm_Uniprot = Q3U5Q1
}}
}}
'''Annexin A5''', also known as '''ANXA5''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The protein encoded by this gene belongs to the annexin family of calcium-dependent phospholipid binding proteins some of which have been implicated in membrane-related events along exocytotic and endocytotic pathways. Annexin 5 is a phospholipase A2 and protein kinase C inhibitory protein with calcium channel activity and a potential role in cellular signal transduction, inflammation, growth and differentiation. Annexin 5 has also been described as placental anticoagulant protein I, vascular anticoagulant-alpha, endonexin II, lipocortin V, placental protein 4 and anchorin CII. The gene spans 29 kb containing 13 exons, and encodes a single transcript of approximately 1.6 kb and a protein product with a molecular weight of about 35 kDa.<ref>{{cite web | title = Entrez Gene: ANXA5 annexin A5| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=308| accessdate = }}</ref>
}}
==References==
{{reflist}}
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on ARF6... {November 7, 2007 1:36:56 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein ARF6 image.jpg {November 7, 2007 1:37:50 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:37:59 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_ARF6_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1e0s.
| PDB = {{PDB2|1e0s}}, {{PDB2|2a5d}}, {{PDB2|2a5f}}, {{PDB2|2a5g}}, {{PDB2|2j5x}}
| Name = ADP-ribosylation factor 6
| HGNCid = 659
| Symbol = ARF6
| AltSymbols =;
| OMIM = 600464
| ECnumber =
| Homologene = 1256
| MGIid = 99435
| GeneAtlas_image1 = PBB_GE_ARF6_203311_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_ARF6_203312_x_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0003924 |text = GTPase activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005525 |text = GTP binding}}
| Component = {{GNF_GO|id=GO:0001726 |text = ruffle}} {{GNF_GO|id=GO:0005622 |text = intracellular}} {{GNF_GO|id=GO:0005624 |text = membrane fraction}} {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005768 |text = endosome}} {{GNF_GO|id=GO:0005886 |text = plasma membrane}} {{GNF_GO|id=GO:0005938 |text = cell cortex}}
| Process = {{GNF_GO|id=GO:0006888 |text = ER to Golgi vesicle-mediated transport}} {{GNF_GO|id=GO:0006928 |text = cell motility}} {{GNF_GO|id=GO:0007155 |text = cell adhesion}} {{GNF_GO|id=GO:0007264 |text = small GTPase mediated signal transduction}} {{GNF_GO|id=GO:0015031 |text = protein transport}} {{GNF_GO|id=GO:0016192 |text = vesicle-mediated transport}} {{GNF_GO|id=GO:0030838 |text = positive regulation of actin filament polymerization}} {{GNF_GO|id=GO:0030866 |text = cortical actin cytoskeleton organization and biogenesis}} {{GNF_GO|id=GO:0031529 |text = ruffle organization and biogenesis}} {{GNF_GO|id=GO:0035020 |text = regulation of Rac protein signal transduction}} {{GNF_GO|id=GO:0048261 |text = negative regulation of receptor mediated endocytosis}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 382
| Hs_Ensembl = ENSG00000165527
| Hs_RefseqProtein = NP_001654
| Hs_RefseqmRNA = NM_001663
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 14
| Hs_GenLoc_start = 49429589
| Hs_GenLoc_end = 49431484
| Hs_Uniprot = P62330
| Mm_EntrezGene = 11845
| Mm_Ensembl = ENSMUSG00000044147
| Mm_RefseqmRNA = NM_007481
| Mm_RefseqProtein = NP_031507
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 12
| Mm_GenLoc_start = 70290755
| Mm_GenLoc_end = 70292386
| Mm_Uniprot = Q3U0D7
}}
}}
'''ADP-ribosylation factor 6''', also known as '''ARF6''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the human ARF gene family, which is part of the RAS superfamily. The ARF genes encode small guanine nucleotide-binding proteins that stimulate the ADP-ribosyltransferase activity of cholera toxin and play a role in vesicular trafficking and as activators of phospholipase D. The product of this gene is localized to the plasma membrane, and regulates vesicular trafficking, remodelling of membrane lipids, and signaling pathways that lead to actin remodeling. A pseudogene of this gene is located on chromosome 7.<ref>{{cite web | title = Entrez Gene: ARF6 ADP-ribosylation factor 6| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=382| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Sabe H |title=Requirement for Arf6 in cell adhesion, migration, and cancer cell invasion. |journal=J. Biochem. |volume=134 |issue= 4 |pages= 485-9 |year= 2004 |pmid= 14607973 |doi= }}
*{{cite journal | author=Joseph AM, Kumar M, Mitra D |title=Nef: "necessary and enforcing factor" in HIV infection. |journal=Curr. HIV Res. |volume=3 |issue= 1 |pages= 87-94 |year= 2005 |pmid= 15638726 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on ATP7A... {November 7, 2007 1:37:59 AM PST}
- UPLOAD: Added new Image to wiki:
 {November 7, 2007 1:38:13 AM PST}
{November 7, 2007 1:38:13 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:38:26 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_ATP7A_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1aw0.
| PDB = {{PDB2|1aw0}}, {{PDB2|1kvi}}, {{PDB2|1kvj}}, {{PDB2|1q8l}}, {{PDB2|1s6o}}, {{PDB2|1s6u}}, {{PDB2|1y3j}}, {{PDB2|1y3k}}, {{PDB2|1yjr}}, {{PDB2|1yjt}}, {{PDB2|1yju}}, {{PDB2|1yjv}}, {{PDB2|2aw0}}, {{PDB2|2g9o}}, {{PDB2|2ga7}}
| Name = ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome)
| HGNCid = 869
| Symbol = ATP7A
| AltSymbols =; MK; MNK
| OMIM = 300011
| ECnumber =
| Homologene = 35
| MGIid = 99400
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0000287 |text = magnesium ion binding}} {{GNF_GO|id=GO:0004008 |text = copper-exporting ATPase activity}} {{GNF_GO|id=GO:0005375 |text = copper ion transmembrane transporter activity}} {{GNF_GO|id=GO:0005507 |text = copper ion binding}} {{GNF_GO|id=GO:0005524 |text = ATP binding}} {{GNF_GO|id=GO:0015097 |text = mercury ion transmembrane transporter activity}} {{GNF_GO|id=GO:0015662 |text = ATPase activity, coupled to transmembrane movement of ions, phosphorylative mechanism}} {{GNF_GO|id=GO:0016787 |text = hydrolase activity}} {{GNF_GO|id=GO:0016820 |text = hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances}} {{GNF_GO|id=GO:0032767 |text = copper-dependent protein binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}} {{GNF_GO|id=GO:0046873 |text = metal ion transmembrane transporter activity}}
| Component = {{GNF_GO|id=GO:0005770 |text = late endosome}} {{GNF_GO|id=GO:0005783 |text = endoplasmic reticulum}} {{GNF_GO|id=GO:0005794 |text = Golgi apparatus}} {{GNF_GO|id=GO:0005802 |text = trans-Golgi network}} {{GNF_GO|id=GO:0005886 |text = plasma membrane}} {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}} {{GNF_GO|id=GO:0016323 |text = basolateral plasma membrane}} {{GNF_GO|id=GO:0030140 |text = trans-Golgi network transport vesicle}} {{GNF_GO|id=GO:0030173 |text = integral to Golgi membrane}} {{GNF_GO|id=GO:0048471 |text = perinuclear region of cytoplasm}}
| Process = {{GNF_GO|id=GO:0006810 |text = transport}} {{GNF_GO|id=GO:0006811 |text = ion transport}} {{GNF_GO|id=GO:0006825 |text = copper ion transport}} {{GNF_GO|id=GO:0006878 |text = cellular copper ion homeostasis}} {{GNF_GO|id=GO:0008152 |text = metabolic process}} {{GNF_GO|id=GO:0015694 |text = mercury ion transport}} {{GNF_GO|id=GO:0030001 |text = metal ion transport}} {{GNF_GO|id=GO:0051353 |text = positive regulation of oxidoreductase activity}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 538
| Hs_Ensembl =
| Hs_RefseqProtein = NP_000043
| Hs_RefseqmRNA = NM_000052
| Hs_GenLoc_db =
| Hs_GenLoc_chr =
| Hs_GenLoc_start =
| Hs_GenLoc_end =
| Hs_Uniprot =
| Mm_EntrezGene = 11977
| Mm_Ensembl = ENSMUSG00000033792
| Mm_RefseqmRNA = NM_009726
| Mm_RefseqProtein = NP_033856
| Mm_GenLoc_db =
| Mm_GenLoc_chr = X
| Mm_GenLoc_start = 102230043
| Mm_GenLoc_end = 102327470
| Mm_Uniprot = Q3T9Y7
}}
}}
'''ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome)''', also known as '''ATP7A''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Harris ED, Reddy MC, Qian Y, ''et al.'' |title=Multiple forms of the Menkes Cu-ATPase. |journal=Adv. Exp. Med. Biol. |volume=448 |issue= |pages= 39-51 |year= 1999 |pmid= 10079814 |doi= }}
*{{cite journal | author=Tümer Z, Møller LB, Horn N |title=Mutation spectrum of ATP7A, the gene defective in Menkes disease. |journal=Adv. Exp. Med. Biol. |volume=448 |issue= |pages= 83-95 |year= 1999 |pmid= 10079817 |doi= }}
*{{cite journal | author=Cox DW, Moore SD |title=Copper transporting P-type ATPases and human disease. |journal=J. Bioenerg. Biomembr. |volume=34 |issue= 5 |pages= 333-8 |year= 2003 |pmid= 12539960 |doi= }}
*{{cite journal | author=Voskoboinik I, Camakaris J |title=Menkes copper-translocating P-type ATPase (ATP7A): biochemical and cell biology properties, and role in Menkes disease. |journal=J. Bioenerg. Biomembr. |volume=34 |issue= 5 |pages= 363-71 |year= 2003 |pmid= 12539963 |doi= }}
*{{cite journal | author=La Fontaine S, Mercer JF |title=Trafficking of the copper-ATPases, ATP7A and ATP7B: role in copper homeostasis. |journal=Arch. Biochem. Biophys. |volume=463 |issue= 2 |pages= 149-67 |year= 2007 |pmid= 17531189 |doi= 10.1016/j.abb.2007.04.021 }}
*{{cite journal | author=Lutsenko S, LeShane ES, Shinde U |title=Biochemical basis of regulation of human copper-transporting ATPases. |journal=Arch. Biochem. Biophys. |volume=463 |issue= 2 |pages= 134-48 |year= 2007 |pmid= 17562324 |doi= 10.1016/j.abb.2007.04.013 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on BMPR2... {November 7, 2007 1:38:26 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein BMPR2 image.jpg {November 7, 2007 1:39:27 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:39:48 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_BMPR2_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 2hlq.
| PDB = {{PDB2|2hlq}}, {{PDB2|2hlr}}
| Name = Bone morphogenetic protein receptor, type II (serine/threonine kinase)
| HGNCid = 1078
| Symbol = BMPR2
| AltSymbols =; BMPR-II; BMPR3; BMR2; BRK-3; T-ALK
| OMIM = 600799
| ECnumber =
| Homologene = 929
| MGIid = 1095407
| GeneAtlas_image1 = PBB_GE_BMPR2_209920_at_tn.png
| GeneAtlas_image2 = PBB_GE_BMPR2_210214_s_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0000287 |text = magnesium ion binding}} {{GNF_GO|id=GO:0004672 |text = protein kinase activity}} {{GNF_GO|id=GO:0004674 |text = protein serine/threonine kinase activity}} {{GNF_GO|id=GO:0004872 |text = receptor activity}} {{GNF_GO|id=GO:0005024 |text = transforming growth factor beta receptor activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005524 |text = ATP binding}} {{GNF_GO|id=GO:0016740 |text = transferase activity}} {{GNF_GO|id=GO:0030145 |text = manganese ion binding}}
| Component = {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}} {{GNF_GO|id=GO:0016021 |text = integral to membrane}}
| Process = {{GNF_GO|id=GO:0000074 |text = regulation of progression through cell cycle}} {{GNF_GO|id=GO:0001501 |text = skeletal development}} {{GNF_GO|id=GO:0006468 |text = protein amino acid phosphorylation}} {{GNF_GO|id=GO:0007178 |text = transmembrane receptor protein serine/threonine kinase signaling pathway}} {{GNF_GO|id=GO:0042127 |text = regulation of cell proliferation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 659
| Hs_Ensembl = ENSG00000204217
| Hs_RefseqProtein = NP_001195
| Hs_RefseqmRNA = NM_001204
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 2
| Hs_GenLoc_start = 202949916
| Hs_GenLoc_end = 203140719
| Hs_Uniprot = Q13873
| Mm_EntrezGene = 12168
| Mm_Ensembl = ENSMUSG00000067336
| Mm_RefseqmRNA = NM_007561
| Mm_RefseqProtein = NP_031587
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 1
| Mm_GenLoc_start = 59709199
| Mm_GenLoc_end = 59815028
| Mm_Uniprot = Q3UER5
}}
}}
'''Bone morphogenetic protein receptor, type II (serine/threonine kinase)''', also known as '''BMPR2''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the bone morphogenetic protein (BMP) receptor family of transmembrane serine/threonine kinases. The ligands of this receptor are BMPs, which are members of the TGF-beta superfamily. BMPs are involved in endochondral bone formation and embryogenesis. These proteins transduce their signals through the formation of heteromeric complexes of 2 different types of serine (threonine) kinase receptors: type I receptors of about 50-55 kD and type II receptors of about 70-80 kD. Type II receptors bind ligands in the absence of type I receptors, but they require their respective type I receptors for signaling, whereas type I receptors require their respective type II receptors for ligand binding. Mutations in this gene have been associated with primary pulmonary hypertension.<ref>{{cite web | title = Entrez Gene: BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=659| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=De Caestecker M, Meyrick B |title=Bone morphogenetic proteins, genetics and the pathophysiology of primary pulmonary hypertension. |journal=Respir. Res. |volume=2 |issue= 4 |pages= 193-7 |year= 2002 |pmid= 11686884 |doi= }}
*{{cite journal | author=Sztrymf B, Yaïci A, Girerd B, Humbert M |title=Genes and pulmonary arterial hypertension. |journal=Respiration; international review of thoracic diseases |volume=74 |issue= 2 |pages= 123-32 |year= 2007 |pmid= 17318011 |doi= 10.1159/000098818 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on CFL1... {November 7, 2007 1:39:48 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein CFL1 image.jpg {November 7, 2007 1:40:26 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:40:37 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_CFL1_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1q8g.
| PDB = {{PDB2|1q8g}}, {{PDB2|1q8x}}
| Name = Cofilin 1 (non-muscle)
| HGNCid = 1874
| Symbol = CFL1
| AltSymbols =; CFL
| OMIM = 601442
| ECnumber =
| Homologene = 39624
| MGIid = 101757
| GeneAtlas_image1 = PBB_GE_CFL1_200021_at_tn.png
| Function = {{GNF_GO|id=GO:0003779 |text = actin binding}} {{GNF_GO|id=GO:0005515 |text = protein binding}}
| Component = {{GNF_GO|id=GO:0005622 |text = intracellular}} {{GNF_GO|id=GO:0005634 |text = nucleus}} {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005856 |text = cytoskeleton}}
| Process = {{GNF_GO|id=GO:0006916 |text = anti-apoptosis}} {{GNF_GO|id=GO:0007266 |text = Rho protein signal transduction}} {{GNF_GO|id=GO:0030036 |text = actin cytoskeleton organization and biogenesis}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 1072
| Hs_Ensembl = ENSG00000172757
| Hs_RefseqProtein = NP_005498
| Hs_RefseqmRNA = NM_005507
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 11
| Hs_GenLoc_start = 65378884
| Hs_GenLoc_end = 65383462
| Hs_Uniprot = P23528
| Mm_EntrezGene = 12631
| Mm_Ensembl = ENSMUSG00000056201
| Mm_RefseqmRNA = XM_979959
| Mm_RefseqProtein = XP_985053
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 19
| Mm_GenLoc_start = 5492497
| Mm_GenLoc_end = 5495201
| Mm_Uniprot = Q544Y7
}}
}}
'''Cofilin 1 (non-muscle)''', also known as '''CFL1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Cofilin is a widely distributed intracellular actin-modulating protein that binds and depolymerizes filamentous F-actin and inhibits the polymerization of monomeric G-actin in a pH-dependent manner. It is involved in the translocation of actin-cofilin complex from cytoplasm to nucleus.[supplied by OMIM]<ref>{{cite web | title = Entrez Gene: CFL1 cofilin 1 (non-muscle)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1072| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Maciver SK, Hussey PJ |title=The ADF/cofilin family: actin-remodeling proteins. |journal=Genome Biol. |volume=3 |issue= 5 |pages= reviews3007 |year= 2002 |pmid= 12049672 |doi= }}
*{{cite journal | author=Samstag Y, Nebl G |title=Interaction of cofilin with the serine phosphatases PP1 and PP2A in normal and neoplastic human T lymphocytes. |journal=Adv. Enzyme Regul. |volume=43 |issue= |pages= 197-211 |year= 2004 |pmid= 12791392 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on CYBB... {November 7, 2007 1:40:37 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:41:32 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Cytochrome b-245, beta polypeptide (chronic granulomatous disease)
| HGNCid = 2578
| Symbol = CYBB
| AltSymbols =; CGD; GP91-1; GP91-PHOX; GP91PHOX; NOX2
| OMIM = 300481
| ECnumber =
| Homologene = 68054
| MGIid = 88574
| GeneAtlas_image1 = PBB_GE_CYBB_203923_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_CYBB_203922_s_at_tn.png
| Function = {{GNF_GO|id=GO:0005244 |text = voltage-gated ion channel activity}} {{GNF_GO|id=GO:0005506 |text = iron ion binding}} {{GNF_GO|id=GO:0016491 |text = oxidoreductase activity}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}} {{GNF_GO|id=GO:0050660 |text = FAD binding}}
| Component = {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005886 |text = plasma membrane}} {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}}
| Process = {{GNF_GO|id=GO:0006118 |text = electron transport}} {{GNF_GO|id=GO:0006811 |text = ion transport}} {{GNF_GO|id=GO:0006954 |text = inflammatory response}} {{GNF_GO|id=GO:0019735 |text = antimicrobial humoral response}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 1536
| Hs_Ensembl = ENSG00000165168
| Hs_RefseqProtein = NP_000388
| Hs_RefseqmRNA = NM_000397
| Hs_GenLoc_db =
| Hs_GenLoc_chr = X
| Hs_GenLoc_start = 37524208
| Hs_GenLoc_end = 37557658
| Hs_Uniprot = P04839
| Mm_EntrezGene = 13058
| Mm_Ensembl = ENSMUSG00000015340
| Mm_RefseqmRNA = NM_007807
| Mm_RefseqProtein = NP_031833
| Mm_GenLoc_db =
| Mm_GenLoc_chr = X
| Mm_GenLoc_start = 8592987
| Mm_GenLoc_end = 8626250
| Mm_Uniprot = Q3TVP4
}}
}}
'''Cytochrome b-245, beta polypeptide (chronic granulomatous disease)''', also known as '''CYBB''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Cytochrome b (-245) is composed of cytochrome b alpha (CYBA) and beta (CYBB) chain. It has been proposed as a primary component of the microbicidal oxidase system of phagocytes. CYBB deficiency is one of five described biochemical defects associated with chronic granulomatous disease (CGD). In this disorder, there is decreased activity of phagocyte NADPH oxidase; neutrophils are able to phagocytize bacteria but cannot kill them in the phagocytic vacuoles. The cause of the killing defect is an inability to increase the cell's respiration and consequent failure to deliver activated oxygen into the phagocytic vacuole.<ref>{{cite web | title = Entrez Gene: CYBB cytochrome b-245, beta polypeptide (chronic granulomatous disease)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1536| accessdate = }}</ref>
}}
==References==
{{reflist}}
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on DNM1... {November 7, 2007 1:41:32 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein DNM1 image.jpg {November 7, 2007 1:42:04 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:42:15 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_DNM1_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1dyn.
| PDB = {{PDB2|1dyn}}, {{PDB2|2aka}}, {{PDB2|2dyn}}
| Name = Dynamin 1
| HGNCid = 2972
| Symbol = DNM1
| AltSymbols =; DNM
| OMIM = 602377
| ECnumber =
| Homologene = 68397
| MGIid = 107384
| GeneAtlas_image1 = PBB_GE_DNM1_215116_s_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0003774 |text = motor activity}} {{GNF_GO|id=GO:0003924 |text = GTPase activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005525 |text = GTP binding}} {{GNF_GO|id=GO:0016301 |text = kinase activity}} {{GNF_GO|id=GO:0016740 |text = transferase activity}} {{GNF_GO|id=GO:0016787 |text = hydrolase activity}}
| Component = {{GNF_GO|id=GO:0005874 |text = microtubule}} {{GNF_GO|id=GO:0005905 |text = coated pit}}
| Process = {{GNF_GO|id=GO:0006897 |text = endocytosis}} {{GNF_GO|id=GO:0006898 |text = receptor-mediated endocytosis}} {{GNF_GO|id=GO:0007268 |text = synaptic transmission}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 1759
| Hs_Ensembl = ENSG00000106976
| Hs_RefseqProtein = NP_001005336
| Hs_RefseqmRNA = NM_001005336
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 9
| Hs_GenLoc_start = 130005479
| Hs_GenLoc_end = 130057348
| Hs_Uniprot = Q05193
| Mm_EntrezGene = 13429
| Mm_Ensembl = ENSMUSG00000026825
| Mm_RefseqmRNA = NM_010065
| Mm_RefseqProtein = NP_034195
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 2
| Mm_GenLoc_start = 32130480
| Mm_GenLoc_end = 32175293
| Mm_Uniprot = Q6PDM5
}}
}}
'''Dynamin 1''', also known as '''DNM1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the dynamin subfamily of GTP-binding proteins. The encoded protein possesses unique mechanochemical properties used to tubulate and sever membranes, and is involved in clathrin-mediated endocytosis and other vesicular trafficking processes. Actin and other cytoskeletal proteins act as binding partners for the encoded protein, which can also self-assemble leading to stimulation of GTPase activity. More than sixty highly conserved copies of the 3' region of this gene are found elsewhere in the genome, particularly on chromosomes Y and 15. Alternatively spliced transcript variants encoding different isoforms have been described.<ref>{{cite web | title = Entrez Gene: DNM1 dynamin 1| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1759| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Sever S |title=Dynamin and endocytosis. |journal=Curr. Opin. Cell Biol. |volume=14 |issue= 4 |pages= 463-7 |year= 2003 |pmid= 12383797 |doi= }}
*{{cite journal | author=Wiejak J, Wyroba E |title=Dynamin: characteristics, mechanism of action and function. |journal=Cell. Mol. Biol. Lett. |volume=7 |issue= 4 |pages= 1073-80 |year= 2003 |pmid= 12511974 |doi= }}
*{{cite journal | author=Orth JD, McNiven MA |title=Dynamin at the actin-membrane interface. |journal=Curr. Opin. Cell Biol. |volume=15 |issue= 1 |pages= 31-9 |year= 2003 |pmid= 12517701 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on EEF1A1... {November 7, 2007 1:42:15 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:44:41 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Eukaryotic translation elongation factor 1 alpha 1
| HGNCid = 3189
| Symbol = EEF1A1
| AltSymbols =; CCS-3; CCS3; EEF-1; EEF1A; EF-Tu; EF1A; FLJ25721; GRAF-1EF; HNGC:16303; LENG7; MGC102687; MGC131894; MGC16224; PTI1; eEF1A-1
| OMIM = 130590
| ECnumber =
| Homologene = 68181
| MGIid = 1096881
| GeneAtlas_image1 = PBB_GE_EEF1A1_204892_x_at_tn.png
| GeneAtlas_image2 = PBB_GE_EEF1A1_206559_x_at_tn.png
| GeneAtlas_image3 = PBB_GE_EEF1A1_213477_x_at_tn.png
| Function = {{GNF_GO|id=GO:0000166 |text = nucleotide binding}} {{GNF_GO|id=GO:0003746 |text = translation elongation factor activity}} {{GNF_GO|id=GO:0003924 |text = GTPase activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0005525 |text = GTP binding}}
| Component = {{GNF_GO|id=GO:0005737 |text = cytoplasm}} {{GNF_GO|id=GO:0005853 |text = eukaryotic translation elongation factor 1 complex}}
| Process = {{GNF_GO|id=GO:0006412 |text = translation}} {{GNF_GO|id=GO:0006414 |text = translational elongation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 1915
| Hs_Ensembl = ENSG00000156508
| Hs_RefseqProtein = NP_001393
| Hs_RefseqmRNA = NM_001402
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 6
| Hs_GenLoc_start = 74282194
| Hs_GenLoc_end = 74288344
| Hs_Uniprot = P68104
| Mm_EntrezGene = 13627
| Mm_Ensembl = ENSMUSG00000037742
| Mm_RefseqmRNA = NM_010106
| Mm_RefseqProtein = NP_034236
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 9
| Mm_GenLoc_start = 78264142
| Mm_GenLoc_end = 78266338
| Mm_Uniprot = Q3TII3
}}
}}
'''Eukaryotic translation elongation factor 1 alpha 1''', also known as '''EEF1A1''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes an isoform of the alpha subunit of the elongation factor-1 complex, which is responsible for the enzymatic delivery of aminoacyl tRNAs to the ribosome. This isoform (alpha 1) is expressed in brain, placenta, lung, liver, kidney, and pancreas, and the other isoform (alpha 2) is expressed in brain, heart and skeletal muscle. This isoform is identified as an autoantigen in 66% of patients with Felty syndrome. This gene has been found to have multiple copies on many chromosomes, some of which, if not all, represent different pseudogenes.<ref>{{cite web | title = Entrez Gene: EEF1A1 eukaryotic translation elongation factor 1 alpha 1| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=1915| accessdate = }}</ref>
}}
==References==
{{reflist}}
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on ELK1... {November 7, 2007 1:44:41 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein ELK1 image.jpg {November 7, 2007 1:45:33 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:45:41 AM PST}
- CREATED: Created new protein page: ELK1 {November 7, 2007 1:45:49 AM PST}
- INFO: Beginning work on GP6... {November 7, 2007 1:59:31 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein GP6 image.jpg {November 7, 2007 2:00:01 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 2:00:14 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_GP6_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 2gi7.
| PDB = {{PDB2|2gi7}}
| Name = Glycoprotein VI (platelet)
| HGNCid = 14388
| Symbol = GP6
| AltSymbols =; GPIV; GPVI; MGC138168
| OMIM = 605546
| ECnumber =
| Homologene = 9488
| MGIid = 1889810
| GeneAtlas_image1 = PBB_GE_GP6_220336_s_at_tn.png
| Function = {{GNF_GO|id=GO:0004888 |text = transmembrane receptor activity}} {{GNF_GO|id=GO:0005518 |text = collagen binding}}
| Component = {{GNF_GO|id=GO:0005887 |text = integral to plasma membrane}} {{GNF_GO|id=GO:0016020 |text = membrane}}
| Process = {{GNF_GO|id=GO:0007167 |text = enzyme linked receptor protein signaling pathway}} {{GNF_GO|id=GO:0030168 |text = platelet activation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 51206
| Hs_Ensembl = ENSG00000088053
| Hs_RefseqProtein = NP_057447
| Hs_RefseqmRNA = NM_016363
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 19
| Hs_GenLoc_start = 60216885
| Hs_GenLoc_end = 60241444
| Hs_Uniprot = Q9HCN6
| Mm_EntrezGene = 243816
| Mm_Ensembl =
| Mm_RefseqmRNA = XM_145298
| Mm_RefseqProtein = XP_145298
| Mm_GenLoc_db =
| Mm_GenLoc_chr =
| Mm_GenLoc_start =
| Mm_GenLoc_end =
| Mm_Uniprot =
}}
}}
'''Glycoprotein VI (platelet)''', also known as '''GP6''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Glycoprotein VI (GP6) is a 58-kD platelet membrane glycoprotein that plays a crucial role in the collagen-induced activation and aggregation of platelets. Upon injury to the vessel wall and subsequent damage to the endothelial lining, exposure of the subendothelial matrix to blood flow results in deposition of platelets. Collagen fibers are the most thrombogenic macromolecular components of the extracellular matrix, with collagen types I, III, and VI being the major forms found in blood vessels. Platelet interaction with collagen occurs as a 2-step procedure: (1) the initial adhesion to collagen is followed by (2) an activation step leading to platelet secretion, recruitment of additional platelets, and aggregation. In physiologic conditions, the resulting platelet plug is the initial hemostatic event limiting blood loss. However, exposure of collagen after rupture of atherosclerotic plaques is a major stimulus of thrombus formation associated with myocardial infarction or stroke (Jandrot-Perrus et al., 2000).[supplied by OMIM]<ref>{{cite web | title = Entrez Gene: GP6 glycoprotein VI (platelet)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=51206| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Nieswandt B, Watson SP |title=Platelet-collagen interaction: is GPVI the central receptor? |journal=Blood |volume=102 |issue= 2 |pages= 449-61 |year= 2003 |pmid= 12649139 |doi= 10.1182/blood-2002-12-3882 }}
*{{cite journal | author=Watson SP, Auger JM, McCarty OJ, Pearce AC |title=GPVI and integrin alphaIIb beta3 signaling in platelets. |journal=J. Thromb. Haemost. |volume=3 |issue= 8 |pages= 1752-62 |year= 2005 |pmid= 16102042 |doi= 10.1111/j.1538-7836.2005.01429.x }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on HMBS... {November 7, 2007 1:45:49 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:47:01 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Hydroxymethylbilane synthase
| HGNCid = 4982
| Symbol = HMBS
| AltSymbols =; PBG-D; PBGD; UPS
| OMIM = 609806
| ECnumber =
| Homologene = 158
| MGIid = 96112
| GeneAtlas_image1 = PBB_GE_HMBS_203040_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_HMBS_213344_s_at_tn.png
| GeneAtlas_image3 = PBB_GE_HMBS_212524_x_at_tn.png
| Function = {{GNF_GO|id=GO:0004418 |text = hydroxymethylbilane synthase activity}} {{GNF_GO|id=GO:0016740 |text = transferase activity}}
| Component =
| Process = {{GNF_GO|id=GO:0006783 |text = heme biosynthetic process}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 3145
| Hs_Ensembl = ENSG00000149397
| Hs_RefseqProtein = NP_000181
| Hs_RefseqmRNA = NM_000190
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 11
| Hs_GenLoc_start = 118460797
| Hs_GenLoc_end = 118469469
| Hs_Uniprot = P08397
| Mm_EntrezGene = 15288
| Mm_Ensembl =
| Mm_RefseqmRNA = NM_013551
| Mm_RefseqProtein = NP_038579
| Mm_GenLoc_db =
| Mm_GenLoc_chr =
| Mm_GenLoc_start =
| Mm_GenLoc_end =
| Mm_Uniprot =
}}
}}
'''Hydroxymethylbilane synthase''', also known as '''HMBS''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a member of the hydroxymethylbilane synthase superfamily. The encoded protein is the third enzyme of the heme biosynthetic pathway and catalyzes the head to tail condensation of four porphobilinogen molecules into the linear hydroxymethylbilane. Mutations in this gene are associated with the autosomal dominant disease acute intermittent porphyria. Alternatively spliced transcript variants encoding different isoforms have been described.<ref>{{cite web | title = Entrez Gene: HMBS hydroxymethylbilane synthase| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3145| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Deybach JC, Puy H |title=Porphobilinogen deaminase gene structure and molecular defects. |journal=J. Bioenerg. Biomembr. |volume=27 |issue= 2 |pages= 197-205 |year= 1995 |pmid= 7592566 |doi= }}
*{{cite journal | author=Astrin KH, Desnick RJ |title=Molecular basis of acute intermittent porphyria: mutations and polymorphisms in the human hydroxymethylbilane synthase gene. |journal=Hum. Mutat. |volume=4 |issue= 4 |pages= 243-52 |year= 1995 |pmid= 7866402 |doi= 10.1002/humu.1380040403 }}
*{{cite journal | author=Helliwell JR, Nieh YP, Habash J, ''et al.'' |title=Time-resolved and static-ensemble structural chemistry of hydroxymethylbilane synthase. |journal=Faraday Discuss. |volume=122 |issue= |pages= 131-44; discussion 171-90 |year= 2003 |pmid= 12555854 |doi= }}
*{{cite journal | author=Hessels J, Voortman G, van der Wagen A, ''et al.'' |title=Homozygous acute intermittent porphyria in a 7-year-old boy with massive excretions of porphyrins and porphyrin precursors. |journal=J. Inherit. Metab. Dis. |volume=27 |issue= 1 |pages= 19-27 |year= 2004 |pmid= 14970743 |doi= 10.1023/B:BOLI.0000016613.75677.05 }}
*{{cite journal | author=Kauppinen R |title=Molecular diagnostics of acute intermittent porphyria. |journal=Expert Rev. Mol. Diagn. |volume=4 |issue= 2 |pages= 243-9 |year= 2004 |pmid= 14995910 |doi= 10.1586/14737159.4.2.243 }}
*{{cite journal | author=Hrdinka M, Puy H, Martasek P |title=May 2006 update in porphobilinogen deaminase gene polymorphisms and mutations causing acute intermittent porphyria: comparison with the situation in Slavic population. |journal=Physiological research / Academia Scientiarum Bohemoslovaca |volume=55 Suppl 2 |issue= |pages= S119-36 |year= 2007 |pmid= 17298216 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on IDS... {November 7, 2007 1:47:01 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:47:15 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Iduronate 2-sulfatase (Hunter syndrome)
| HGNCid = 5389
| Symbol = IDS
| AltSymbols =; MPS2; SIDS
| OMIM = 309900
| ECnumber =
| Homologene = 169
| MGIid = 96417
| Function = {{GNF_GO|id=GO:0004423 |text = iduronate-2-sulfatase activity}} {{GNF_GO|id=GO:0005509 |text = calcium ion binding}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0008484 |text = sulfuric ester hydrolase activity}} {{GNF_GO|id=GO:0016787 |text = hydrolase activity}}
| Component = {{GNF_GO|id=GO:0005764 |text = lysosome}} {{GNF_GO|id=GO:0005783 |text = endoplasmic reticulum}}
| Process = {{GNF_GO|id=GO:0008152 |text = metabolic process}} {{GNF_GO|id=GO:0030203 |text = glycosaminoglycan metabolic process}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 3423
| Hs_Ensembl =
| Hs_RefseqProtein = NP_000193
| Hs_RefseqmRNA = NM_000202
| Hs_GenLoc_db =
| Hs_GenLoc_chr =
| Hs_GenLoc_start =
| Hs_GenLoc_end =
| Hs_Uniprot =
| Mm_EntrezGene = 15931
| Mm_Ensembl = ENSMUSG00000035847
| Mm_RefseqmRNA = NM_001038990
| Mm_RefseqProtein = NP_001034079
| Mm_GenLoc_db =
| Mm_GenLoc_chr = X
| Mm_GenLoc_start = 66603734
| Mm_GenLoc_end = 66625640
| Mm_Uniprot = Q8CJ15
}}
}}
'''Iduronate 2-sulfatase (Hunter syndrome)''', also known as '''IDS''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Iduronate-2-sulfatase is required for the lysosomal degradation of heparan sulfate and dermatan sulfate. Mutations in this X-chromosome gene that result in enzymatic deficiency lead to the sex-linked Mucopolysaccharidosis Type II, also known as Hunter Syndrome. Iduronate-2-sulfatase has a strong sequence homology with human arylsulfatases A, B, and C, and human glucosamine-6-sulfatase. A splice variant of this gene has been described.<ref>{{cite web | title = Entrez Gene: IDS iduronate 2-sulfatase (Hunter syndrome)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3423| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Hopwood JJ, Bunge S, Morris CP, ''et al.'' |title=Molecular basis of mucopolysaccharidosis type II: mutations in the iduronate-2-sulphatase gene. |journal=Hum. Mutat. |volume=2 |issue= 6 |pages= 435-42 |year= 1994 |pmid= 8111411 |doi= 10.1002/humu.1380020603 }}
*{{cite journal | author=Gort L, Chabás A, Coll MJ |title=Hunter disease in the Spanish population: molecular analysis in 31 families. |journal=J. Inherit. Metab. Dis. |volume=21 |issue= 6 |pages= 655-61 |year= 1998 |pmid= 9762601 |doi= }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on IGFBP2... {November 7, 2007 1:47:15 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein IGFBP2 image.jpg {November 7, 2007 1:47:47 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:48:00 AM PST}
- CREATED: Created new protein page: IGFBP2 {November 7, 2007 1:48:07 AM PST}
- INFO: Beginning work on IL7... {November 7, 2007 1:48:07 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:48:40 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Interleukin 7
| HGNCid = 6023
| Symbol = IL7
| AltSymbols =; IL-7
| OMIM = 146660
| ECnumber =
| Homologene = 680
| MGIid = 96561
| GeneAtlas_image1 = PBB_GE_IL7_206693_at_tn.png
| Function = {{GNF_GO|id=GO:0005139 |text = interleukin-7 receptor binding}} {{GNF_GO|id=GO:0005515 |text = protein binding}}
| Component = {{GNF_GO|id=GO:0005576 |text = extracellular region}} {{GNF_GO|id=GO:0005615 |text = extracellular space}}
| Process = {{GNF_GO|id=GO:0006955 |text = immune response}} {{GNF_GO|id=GO:0006959 |text = humoral immune response}} {{GNF_GO|id=GO:0007267 |text = cell-cell signaling}} {{GNF_GO|id=GO:0008284 |text = positive regulation of cell proliferation}} {{GNF_GO|id=GO:0009887 |text = organ morphogenesis}} {{GNF_GO|id=GO:0030890 |text = positive regulation of B cell proliferation}} {{GNF_GO|id=GO:0043066 |text = negative regulation of apoptosis}} {{GNF_GO|id=GO:0045453 |text = bone resorption}} {{GNF_GO|id=GO:0045582 |text = positive regulation of T cell differentiation}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 3574
| Hs_Ensembl = ENSG00000104432
| Hs_RefseqProtein = NP_000871
| Hs_RefseqmRNA = NM_000880
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 8
| Hs_GenLoc_start = 79807564
| Hs_GenLoc_end = 79880313
| Hs_Uniprot = P13232
| Mm_EntrezGene = 16196
| Mm_Ensembl = ENSMUSG00000040329
| Mm_RefseqmRNA = NM_008371
| Mm_RefseqProtein = NP_032397
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 3
| Mm_GenLoc_start = 7556913
| Mm_GenLoc_end = 7587247
| Mm_Uniprot = Q3UT18
}}
}}
'''Interleukin 7''', also known as '''IL7''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = The protein encoded by this gene is a cytokine important for B and T cell development. This cytokine and the hepatocyte growth factor (HGF) form a heterodimer that functions as a pre-pro-B cell growth-stimulating factor. This cytokine is found to be a cofactor for V(D)J rearrangement of the T cell receptor beta (TCRB) during early T cell development. This cytokine can be produced locally by intestinal epithelial and epithelial goblet cells, and may serve as a regulatory factor for intestinal mucosal lymphocytes. Knockout studies in mice suggested that this cytokine plays an essential role in lymphoid cell survival.<ref>{{cite web | title = Entrez Gene: IL7 interleukin 7| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3574| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Möller P, Böhm M, Czarnetszki BM, Schadendorf D |title=Interleukin-7. Biology and implications for dermatology. |journal=Exp. Dermatol. |volume=5 |issue= 3 |pages= 129-37 |year= 1997 |pmid= 8840152 |doi= }}
*{{cite journal | author=Appasamy PM |title=Biological and clinical implications of interleukin-7 and lymphopoiesis. |journal=Cytokines Cell. Mol. Ther. |volume=5 |issue= 1 |pages= 25-39 |year= 1999 |pmid= 10390077 |doi= }}
*{{cite journal | author=Fry TJ, Mackall CL |title=Interleukin-7: from bench to clinic. |journal=Blood |volume=99 |issue= 11 |pages= 3892-904 |year= 2002 |pmid= 12010786 |doi= }}
*{{cite journal | author=Fry TJ, Mackall CL |title=Interleukin-7 and immunorestoration in HIV: beyond the thymus. |journal=J. Hematother. Stem Cell Res. |volume=11 |issue= 5 |pages= 803-7 |year= 2003 |pmid= 12427286 |doi= 10.1089/152581602760404603 }}
*{{cite journal | author=Al-Rawi MA, Mansel RE, Jiang WG |title=Interleukin-7 (IL-7) and IL-7 receptor (IL-7R) signalling complex in human solid tumours. |journal=Histol. Histopathol. |volume=18 |issue= 3 |pages= 911-23 |year= 2004 |pmid= 12792903 |doi= }}
*{{cite journal | author=Aspinall R, Henson S, Pido-Lopez J, Ngom PT |title=Interleukin-7: an interleukin for rejuvenating the immune system. |journal=Ann. N. Y. Acad. Sci. |volume=1019 |issue= |pages= 116-22 |year= 2004 |pmid= 15247003 |doi= 10.1196/annals.1297.021 }}
*{{cite journal | author=Snyder KM, Mackall CL, Fry TJ |title=IL-7 in allogeneic transplant: clinical promise and potential pitfalls. |journal=Leuk. Lymphoma |volume=47 |issue= 7 |pages= 1222-8 |year= 2007 |pmid= 16923550 |doi= 10.1080/10428190600555876 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on INDO... {November 7, 2007 1:48:40 AM PST}
- UPLOAD: Added new Image to wiki:
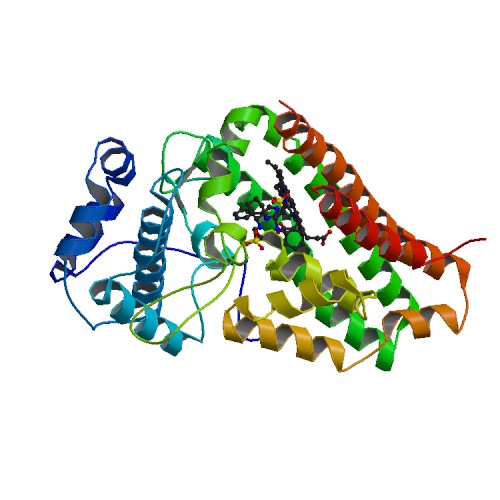 {November 7, 2007 1:49:12 AM PST}
{November 7, 2007 1:49:12 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:49:23 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_INDO_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 2d0t.
| PDB = {{PDB2|2d0t}}, {{PDB2|2d0u}}
| Name = Indoleamine-pyrrole 2,3 dioxygenase
| HGNCid = 6059
| Symbol = INDO
| AltSymbols =; CD107B; IDO
| OMIM = 147435
| ECnumber =
| Homologene = 48082
| MGIid = 96416
| GeneAtlas_image1 = PBB_GE_INDO_210029_at_tn.png
| Function = {{GNF_GO|id=GO:0004426 |text = tryptophan 2,3-dioxygenase activity}} {{GNF_GO|id=GO:0005506 |text = iron ion binding}} {{GNF_GO|id=GO:0009055 |text = electron carrier activity}} {{GNF_GO|id=GO:0020037 |text = heme binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}}
| Component =
| Process = {{GNF_GO|id=GO:0006955 |text = immune response}} {{GNF_GO|id=GO:0007565 |text = female pregnancy}} {{GNF_GO|id=GO:0019441 |text = tryptophan catabolic process to kynurenine}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 3620
| Hs_Ensembl = ENSG00000131203
| Hs_RefseqProtein = NP_002155
| Hs_RefseqmRNA = NM_002164
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 8
| Hs_GenLoc_start = 39890545
| Hs_GenLoc_end = 39905120
| Hs_Uniprot = P14902
| Mm_EntrezGene = 15930
| Mm_Ensembl = ENSMUSG00000031551
| Mm_RefseqmRNA = NM_008324
| Mm_RefseqProtein = NP_032350
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 8
| Mm_GenLoc_start = 26049686
| Mm_GenLoc_end = 26062554
| Mm_Uniprot = P28776
}}
}}
'''Indoleamine-pyrrole 2,3 dioxygenase''', also known as '''INDO''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Gamma-interferon (IFNG; MIM 147570) has an antiproliferative effect on many tumor cells and inhibits intracellular pathogens such as Toxoplasma and Chlamydia, at least partly because of the induction of indoleamine 2,3-dioxygenase (INDO; EC 1.13.11.42). This enzyme catalyzes the degradation of the essential amino acid L-tryptophan to N-formylkynurenine.[supplied by OMIM]<ref>{{cite web | title = Entrez Gene: INDO indoleamine-pyrrole 2,3 dioxygenase| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3620| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Grohmann U, Fallarino F, Puccetti P |title=Tolerance, DCs and tryptophan: much ado about IDO. |journal=Trends Immunol. |volume=24 |issue= 5 |pages= 242-8 |year= 2004 |pmid= 12738417 |doi= }}
*{{cite journal | author=Takikawa O |title=Biochemical and medical aspects of the indoleamine 2,3-dioxygenase-initiated L-tryptophan metabolism. |journal=Biochem. Biophys. Res. Commun. |volume=338 |issue= 1 |pages= 12-9 |year= 2005 |pmid= 16176799 |doi= 10.1016/j.bbrc.2005.09.032 }}
*{{cite journal | author=Puccetti P |title=On watching the watchers: IDO and type I/II IFN. |journal=Eur. J. Immunol. |volume=37 |issue= 4 |pages= 876-9 |year= 2007 |pmid= 17393386 |doi= 10.1002/eji.200737184 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on LIPC... {November 7, 2007 1:49:23 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:49:58 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Lipase, hepatic
| HGNCid = 6619
| Symbol = LIPC
| AltSymbols =; HL; HTGL; LIPH
| OMIM = 151670
| ECnumber =
| Homologene = 199
| MGIid = 96216
| GeneAtlas_image1 = PBB_GE_LIPC_206606_at_tn.png
| Function = {{GNF_GO|id=GO:0004806 |text = triacylglycerol lipase activity}} {{GNF_GO|id=GO:0005319 |text = lipid transporter activity}} {{GNF_GO|id=GO:0008201 |text = heparin binding}} {{GNF_GO|id=GO:0016787 |text = hydrolase activity}}
| Component =
| Process = {{GNF_GO|id=GO:0006487 |text = protein amino acid N-linked glycosylation}} {{GNF_GO|id=GO:0016042 |text = lipid catabolic process}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 3990
| Hs_Ensembl = ENSG00000166035
| Hs_RefseqProtein = NP_000227
| Hs_RefseqmRNA = NM_000236
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 15
| Hs_GenLoc_start = 56511467
| Hs_GenLoc_end = 56648364
| Hs_Uniprot = P11150
| Mm_EntrezGene = 15450
| Mm_Ensembl = ENSMUSG00000032207
| Mm_RefseqmRNA = NM_008280
| Mm_RefseqProtein = NP_032306
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 9
| Mm_GenLoc_start = 70597343
| Mm_GenLoc_end = 70734025
| Mm_Uniprot = Q3TYU0
}}
}}
'''Lipase, hepatic''', also known as '''LIPC''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = LIPC encodes hepatic triglyceride lipase, which is expressed in liver. LIPC has the dual functions of triglyceride hydrolase and ligand/bridging factor for receptor-mediated lipoprotein uptake.<ref>{{cite web | title = Entrez Gene: LIPC lipase, hepatic| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=3990| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Santamarina-Fojo S, Haudenschild C, Amar M |title=The role of hepatic lipase in lipoprotein metabolism and atherosclerosis. |journal=Curr. Opin. Lipidol. |volume=9 |issue= 3 |pages= 211-9 |year= 1998 |pmid= 9645503 |doi= }}
*{{cite journal | author=Jansen H, Verhoeven AJ, Sijbrands EJ |title=Hepatic lipase: a pro- or anti-atherogenic protein? |journal=J. Lipid Res. |volume=43 |issue= 9 |pages= 1352-62 |year= 2003 |pmid= 12235167 |doi= }}
*{{cite journal | author=Zambon A, Deeb SS, Pauletto P, ''et al.'' |title=Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. |journal=Curr. Opin. Lipidol. |volume=14 |issue= 2 |pages= 179-89 |year= 2003 |pmid= 12642787 |doi= 10.1097/01.mol.0000064055.08840.77 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on MEFV... {November 7, 2007 1:49:58 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:50:34 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Mediterranean fever
| HGNCid = 6998
| Symbol = MEFV
| AltSymbols =; MEF; FMF; MGC126560; MGC126586; TRIM20
| OMIM = 608107
| ECnumber =
| Homologene = 32441
| MGIid = 1859396
| GeneAtlas_image1 = PBB_GE_MEFV_208262_x_at_tn.png
| Function = {{GNF_GO|id=GO:0003779 |text = actin binding}} {{GNF_GO|id=GO:0008270 |text = zinc ion binding}} {{GNF_GO|id=GO:0046872 |text = metal ion binding}}
| Component = {{GNF_GO|id=GO:0005622 |text = intracellular}} {{GNF_GO|id=GO:0005634 |text = nucleus}} {{GNF_GO|id=GO:0005856 |text = cytoskeleton}} {{GNF_GO|id=GO:0005874 |text = microtubule}} {{GNF_GO|id=GO:0005875 |text = microtubule associated complex}}
| Process = {{GNF_GO|id=GO:0006954 |text = inflammatory response}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 4210
| Hs_Ensembl = ENSG00000103313
| Hs_RefseqProtein = NP_000234
| Hs_RefseqmRNA = NM_000243
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 16
| Hs_GenLoc_start = 3232029
| Hs_GenLoc_end = 3246628
| Hs_Uniprot = O15553
| Mm_EntrezGene = 54483
| Mm_Ensembl = ENSMUSG00000022534
| Mm_RefseqmRNA = NM_019453
| Mm_RefseqProtein = NP_062326
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 16
| Mm_GenLoc_start = 3622450
| Mm_GenLoc_end = 3633325
| Mm_Uniprot = Q32MT1
}}
}}
'''Mediterranean fever''', also known as '''MEFV''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = MEFV was identified as the gene that when mutated causes Mediterranean fever, a hereditary periodic fever syndrome. MEFV is expressed in granulocytes and myeloid bone marrow precursors.<ref>{{cite web | title = Entrez Gene: MEFV Mediterranean fever| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=4210| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Pras M |title=Familial Mediterranean fever: from the clinical syndrome to the cloning of the pyrin gene. |journal=Scand. J. Rheumatol. |volume=27 |issue= 2 |pages= 92-7 |year= 1998 |pmid= 9572633 |doi= }}
*{{cite journal | author=Centola M, Aksentijevich I, Kastner DL |title=The hereditary periodic fever syndromes: molecular analysis of a new family of inflammatory diseases. |journal=Hum. Mol. Genet. |volume=7 |issue= 10 |pages= 1581-8 |year= 1998 |pmid= 9735379 |doi= }}
*{{cite journal | author=Touitou I |title=The spectrum of Familial Mediterranean Fever (FMF) mutations. |journal=Eur. J. Hum. Genet. |volume=9 |issue= 7 |pages= 473-83 |year= 2001 |pmid= 11464238 |doi= 10.1038/sj.ejhg.5200658 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on NES... {November 7, 2007 1:58:03 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:58:39 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Nestin
| HGNCid = 7756
| Symbol = NES
| AltSymbols =; FLJ21841; Nbla00170
| OMIM = 600915
| ECnumber =
| Homologene = 31391
| MGIid = 101784
| GeneAtlas_image1 = PBB_GE_NES_218678_at_tn.png
| Function = {{GNF_GO|id=GO:0005198 |text = structural molecule activity}}
| Component = {{GNF_GO|id=GO:0005882 |text = intermediate filament}}
| Process = {{GNF_GO|id=GO:0007417 |text = central nervous system development}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 10763
| Hs_Ensembl = ENSG00000132688
| Hs_RefseqProtein = NP_006608
| Hs_RefseqmRNA = NM_006617
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 1
| Hs_GenLoc_start = 154905179
| Hs_GenLoc_end = 154913813
| Hs_Uniprot = P48681
| Mm_EntrezGene = 18008
| Mm_Ensembl = ENSMUSG00000004891
| Mm_RefseqmRNA = NM_016701
| Mm_RefseqProtein = NP_057910
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 3
| Mm_GenLoc_start = 88057020
| Mm_GenLoc_end = 88066378
| Mm_Uniprot =
}}
}}
'''Nestin''', also known as '''NES''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = Nestin is an intermediate filament protein that was first identified with a monoclonal antibody by Hockfield and McKay (1985). It is expressed predominantly in stem cells of the central nervous system in the neural tube. Upon terminal neural differentiation, nestin is downregulated and replaced by neurofilaments.[supplied by OMIM]<ref>{{cite web | title = Entrez Gene: NES nestin| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=10763| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Wiese C, Rolletschek A, Kania G, ''et al.'' |title=Nestin expression--a property of multi-lineage progenitor cells? |journal=Cell. Mol. Life Sci. |volume=61 |issue= 19-20 |pages= 2510-22 |year= 2004 |pmid= 15526158 |doi= 10.1007/s00018-004-4144-6 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on NTRK2... {November 7, 2007 1:50:34 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein NTRK2 image.jpg {November 7, 2007 1:52:12 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:52:24 AM PST}
- CREATED: Created new protein page: NTRK2 {November 7, 2007 1:52:31 AM PST}
- INFO: Beginning work on PAK2... {November 7, 2007 1:52:31 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PAK2 image.jpg {November 7, 2007 1:54:06 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:54:18 AM PST}
- CREATED: Created new protein page: PAK2 {November 7, 2007 1:54:26 AM PST}
- INFO: Beginning work on PARK7... {November 7, 2007 1:58:39 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PARK7 image.jpg {November 7, 2007 1:59:10 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:59:23 AM PST}
- CREATED: Created new protein page: PARK7 {November 7, 2007 1:59:31 AM PST}
- INFO: Beginning work on POLG... {November 7, 2007 1:54:26 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:55:28 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image =
| image_source =
| PDB =
| Name = Polymerase (DNA directed), gamma
| HGNCid = 9179
| Symbol = POLG
| AltSymbols =; FLJ27114; PEO; POLG1; POLGA; SANDO; SCAE
| OMIM = 174763
| ECnumber =
| Homologene = 2016
| MGIid = 1196389
| GeneAtlas_image1 = PBB_GE_POLG_203366_at_tn.png
| GeneAtlas_image2 = PBB_GE_POLG_217635_s_at_tn.png
| Function = {{GNF_GO|id=GO:0000287 |text = magnesium ion binding}} {{GNF_GO|id=GO:0003677 |text = DNA binding}} {{GNF_GO|id=GO:0003891 |text = delta DNA polymerase activity}} {{GNF_GO|id=GO:0003895 |text = gamma DNA-directed DNA polymerase activity}} {{GNF_GO|id=GO:0004527 |text = exonuclease activity}} {{GNF_GO|id=GO:0005515 |text = protein binding}} {{GNF_GO|id=GO:0016740 |text = transferase activity}}
| Component = {{GNF_GO|id=GO:0005739 |text = mitochondrion}} {{GNF_GO|id=GO:0005743 |text = mitochondrial inner membrane}} {{GNF_GO|id=GO:0005760 |text = gamma DNA polymerase complex}}
| Process = {{GNF_GO|id=GO:0006264 |text = mitochondrial DNA replication}} {{GNF_GO|id=GO:0006287 |text = base-excision repair, gap-filling}} {{GNF_GO|id=GO:0007568 |text = aging}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5428
| Hs_Ensembl = ENSG00000140521
| Hs_RefseqProtein = NP_002684
| Hs_RefseqmRNA = NM_002693
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 15
| Hs_GenLoc_start = 87660577
| Hs_GenLoc_end = 87679030
| Hs_Uniprot = P54098
| Mm_EntrezGene = 18975
| Mm_Ensembl = ENSMUSG00000039176
| Mm_RefseqmRNA = NM_017462
| Mm_RefseqProtein = NP_059490
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 7
| Mm_GenLoc_start = 79322896
| Mm_GenLoc_end = 79339786
| Mm_Uniprot = P54099
}}
}}
'''Polymerase (DNA directed), gamma''', also known as '''POLG''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text =
}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Graziewicz MA, Longley MJ, Copeland WC |title=DNA polymerase gamma in mitochondrial DNA replication and repair. |journal=Chem. Rev. |volume=106 |issue= 2 |pages= 383-405 |year= 2006 |pmid= 16464011 |doi= 10.1021/cr040463d }}
*{{cite journal | author=Hudson G, Chinnery PF |title=Mitochondrial DNA polymerase-gamma and human disease. |journal=Hum. Mol. Genet. |volume=15 Spec No 2 |issue= |pages= R244-52 |year= 2006 |pmid= 16987890 |doi= 10.1093/hmg/ddl233 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on POLR2J... {November 7, 2007 1:55:28 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:56:09 AM PST}
- CREATED: Created new protein page: POLR2J {November 7, 2007 1:56:16 AM PST}
- INFO: Beginning work on PSAP... {November 7, 2007 1:56:16 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein PSAP image.jpg {November 7, 2007 1:57:06 AM PST}
- AMBIGUITY: Did not locate an acceptable page to update. {November 7, 2007 1:57:24 AM PST}
<!-- The PBB_Controls template provides controls for Protein Box Bot, please see Template:PBB_Controls for details. -->
{{PBB_Controls
| update_page = yes
| require_manual_inspection = no
| update_protein_box = yes
| update_summary = yes
| update_citations = yes
}}
<!-- The GNF_Protein_box is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{GNF_Protein_box
| image = PBB_Protein_PSAP_image.jpg
| image_source = [[Protein_Data_Bank|PDB]] rendering based on 1m12.
| PDB = {{PDB2|1m12}}, {{PDB2|1n69}}, {{PDB2|1sn6}}, {{PDB2|2dob}}, {{PDB2|2gtg}}
| Name = Prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy)
| HGNCid = 9498
| Symbol = PSAP
| AltSymbols =; SAP1; FLJ00245; GLBA; MGC110993
| OMIM = 176801
| ECnumber =
| Homologene = 37680
| MGIid = 97783
| GeneAtlas_image1 = PBB_GE_PSAP_200866_s_at_tn.png
| GeneAtlas_image2 = PBB_GE_PSAP_200871_s_at_tn.png
| Function = {{GNF_GO|id=GO:0004098 |text = cerebroside-sulfatase activity}} {{GNF_GO|id=GO:0004336 |text = galactosylceramidase activity}} {{GNF_GO|id=GO:0004348 |text = glucosylceramidase activity}} {{GNF_GO|id=GO:0004557 |text = alpha-galactosidase activity}} {{GNF_GO|id=GO:0004565 |text = beta-galactosidase activity}} {{GNF_GO|id=GO:0004767 |text = sphingomyelin phosphodiesterase activity}} {{GNF_GO|id=GO:0008047 |text = enzyme activator activity}} {{GNF_GO|id=GO:0008289 |text = lipid binding}}
| Component = {{GNF_GO|id=GO:0005615 |text = extracellular space}} {{GNF_GO|id=GO:0005764 |text = lysosome}} {{GNF_GO|id=GO:0016021 |text = integral to membrane}}
| Process = {{GNF_GO|id=GO:0006629 |text = lipid metabolic process}} {{GNF_GO|id=GO:0006665 |text = sphingolipid metabolic process}} {{GNF_GO|id=GO:0006687 |text = glycosphingolipid metabolic process}} {{GNF_GO|id=GO:0006869 |text = lipid transport}} {{GNF_GO|id=GO:0007040 |text = lysosome organization and biogenesis}}
| Orthologs = {{GNF_Ortholog_box
| Hs_EntrezGene = 5660
| Hs_Ensembl = ENSG00000197746
| Hs_RefseqProtein = NP_001035930
| Hs_RefseqmRNA = NM_001042465
| Hs_GenLoc_db =
| Hs_GenLoc_chr = 10
| Hs_GenLoc_start = 73246061
| Hs_GenLoc_end = 73281132
| Hs_Uniprot = P07602
| Mm_EntrezGene = 19156
| Mm_Ensembl = ENSMUSG00000004207
| Mm_RefseqmRNA = NM_011179
| Mm_RefseqProtein = NP_035309
| Mm_GenLoc_db =
| Mm_GenLoc_chr = 10
| Mm_GenLoc_start = 59673071
| Mm_GenLoc_end = 59697954
| Mm_Uniprot = Q3TID4
}}
}}
'''Prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy)''', also known as '''PSAP''', is a human [[gene]].
<!-- The PBB_Summary template is automatically maintained by Protein Box Bot. See Template:PBB_Controls to Stop updates. -->
{{PBB_Summary
| section_title =
| summary_text = This gene encodes a highly conserved glycoprotein which is a precursor for 4 cleavage products: saposins A, B, C, and D. Each domain of the precursor protein is approximately 80 amino acid residues long with nearly identical placement of cysteine residues and glycosylation sites. Saposins A-D localize primarily to the lysosomal compartment where they facilitate the catabolism of glycosphingolipids with short oligosaccharide groups. The precursor protein exists both as a secretory protein and as an integral membrane protein and has neurotrophic activities. Mutations in this gene have been associated with Gaucher disease, Tay-Sachs disease, and metachromatic leukodystrophy. Alternative splicing results in multiple transcript variants encoding different isoforms.<ref>{{cite web | title = Entrez Gene: PSAP prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy)| url = http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5660| accessdate = }}</ref>
}}
==References==
{{reflist}}
==Further reading==
{{refbegin | 2}}
{{PBB_Further_reading
| citations =
*{{cite journal | author=Gieselmann V, Zlotogora J, Harris A, ''et al.'' |title=Molecular genetics of metachromatic leukodystrophy. |journal=Hum. Mutat. |volume=4 |issue= 4 |pages= 233-42 |year= 1995 |pmid= 7866401 |doi= 10.1002/humu.1380040402 }}
}}
{{refend}}
{{protein-stub}}
- INFO: Beginning work on RPA1... {November 7, 2007 1:57:24 AM PST}
- UPLOAD: Added new Image to wiki: File:PBB Protein RPA1 image.jpg {November 7, 2007 1:57:38 AM PST}
- CREATE: Found no pages, creating new page. {November 7, 2007 1:57:57 AM PST}
- CREATED: Created new protein page: RPA1 {November 7, 2007 1:58:03 AM PST}
end log.
 {November 7, 2007 1:38:13 AM PST}
{November 7, 2007 1:38:13 AM PST}  {November 7, 2007 1:49:12 AM PST}
{November 7, 2007 1:49:12 AM PST} 

