Methicillin-resistant Staphylococcus aureus: Difference between revisions
Citation bot (talk | contribs) |
|||
| Line 27: | Line 27: | ||
*People staying or working in a health care facility for an extended period of time<ref name="riskfactors"/> |
*People staying or working in a health care facility for an extended period of time<ref name="riskfactors"/> |
||
*People who spend time in coastal waters where MRSA is present, such as some beaches in [[Florida]] and the [[west coast of the United States]]<ref name="florida">{{cite news|url=http://www.foxnews.com/story/0,2933,493604,00.html|author=Reuters|publisher=[[Fox Entertainment Group|FoxNews.com]]|title=Study: Beachgoers More Likely to Catch MRSA | date=2009-02-16}}</ref><ref name="AP">{{cite news|url=http://www.foxnews.com/story/0,2933,549601,00.html|author=Marilynn Marchione|publisher=AP|title=Dangerous staph germs found at West Coast beaches | date=2009-09-12}}</ref> |
*People who spend time in coastal waters where MRSA is present, such as some beaches in [[Florida]] and the [[west coast of the United States]]<ref name="florida">{{cite news|url=http://www.foxnews.com/story/0,2933,493604,00.html|author=Reuters|publisher=[[Fox Entertainment Group|FoxNews.com]]|title=Study: Beachgoers More Likely to Catch MRSA | date=2009-02-16}}</ref><ref name="AP">{{cite news|url=http://www.foxnews.com/story/0,2933,549601,00.html|author=Marilynn Marchione|publisher=AP|title=Dangerous staph germs found at West Coast beaches | date=2009-09-12}}</ref> |
||
*People who spend time in confined spaces with other people, including occupants of homeless shelters and [[warming centers]], [[prison]] inmates, military recruits in [[Recruit training|basic training]],<ref name="soldiers">{{Cite journal | last1 = Zinderman | first1 = C. | last2 = Conner | first2 = B. | last3 = Malakooti | first3 = M. | last4 = LaMar | first4 = J. | last5 = Armstrong | first5 = A. | last6 = Bohnker | first6 = A. | title = Community-Acquired Methicillin-Resistant ''Staphylococcus aureus'' Among Military Recruits | journal = Emerging Infectious Diseases | month = May | year = 2004 | url=http://www.medscape.com/viewarticle/474843 }}</ref> and individuals who spend considerable time in [[changeroom]]s or [[gym]]s.{{Citation needed|date=May 2012}} |
*People who spend time in confined spaces with other people, including occupants of homeless shelters and [[warming centers]], [[prison]] inmates, military recruits in [[Recruit training|basic training]],<ref name="soldiers">{{Cite journal | last1 = Zinderman | first1 = C. | last2 = Conner | first2 = B. | last3 = Malakooti | first3 = M. | last4 = LaMar | first4 = J. | last5 = Armstrong | first5 = A. | last6 = Bohnker | first6 = A. | title = Community-Acquired Methicillin-Resistant ''Staphylococcus aureus'' Among Military Recruits | journal = Emerging Infectious Diseases | month = May | year = 2004 | url=http://www.medscape.com/viewarticle/474843 | doi = 10.3201/eid1005.030604 | volume = 10 | issue = 5 | pages = 941–4 | pmid = 15200838 | pmc = 3323224 }}</ref> and individuals who spend considerable time in [[changeroom]]s or [[gym]]s.{{Citation needed|date=May 2012}} |
||
*Urban under-served<ref name="cmr">{{cite journal | author = David, Micheal Z. and Daum, Robert S. | title = Community-Associated Methicillin-Resistant ''Staphylococcus aureus'': Epidemiology and Clinical Consequences of an Emerging Epidemic | publisher = American Society for Microbiology | volume = 23 | issue = 6 | year = 2010 | pages = 616–687 | url = http://cmr.asm.org/content/23/3/616.full.pdf+html}}</ref> |
*Urban under-served<ref name="cmr">{{cite journal | author = David, Micheal Z. and Daum, Robert S. | title = Community-Associated Methicillin-Resistant ''Staphylococcus aureus'': Epidemiology and Clinical Consequences of an Emerging Epidemic | publisher = American Society for Microbiology | volume = 23 | issue = 6 | year = 2010 | pages = 616–687 | url = http://cmr.asm.org/content/23/3/616.full.pdf+html | pmid = 20610826 | doi = 10.1128/CMR.00081-09 | pmc = 2901661 | journal = Clinical microbiology reviews}}</ref> |
||
*Indigenous populations, including Native Americans, Native Alaskans, and Australian Aborigines<ref name="cmr"/> |
*Indigenous populations, including Native Americans, Native Alaskans, and Australian Aborigines<ref name="cmr"/> |
||
*Veterinarians, Livestock handlers, and Pet owners<ref name="cmr"/> |
*Veterinarians, Livestock handlers, and Pet owners<ref name="cmr"/> |
||
| Line 43: | Line 43: | ||
Cases of MRSA have increased in livestock animals. CC398 is a new variant of MRSA that has emerged in animals and is found in intensively reared production animals (primarily pigs, but also cattle and poultry), where it can be transmitted to humans. Though dangerous to humans, CC398 is often asymptomatic in food-producing animals.<ref>{{cite web|url=http://www.efsa.europa.eu/EFSA/Report/biohaz_report_301_joint_mrsa_en,0.pdf|title= Joint scientific report of ECDC, EFSA and EMEA on meticillin resistant ''Staphylococcus aureus'' (MRSA) in livestock, companion animals and food|date=2009-06-16|accessdate=2009-09-19}}</ref> |
Cases of MRSA have increased in livestock animals. CC398 is a new variant of MRSA that has emerged in animals and is found in intensively reared production animals (primarily pigs, but also cattle and poultry), where it can be transmitted to humans. Though dangerous to humans, CC398 is often asymptomatic in food-producing animals.<ref>{{cite web|url=http://www.efsa.europa.eu/EFSA/Report/biohaz_report_301_joint_mrsa_en,0.pdf|title= Joint scientific report of ECDC, EFSA and EMEA on meticillin resistant ''Staphylococcus aureus'' (MRSA) in livestock, companion animals and food|date=2009-06-16|accessdate=2009-09-19}}</ref> |
||
A 2011 study reported 47% of the meat and poultry sold in surveyed U.S. grocery stores was contaminated with S. aureus and, of those, 52%—or 24.4% of the total—were resistant to at least three classes of antibiotics. "Now we need to determine what this means in terms of risk to the consumer," said Dr. Keim, a co-author of the paper.<ref>[http://www.sciencedaily.com/releases/2011/04/110415083153.htm US meat and poultry is widely contaminated with drug-resistant Staph bacteria, study finds<!-- Bot generated title -->]</ref> Some samples of commercially sold meat products in Japan were also found to harbor MRSA strains.<ref>{{cite journal|last=Ogata|first=K|coauthors=Narimatsu, H, Suzuki, M, Higuchi, W, Yamamoto, T, Taniguchi, H|title=Commercially distributed meat as a potential vehicle for community-acquired methicillin-resistant ''Staphylococcus aureus'' |journal=Applied and environmental microbiology|date=2012-02-03|doi=10.1128/AEM.07470-11|pmid=22307310|volume=78|issue=8|pages=2797–802}}</ref> |
A 2011 study reported 47% of the meat and poultry sold in surveyed U.S. grocery stores was contaminated with S. aureus and, of those, 52%—or 24.4% of the total—were resistant to at least three classes of antibiotics. "Now we need to determine what this means in terms of risk to the consumer," said Dr. Keim, a co-author of the paper.<ref>[http://www.sciencedaily.com/releases/2011/04/110415083153.htm US meat and poultry is widely contaminated with drug-resistant Staph bacteria, study finds<!-- Bot generated title -->]</ref> Some samples of commercially sold meat products in Japan were also found to harbor MRSA strains.<ref>{{cite journal|last=Ogata|first=K|coauthors=Narimatsu, H, Suzuki, M, Higuchi, W, Yamamoto, T, Taniguchi, H|title=Commercially distributed meat as a potential vehicle for community-acquired methicillin-resistant ''Staphylococcus aureus'' |journal=Applied and environmental microbiology|date=2012-02-03|doi=10.1128/AEM.07470-11|pmid=22307310|volume=78|issue=8|pages=2797–802|pmc=3318828}}</ref> |
||
=== Athletes === |
=== Athletes === |
||
In the United States, there have been increasing numbers of reports of outbreaks of MRSA colonization and infection through skin contact in [[changeroom|locker rooms]] and [[gym]]s, even among healthy populations.{{Citation needed|date=February 2012}} A study published in the ''New England Journal of Medicine'' linked MRSA to the abrasions caused by artificial turf.<ref>{{cite journal|last=Kazakova|first=SV|coauthors=Hageman, JC, Matava, M, Srinivasan, A, Phelan, L, Garfinkel, B, Boo, T, McAllister, S, Anderson, J, Jensen, B, Dodson, D, Lonsway, D, McDougal, LK, Arduino, M, Fraser, VJ, Killgore, G, Tenover, FC, Cody, S, Jernigan, DB|title=A clone of methicillin-resistant ''Staphylococcus aureus'' among professional football players |
In the United States, there have been increasing numbers of reports of outbreaks of MRSA colonization and infection through skin contact in [[changeroom|locker rooms]] and [[gym]]s, even among healthy populations.{{Citation needed|date=February 2012}} A study published in the ''New England Journal of Medicine'' linked MRSA to the abrasions caused by artificial turf.<ref>{{cite journal|last=Kazakova|first=SV|coauthors=Hageman, JC, Matava, M, Srinivasan, A, Phelan, L, Garfinkel, B, Boo, T, McAllister, S, Anderson, J, Jensen, B, Dodson, D, Lonsway, D, McDougal, LK, Arduino, M, Fraser, VJ, Killgore, G, Tenover, FC, Cody, S, Jernigan, DB|title=A clone of methicillin-resistant ''Staphylococcus aureus'' among professional football players|journal=The New England Journal of Medicine|date=2005-02-03|volume=352|issue=5|pages=468–75|pmid=15689585|url=http://www.nejm.org/doi/full/10.1056/NEJMoa042859|doi=10.1056/NEJMoa042859}}</ref> Three studies by the Texas State Department of Health found that the infection rate among football players was 16 times the national average. In October 2006, a high school football player was temporarily paralyzed from MRSA-infected turf burns. His infection returned in January 2007 and required three surgeries to remove infected tissue, as well as three weeks of hospital stay.<ref name="Epstein">{{cite news|url=http://www.bloomberg.com/apps/news?pid=newsarchive&sid=alxhrJDn.cdc|title=Texas Football Succumbs to Virulent Staph Infection From Turf|last=Epstein|first=Victor|date=21 December 2007|publisher=[[Bloomberg L.P.|Bloomberg]]|accessdate=10 June 2010}}</ref> |
||
=== Children === |
=== Children === |
||
MRSA is also becoming a problem in pediatric settings,<ref>{{cite journal |author=Gray JW |title=MRSA: the problem reaches paediatrics |journal=Arch. Dis. Child. |volume=89 |issue=4 |pages=297–8 |year=2004 |month=April |pmid=15033832 |pmc=1719885 |doi=10.1136/adc.2003.045534 |url=http://adc.bmjjournals.com/cgi/content/full/89/4/297}}</ref> including hospital nurseries.<ref>{{cite journal |author=Bratu S, Eramo A, Kopec R, ''et al.'' |title=Community-associated methicillin-resistant ''Staphylococcus aureus'' in hospital nursery and maternity units |journal=Emerging Infect. Dis. |volume=11 |issue=6 |pages=808–13 |year=2005 |month=June |pmid=15963273 |url=http://www.cdc.gov/ncidod/EID/vol11no06/04-0885.htm}}</ref> A 2007 study found that 4.6% of patients in U.S. health care facilities were infected or colonized with MRSA.<ref name="APIC"> |
MRSA is also becoming a problem in pediatric settings,<ref>{{cite journal |author=Gray JW |title=MRSA: the problem reaches paediatrics |journal=Arch. Dis. Child. |volume=89 |issue=4 |pages=297–8 |year=2004 |month=April |pmid=15033832 |pmc=1719885 |doi=10.1136/adc.2003.045534 |url=http://adc.bmjjournals.com/cgi/content/full/89/4/297}}</ref> including hospital nurseries.<ref>{{cite journal |author=Bratu S, Eramo A, Kopec R, ''et al.'' |title=Community-associated methicillin-resistant ''Staphylococcus aureus'' in hospital nursery and maternity units |journal=Emerging Infect. Dis. |volume=11 |issue=6 |pages=808–13 |year=2005 |month=June |pmid=15963273 |url=http://www.cdc.gov/ncidod/EID/vol11no06/04-0885.htm |doi=10.3201/eid1106.040885 |pmc=3367583}}</ref> A 2007 study found that 4.6% of patients in U.S. health care facilities were infected or colonized with MRSA.<ref name="APIC"> |
||
{{cite web |
{{cite web |
||
| url = http://www.apic.org/Content/NavigationMenu/ResearchFoundation/NationalMRSAPrevalenceStudy/MRSA_Study_Results.htm |
| url = http://www.apic.org/Content/NavigationMenu/ResearchFoundation/NationalMRSAPrevalenceStudy/MRSA_Study_Results.htm |
||
| Line 70: | Line 70: | ||
=== SCC''mec'' === |
=== SCC''mec'' === |
||
Staphylococcal [[Gene cassette|cassette chromosome]] ''mec'' ([[SCCmec|SCC''mec'']]) is a genomic island of unknown origin containing the antibiotic resistance gene ''mecA''.<ref name="lowy">{{cite journal |author=Lowy FD |title=Antimicrobial resistance: the example of ''Staphylococcus aureus'' |journal=J. Clin. Invest. |volume=111 |issue=9 |pages=1265–73 |year=2003 |month=May |pmid=12727914 |pmc=154455 |doi=10.1172/JCI18535 }}</ref><ref name="monaco">{{cite journal |author=Pantosti A, Sanchini A, Monaco M |title=Mechanisms of antibiotic resistance in ''Staphylococcus aureus'' |journal=Future Microbiol |volume=2 |issue=3 |pages=323–34 |year=2007 |month=June |pmid=17661706 |doi=10.2217/17460913.2.3.323 |url=http://www.futuremedicine.com/doi/abs/10.2217/17460913.2.3.323?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed}}</ref> SCC''mec'' contains additional genes beyond ''mecA'', including the [[cytolysin]] gene ''psm-mec'', which may suppress virulence in hospital-acquired MRSA strains.<ref>{{cite journal |author=Kaito C, Saito Y, Nagano G, ''et al.'' |title=Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate ''Staphylococcus aureus'' virulence |journal=PLoS Pathog. |volume=7 |issue=2 |pages=e1001267 |year=2011 |pmid=21304931 |pmc=3033363 |doi=10.1371/journal.ppat.1001267 |url=http://dx.plos.org/10.1371/journal.ppat.1001267}}</ref> SCC''mec'' also contains ''ccrA'' and ''ccrB''; both genes encode recombinases that mediate the site-specific integration and excision of the SCC''mec'' element from the ''S. aureus'' chromosome.<ref name="lowy"/><ref name="monaco"/> Currently, six unique SCC''mec'' types ranging in size from 21–67 kb have been identified;<ref name="lowy"/> they are designated types I-VI and are distinguished by variation in ''mec'' and ''ccr'' gene complexes.<ref name="jenson"/> Owing to the size of the SCC''mec'' element and the constraints of horizontal gene transfer, a limited number of clones is thought to be responsible for the spread of MRSA infections.<ref name="lowy"/> |
Staphylococcal [[Gene cassette|cassette chromosome]] ''mec'' ([[SCCmec|SCC''mec'']]) is a genomic island of unknown origin containing the antibiotic resistance gene ''mecA''.<ref name="lowy">{{cite journal |author=Lowy FD |title=Antimicrobial resistance: the example of ''Staphylococcus aureus'' |journal=J. Clin. Invest. |volume=111 |issue=9 |pages=1265–73 |year=2003 |month=May |pmid=12727914 |pmc=154455 |doi=10.1172/JCI18535 }}</ref><ref name="monaco">{{cite journal |author=Pantosti A, Sanchini A, Monaco M |title=Mechanisms of antibiotic resistance in ''Staphylococcus aureus'' |journal=Future Microbiol |volume=2 |issue=3 |pages=323–34 |year=2007 |month=June |pmid=17661706 |doi=10.2217/17460913.2.3.323 |url=http://www.futuremedicine.com/doi/abs/10.2217/17460913.2.3.323?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dpubmed}}</ref> SCC''mec'' contains additional genes beyond ''mecA'', including the [[cytolysin]] gene ''psm-mec'', which may suppress virulence in hospital-acquired MRSA strains.<ref>{{cite journal |author=Kaito C, Saito Y, Nagano G, ''et al.'' |title=Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate ''Staphylococcus aureus'' virulence |journal=PLoS Pathog. |volume=7 |issue=2 |pages=e1001267 |year=2011 |pmid=21304931 |pmc=3033363 |doi=10.1371/journal.ppat.1001267 |url=http://dx.plos.org/10.1371/journal.ppat.1001267 |editor1-last=Cheung |editor1-first=Ambrose}}</ref> SCC''mec'' also contains ''ccrA'' and ''ccrB''; both genes encode recombinases that mediate the site-specific integration and excision of the SCC''mec'' element from the ''S. aureus'' chromosome.<ref name="lowy"/><ref name="monaco"/> Currently, six unique SCC''mec'' types ranging in size from 21–67 kb have been identified;<ref name="lowy"/> they are designated types I-VI and are distinguished by variation in ''mec'' and ''ccr'' gene complexes.<ref name="jenson"/> Owing to the size of the SCC''mec'' element and the constraints of horizontal gene transfer, a limited number of clones is thought to be responsible for the spread of MRSA infections.<ref name="lowy"/> |
||
Different SCC''mec'' genotypes confer different microbiological characteristics, such as different antimicrobial resistance rates.<ref name="kuo">{{cite journal |author=Kuo SC, Chiang MC, Lee WS, ''et al.'' |title=Comparison of microbiological and clinical characteristics based on SCCmec typing in patients with community-onset meticillin-resistant ''Staphylococcus aureus'' (MRSA) bacteraemia |journal=Int. J. Antimicrob. Agents |volume=39 |issue=1 |pages=22–6 |year=2012 |month=January |pmid=21982834 |doi=10.1016/j.ijantimicag.2011.08.014 |url=http://linkinghub.elsevier.com/retrieve/pii/S0924-8579(11)00359-1}}</ref> Different genotypes are also associated with different types of infections. Types I-III SCC''mec'' are large elements that typically contain additional resistance genes and are characteristically isolated from HA-MRSA strains.<ref name="monaco"/><ref name="kuo"/> Conversely, CA-MRSA is associated with types IV and V, which are smaller and lack resistance genes other than ''mecA''.<ref name="monaco"/><ref name="kuo"/> |
Different SCC''mec'' genotypes confer different microbiological characteristics, such as different antimicrobial resistance rates.<ref name="kuo">{{cite journal |author=Kuo SC, Chiang MC, Lee WS, ''et al.'' |title=Comparison of microbiological and clinical characteristics based on SCCmec typing in patients with community-onset meticillin-resistant ''Staphylococcus aureus'' (MRSA) bacteraemia |journal=Int. J. Antimicrob. Agents |volume=39 |issue=1 |pages=22–6 |year=2012 |month=January |pmid=21982834 |doi=10.1016/j.ijantimicag.2011.08.014 |url=http://linkinghub.elsevier.com/retrieve/pii/S0924-8579(11)00359-1}}</ref> Different genotypes are also associated with different types of infections. Types I-III SCC''mec'' are large elements that typically contain additional resistance genes and are characteristically isolated from HA-MRSA strains.<ref name="monaco"/><ref name="kuo"/> Conversely, CA-MRSA is associated with types IV and V, which are smaller and lack resistance genes other than ''mecA''.<ref name="monaco"/><ref name="kuo"/> |
||
| Line 77: | Line 77: | ||
''mecA'' is responsible for resistance to methicillin and other β-lactam antibiotics. After acquisition of ''mecA'', the gene must be integrated and localized in the S. aureus chromosome.<ref name="lowy"/> ''mecA'' encodes penicillin-binding protein 2a (PBP2a), which differs from other penicillin-binding proteins as its active site does not bind methicillin or other β-lactam antibiotics.<ref name="lowy"/> As such, PBP2a can continue to catalyze the transpeptidation reaction required for [[peptidoglycan]] cross-linking, enabling cell wall synthesis in the presence of antibiotics. As a consequence of the inability of PBP2a to interact with β-lactam moieties, acquisition of ''mecA'' confers resistance to all β-lactam antibiotics in addition to methicillin.<ref name="lowy"/><ref>Sahebnasagh R, Saderi H, Owlia P. Detection of methicillin-resistant Staphylococcus aureus strains from clinical samples in Tehran by detection of the mecA and nuc genes. The First Iranian International Congress of Medical Bacteriology; 4-7 September; Tabriz, Iran. 2011. 195 pp.</ref> |
''mecA'' is responsible for resistance to methicillin and other β-lactam antibiotics. After acquisition of ''mecA'', the gene must be integrated and localized in the S. aureus chromosome.<ref name="lowy"/> ''mecA'' encodes penicillin-binding protein 2a (PBP2a), which differs from other penicillin-binding proteins as its active site does not bind methicillin or other β-lactam antibiotics.<ref name="lowy"/> As such, PBP2a can continue to catalyze the transpeptidation reaction required for [[peptidoglycan]] cross-linking, enabling cell wall synthesis in the presence of antibiotics. As a consequence of the inability of PBP2a to interact with β-lactam moieties, acquisition of ''mecA'' confers resistance to all β-lactam antibiotics in addition to methicillin.<ref name="lowy"/><ref>Sahebnasagh R, Saderi H, Owlia P. Detection of methicillin-resistant Staphylococcus aureus strains from clinical samples in Tehran by detection of the mecA and nuc genes. The First Iranian International Congress of Medical Bacteriology; 4-7 September; Tabriz, Iran. 2011. 195 pp.</ref> |
||
''mecA'' is under the control of two [[regulatory genes]], ''mecI'' and ''mecR1''. MecI is usually bound to the ''mecA'' promoter and functions as a repressor.<ref name="jenson"/><ref name="monaco"/> In the presence of a β-lactam antibiotic, MecR1 initiates a [[signal transduction cascade]] that leads to transcriptional activation of ''mecA''.<ref name="jenson"/><ref name="monaco"/> This is achieved by MecR1-mediated cleavage of MecI, which alleviates MecI repression.<ref name="jenson"/> ''mecA'' is further controlled by two co-repressors, BlaI and BlaR1. ''blaI'' and ''blaR1'' are homologous to ''mecI'' and ''mecR1'', respectively, and normally function as regulators of ''blaZ'', which is responsible for penicillin resistance.<ref name="lowy"/><ref name="berger-bachi">{{cite journal |author=Berger-Bächi B |title=Genetic basis of methicillin resistance in ''Staphylococcus aureus'' |journal=Cell. Mol. Life Sci. |volume=56 |issue= |
''mecA'' is under the control of two [[regulatory genes]], ''mecI'' and ''mecR1''. MecI is usually bound to the ''mecA'' promoter and functions as a repressor.<ref name="jenson"/><ref name="monaco"/> In the presence of a β-lactam antibiotic, MecR1 initiates a [[signal transduction cascade]] that leads to transcriptional activation of ''mecA''.<ref name="jenson"/><ref name="monaco"/> This is achieved by MecR1-mediated cleavage of MecI, which alleviates MecI repression.<ref name="jenson"/> ''mecA'' is further controlled by two co-repressors, BlaI and BlaR1. ''blaI'' and ''blaR1'' are homologous to ''mecI'' and ''mecR1'', respectively, and normally function as regulators of ''blaZ'', which is responsible for penicillin resistance.<ref name="lowy"/><ref name="berger-bachi">{{cite journal |author=Berger-Bächi B |title=Genetic basis of methicillin resistance in ''Staphylococcus aureus'' |journal=Cell. Mol. Life Sci. |volume=56 |issue=9–10 |pages=764–70 |year=1999 |month=November |pmid=11212336 |url=http://link.springer.de/link/service/journals/00018/bibs/90569-10/90560764.htm |doi=10.1007/s000180050023}}</ref> The DNA sequences bound by MecI and BlaI are identical;<ref name="lowy"/> therefore, BlaI can also bind the ''mecA'' operator to repress transcription of ''mecA''.<ref name="berger-bachi"/> |
||
=== Strains === |
=== Strains === |
||
| Line 84: | Line 84: | ||
Acquisition of SCC''mec'' in methicillin-sensitive staphylococcus aureus (''MSSA'') gives rise to a number of genetically different MRSA lineages. These genetic variations within different MRSA strains possibly explain the variability in virulence and associated MRSA infections.<ref name="GordonLowy2008">{{cite journal |author=Gordon RJ, Lowy FD |title=Pathogenesis of methicillin-resistant ''Staphylococcus aureus'' infection |journal=Clin. Infect. Dis. |volume=46 |issue=Suppl 5|pages=S350–9 |year=2008 |month=June |pmid=18462090 |pmc=2474459 |doi=10.1086/533591 |url=http://www.cid.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=18462090}}</ref> The first MRSA strain, ST250 MRSA-1 originated from SCC''mec'' and ST250-MSSA integration.<ref name="GordonLowy2008"/> Historically, major MRSA clones: ST2470-MRSA-I, ST239-MRSA-III, ST5-MRSA-II, and ST5-MRSA-IV were responsible for causing hospital-acquired MRSA (HA-MRSA) infections.<ref name="GordonLowy2008"/> ST239-MRSA-III, known as the Brazilian clone, was highly transmissible compared to others and distributed in Argentina, Czech Republic, and Portugal.<ref name="GordonLowy2008"/> |
Acquisition of SCC''mec'' in methicillin-sensitive staphylococcus aureus (''MSSA'') gives rise to a number of genetically different MRSA lineages. These genetic variations within different MRSA strains possibly explain the variability in virulence and associated MRSA infections.<ref name="GordonLowy2008">{{cite journal |author=Gordon RJ, Lowy FD |title=Pathogenesis of methicillin-resistant ''Staphylococcus aureus'' infection |journal=Clin. Infect. Dis. |volume=46 |issue=Suppl 5|pages=S350–9 |year=2008 |month=June |pmid=18462090 |pmc=2474459 |doi=10.1086/533591 |url=http://www.cid.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=18462090}}</ref> The first MRSA strain, ST250 MRSA-1 originated from SCC''mec'' and ST250-MSSA integration.<ref name="GordonLowy2008"/> Historically, major MRSA clones: ST2470-MRSA-I, ST239-MRSA-III, ST5-MRSA-II, and ST5-MRSA-IV were responsible for causing hospital-acquired MRSA (HA-MRSA) infections.<ref name="GordonLowy2008"/> ST239-MRSA-III, known as the Brazilian clone, was highly transmissible compared to others and distributed in Argentina, Czech Republic, and Portugal.<ref name="GordonLowy2008"/> |
||
In the UK, where MRSA is commonly called "Golden Staph", the most common strains of MRSA are EMRSA15 and EMRSA16.<ref name="JAntimicrobChemother2001-Johnson">{{cite journal | author=Johnson AP, Aucken HM, Cavendish S, ''et al.'' | title=Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) | journal=J Antimicrob Chemother | year=2001 | pages=143–4 | volume=48 | issue=1 | pmid = 11418528 | url =http://jac.oxfordjournals.org/cgi/content/full/48/1/143 | doi=10.1093/jac/48.1.143}}</ref> EMRSA16 is the best described epidemiologically: it originated in [[Kettering]], England, and the full genomic sequence of this strain has been published.<ref>{{cite journal | author=Holden MTG, Feil EJ, Lindsay JA, ''et al.'' | title=Complete genomes of two clinical ''Staphylococcus aureus'' strains: Evidence for the rapid evolution of virulence and drug resistance | journal=Proc Natl Acad Sci USA | year=2004 | volume=101 | pages=9786–91| doi=10.1073/pnas.0402521101 | pmid = 15213324 | issue=26 | pmc=470752}}</ref> EMRSA16 has been found to be identical to the [[MLST|ST]]36:USA200 strain, which circulates in the United States, and to carry the SCC''mec'' type II, [[enterotoxin|enterotoxin A]] and [[toxic shock syndrome]] toxin 1 genes.<ref name="Diep2006" /> Under the new international typing system, this strain is now called MRSA252. EMRSA 15 is also found to be one of the common MRSA strains in Asia. Other common strains include ST5:USA100 and EMRSA 1.<ref name="StefaniChung2012">{{cite journal|last1=Stefani|first1=Stefania|last2=Chung|first2=Doo Ryeon|last3=Lindsay|first3=Jodi A.|last4=Friedrich|first4=Alex W.|last5=Kearns|first5=Angela M.|last6=Westh|first6=Henrik|last7=MacKenzie|first7=Fiona M.|title=Meticillin-resistant ''Staphylococcus aureus'' (MRSA): global epidemiology and harmonisation of typing methods|journal=International Journal of Antimicrobial Agents|year=2012|issn=09248579|doi=10.1016/j.ijantimicag.2011.09.030}}</ref> These strains are genetic characteristics of HA-MRSA.<ref name="Calfee2011">{{cite journal |author=Calfee DP |title=The epidemiology, treatment, and prevention of transmission of methicillin-resistant ''Staphylococcus aureus'' |journal=J Infus Nurs |volume=34 |issue=6 |pages=359–64 |year=2011 |pmid=22101629 |doi=10.1097/NAN.0b013e31823061d6 |url=http://meta.wkhealth.com/pt/pt-core/template-journal/lwwgateway/media/landingpage.htm?issn=1533-1458&volume=34&issue=6&spage=359}}</ref> |
In the UK, where MRSA is commonly called "Golden Staph", the most common strains of MRSA are EMRSA15 and EMRSA16.<ref name="JAntimicrobChemother2001-Johnson">{{cite journal | author=Johnson AP, Aucken HM, Cavendish S, ''et al.'' | title=Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) | journal=J Antimicrob Chemother | year=2001 | pages=143–4 | volume=48 | issue=1 | pmid = 11418528 | url =http://jac.oxfordjournals.org/cgi/content/full/48/1/143 | doi=10.1093/jac/48.1.143}}</ref> EMRSA16 is the best described epidemiologically: it originated in [[Kettering]], England, and the full genomic sequence of this strain has been published.<ref>{{cite journal | author=Holden MTG, Feil EJ, Lindsay JA, ''et al.'' | title=Complete genomes of two clinical ''Staphylococcus aureus'' strains: Evidence for the rapid evolution of virulence and drug resistance | journal=Proc Natl Acad Sci USA | year=2004 | volume=101 | pages=9786–91| doi=10.1073/pnas.0402521101 | pmid = 15213324 | issue=26 | pmc=470752}}</ref> EMRSA16 has been found to be identical to the [[MLST|ST]]36:USA200 strain, which circulates in the United States, and to carry the SCC''mec'' type II, [[enterotoxin|enterotoxin A]] and [[toxic shock syndrome]] toxin 1 genes.<ref name="Diep2006" /> Under the new international typing system, this strain is now called MRSA252. EMRSA 15 is also found to be one of the common MRSA strains in Asia. Other common strains include ST5:USA100 and EMRSA 1.<ref name="StefaniChung2012">{{cite journal|last1=Stefani|first1=Stefania|last2=Chung|first2=Doo Ryeon|last3=Lindsay|first3=Jodi A.|last4=Friedrich|first4=Alex W.|last5=Kearns|first5=Angela M.|last6=Westh|first6=Henrik|last7=MacKenzie|first7=Fiona M.|title=Meticillin-resistant ''Staphylococcus aureus'' (MRSA): global epidemiology and harmonisation of typing methods|journal=International Journal of Antimicrobial Agents|year=2012|issn=09248579|doi=10.1016/j.ijantimicag.2011.09.030|volume=39|issue=4|pages=273–82|pmid=22230333}}</ref> These strains are genetic characteristics of HA-MRSA.<ref name="Calfee2011">{{cite journal |author=Calfee DP |title=The epidemiology, treatment, and prevention of transmission of methicillin-resistant ''Staphylococcus aureus'' |journal=J Infus Nurs |volume=34 |issue=6 |pages=359–64 |year=2011 |pmid=22101629 |doi=10.1097/NAN.0b013e31823061d6 |url=http://meta.wkhealth.com/pt/pt-core/template-journal/lwwgateway/media/landingpage.htm?issn=1533-1458&volume=34&issue=6&spage=359}}</ref> |
||
It is not entirely certain why some strains are highly transmissible and persistent in healthcare facilities.<ref name="GordonLowy2008"/> One explanation is the characteristic pattern of antibiotic susceptibility. Both the EMRSA15 and EMRSA16 strains are resistant to [[erythromycin]] and [[ciprofloxacin]]. It is known that ''Staphylococcus aureus'' can survive intracellularly,<ref>{{cite journal | author=von Eiff C, Becker K, Metze D, ''et al.'' | title=Intracellular persistence of ''Staphylococcus aureus'' small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier's disease | journal=Clin Infect Dis | year=2001 | volume=32 | issue=11 | pages=1643–7 | pmid = 11340539 | doi=10.1086/320519}}</ref> for example in the nasal mucosa <ref>{{cite journal | author=Clement S, Vaudaux P, François P, ''et al.'' | title=Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent ''Staphylococcus aureus'' rhinosinusitis | journal=J Infect Dis | year=2005 | volume=192 | issue=6 | pages=1023–8 | pmid = 16107955 | doi=10.1086/432735}}</ref> and in the tonsil tissue, .<ref>{{cite journal | author=Zautner AE, Krause M, Stropahl G, ''et al.'' | editor1-last=Bereswill | editor1-first=Stefan | title=Intracellular persisting ''Staphylococcus aureus'' is the major pathogen in recurrent tonsillitis | journal=PloS One | year=2010 | volume=5 | issue=3 | pages=e9452 | pmid = 20209109 | pmc=2830486 | doi=10.1371/journal.pone.0009452}}</ref> Erythromycin and Ciprofloxacin are precisely the antibiotics that best penetrate intracellularly; it may be that these strains of ''S. aureus'' are therefore able to exploit an intracellular niche. |
It is not entirely certain why some strains are highly transmissible and persistent in healthcare facilities.<ref name="GordonLowy2008"/> One explanation is the characteristic pattern of antibiotic susceptibility. Both the EMRSA15 and EMRSA16 strains are resistant to [[erythromycin]] and [[ciprofloxacin]]. It is known that ''Staphylococcus aureus'' can survive intracellularly,<ref>{{cite journal | author=von Eiff C, Becker K, Metze D, ''et al.'' | title=Intracellular persistence of ''Staphylococcus aureus'' small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier's disease | journal=Clin Infect Dis | year=2001 | volume=32 | issue=11 | pages=1643–7 | pmid = 11340539 | doi=10.1086/320519}}</ref> for example in the nasal mucosa <ref>{{cite journal | author=Clement S, Vaudaux P, François P, ''et al.'' | title=Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent ''Staphylococcus aureus'' rhinosinusitis | journal=J Infect Dis | year=2005 | volume=192 | issue=6 | pages=1023–8 | pmid = 16107955 | doi=10.1086/432735}}</ref> and in the tonsil tissue, .<ref>{{cite journal | author=Zautner AE, Krause M, Stropahl G, ''et al.'' | editor1-last=Bereswill | editor1-first=Stefan | title=Intracellular persisting ''Staphylococcus aureus'' is the major pathogen in recurrent tonsillitis | journal=PloS One | year=2010 | volume=5 | issue=3 | pages=e9452 | pmid = 20209109 | pmc=2830486 | doi=10.1371/journal.pone.0009452}}</ref> Erythromycin and Ciprofloxacin are precisely the antibiotics that best penetrate intracellularly; it may be that these strains of ''S. aureus'' are therefore able to exploit an intracellular niche. |
||
| Line 154: | Line 154: | ||
On January 6, 2008, half of 64 non-Chinese cases of MRSA infections in [[Hong Kong]] in 2007 were [[Filipino people|Filipino]] domestic helpers. Ho Pak-leung, professor of microbiology at the [[University of Hong Kong]], traced the cause to high use of antibiotics. In 2007, there were 166 community cases in Hong Kong compared with 8,000 hospital-acquired MRSA case (155 recorded cases—91 involved Chinese locals, 33 Filipinos, 5 each for Americans and Indians, and 2 each from Nepal, Australia, Denmark and England).<ref>[http://globalnation.inquirer.net/news/breakingnews/view_article.php?article_id=110923 Inquirer.net, Cases of RP maids with 'superbug' infection growing in HK]</ref> |
On January 6, 2008, half of 64 non-Chinese cases of MRSA infections in [[Hong Kong]] in 2007 were [[Filipino people|Filipino]] domestic helpers. Ho Pak-leung, professor of microbiology at the [[University of Hong Kong]], traced the cause to high use of antibiotics. In 2007, there were 166 community cases in Hong Kong compared with 8,000 hospital-acquired MRSA case (155 recorded cases—91 involved Chinese locals, 33 Filipinos, 5 each for Americans and Indians, and 2 each from Nepal, Australia, Denmark and England).<ref>[http://globalnation.inquirer.net/news/breakingnews/view_article.php?article_id=110923 Inquirer.net, Cases of RP maids with 'superbug' infection growing in HK]</ref> |
||
Worldwide, an estimated 2 billion people carry some form of ''S. aureus''; of these, up to 53 million (2.7% of carriers) are thought to carry MRSA.<ref>{{cite web | url = http://www.keepkidshealthy.com/welcome/infectionsguide/mrsa.html | title = MRSA Infections | publisher = Keep Kids Healthy}}</ref> In the United States, 95 million carry ''S. aureus'' in their noses; of these, 2.5 million (2.6% of carriers) carry MRSA.<ref>{{cite journal |author=Graham P, Lin S, Larson E |title=A U.S. population-based survey of ''Staphylococcus aureus'' colonization |journal=Ann Intern Med |volume=144 |issue=5 |pages=318–25 |year=2006 |pmid=16520472}}</ref> A population review conducted in three U.S. communities showed the annual incidence of CA-MRSA during 2001–2002 to be 18–25.7/100,000; most CA-MRSA isolates were associated with clinically relevant infections, and 23% of patients required hospitalization.<ref>{{cite conference | author = Jernigan JA, Arnold K, Heilpern K, Kainer M, Woods C, Hughes JM | title = Methicillin-resistant ''Staphylococcus aureus'' as community pathogen | booktitle = Symposium on Community-Associated Methicillin-resistant ''Staphylococcus aureus'' (Atlanta, Georgia, U.S.). Cited in Emerg Infect Dis | publisher = Centers for Disease Control and Prevention |date=2006-05-12 | url = http://www.cdc.gov/ncidod/EID/vol12no11/06-0911.htm | accessdate = 2007-01-27}}</ref> |
Worldwide, an estimated 2 billion people carry some form of ''S. aureus''; of these, up to 53 million (2.7% of carriers) are thought to carry MRSA.<ref>{{cite web | url = http://www.keepkidshealthy.com/welcome/infectionsguide/mrsa.html | title = MRSA Infections | publisher = Keep Kids Healthy}}</ref> In the United States, 95 million carry ''S. aureus'' in their noses; of these, 2.5 million (2.6% of carriers) carry MRSA.<ref>{{cite journal |author=Graham P, Lin S, Larson E |title=A U.S. population-based survey of ''Staphylococcus aureus'' colonization |journal=Ann Intern Med |volume=144 |issue=5 |pages=318–25 |year=2006 |pmid=16520472 |doi=10.7326/0003-4819-144-5-200603070-00006}}</ref> A population review conducted in three U.S. communities showed the annual incidence of CA-MRSA during 2001–2002 to be 18–25.7/100,000; most CA-MRSA isolates were associated with clinically relevant infections, and 23% of patients required hospitalization.<ref>{{cite conference | author = Jernigan JA, Arnold K, Heilpern K, Kainer M, Woods C, Hughes JM | title = Methicillin-resistant ''Staphylococcus aureus'' as community pathogen | booktitle = Symposium on Community-Associated Methicillin-resistant ''Staphylococcus aureus'' (Atlanta, Georgia, U.S.). Cited in Emerg Infect Dis | publisher = Centers for Disease Control and Prevention |date=2006-05-12 | url = http://www.cdc.gov/ncidod/EID/vol12no11/06-0911.htm | accessdate = 2007-01-27}}</ref> |
||
One possible contribution to the increased spread of MRSA infections comes from the use of antibiotics in [[intensive pig farming]]. A 2008 study in Canada found MRSA in 10% of tested pork chops and ground pork; a U.S. study in the same year found MRSA in the noses of 70% of the tested farm pigs and in 45% of the tested pig farm workers.<ref>[http://blog.seattlepi.com/secretingredients/archives/140336.asp First study finds MRSA in U.S. pigs and farmers], ''seattlepi.com'', 4 June 2008</ref> There have also been anecdotal reports of increased MRSA infection rates in rural communities with pig farms.<ref>[http://www.nytimes.com/2009/03/12/opinion/12kristof.html?_r=1 Our Pigs, Our Food, Our Health], ''The New York Times'', 12 March 2009</ref> |
One possible contribution to the increased spread of MRSA infections comes from the use of antibiotics in [[intensive pig farming]]. A 2008 study in Canada found MRSA in 10% of tested pork chops and ground pork; a U.S. study in the same year found MRSA in the noses of 70% of the tested farm pigs and in 45% of the tested pig farm workers.<ref>[http://blog.seattlepi.com/secretingredients/archives/140336.asp First study finds MRSA in U.S. pigs and farmers], ''seattlepi.com'', 4 June 2008</ref> There have also been anecdotal reports of increased MRSA infection rates in rural communities with pig farms.<ref>[http://www.nytimes.com/2009/03/12/opinion/12kristof.html?_r=1 Our Pigs, Our Food, Our Health], ''The New York Times'', 12 March 2009</ref> |
||
| Line 161: | Line 161: | ||
=== Decolonization === |
=== Decolonization === |
||
Care should be taken when trying to drain boils, as disruption of surrounding tissue can lead to larger infections, or even infection of the blood stream (often with fatal consequences).<ref name="nih">{{cite web|title=PubMed Health|url=http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0004520/|publisher=US National Institutes of Health|accessdate=20 November 2011}}</ref> Any drainage should be disposed of very carefully. After the drainage of boils or other treatment for MRSA, patients can shower at home using [[chlorhexidine]] (Hibiclens) or [[hexachlorophene]] (Phisohex) antiseptic soap (available over-the-counter at many pharmacies) from head to toe. Alternatively, a dilute bleach bath can be taken at a concentration of 1/2 cup bleach per 1/4-full bathtub of water.<ref>{{cite web|title=Optimal Bleach Concentration Required to Kill MRSA in Bath Water|publisher=American Academy of Pediatrics |
Care should be taken when trying to drain boils, as disruption of surrounding tissue can lead to larger infections, or even infection of the blood stream (often with fatal consequences).<ref name="nih">{{cite web|title=PubMed Health|url=http://www.ncbi.nlm.nih.gov/pubmedhealth/PMH0004520/|publisher=US National Institutes of Health|accessdate=20 November 2011}}</ref> Any drainage should be disposed of very carefully. After the drainage of boils or other treatment for MRSA, patients can shower at home using [[chlorhexidine]] (Hibiclens) or [[hexachlorophene]] (Phisohex) antiseptic soap (available over-the-counter at many pharmacies) from head to toe. Alternatively, a dilute bleach bath can be taken at a concentration of 1/2 cup bleach per 1/4-full bathtub of water.<ref>{{cite web|title=Optimal Bleach Concentration Required to Kill MRSA in Bath Water|publisher=American Academy of Pediatrics}}</ref> Care should be taken to use a clean towel, and to ensure that nasal discharge doesn't infect the towel (see below). |
||
All infectious lesions should be kept covered with a dressing.<ref name="nih" /> [[Mupirocin]] (Bactroban) 2% ointment can be effective at reducing the size of lesions. A secondary covering of clothing is preferred.<ref name="tpchd">{{cite web|title=Living With MRSA|url=http://www.tpchd.org/files/library/2357adf2a147d1aa.pdf|publisher=Group Health Cooperative/Tacoma-Pierce County Health Dept./Washington State Dept. of Health|accessdate=20 November 2011}}</ref> As shown in an animal study with diabetic mice, the topical application of a mixture of sugar (70%) and 3% povidone-iodine paste is an effective agent for the treatment of diabetic ulcers with MRSA infection.<ref>{{cite journal |author=Shi CM, Nakao H, Yamazaki M, Tsuboi R, Ogawa H |title=Mixture of sugar and povidone-iodine stimulates healing of MRSA-infected skin ulcers on db/db mice |journal=Arch. Dermatol. Res. |volume=299 |issue=9 |pages=449–56 |year=2007 |month=November |pmid=17680256 |doi=10.1007/s00403-007-0776-3 }}</ref> |
All infectious lesions should be kept covered with a dressing.<ref name="nih" /> [[Mupirocin]] (Bactroban) 2% ointment can be effective at reducing the size of lesions. A secondary covering of clothing is preferred.<ref name="tpchd">{{cite web|title=Living With MRSA|url=http://www.tpchd.org/files/library/2357adf2a147d1aa.pdf|publisher=Group Health Cooperative/Tacoma-Pierce County Health Dept./Washington State Dept. of Health|accessdate=20 November 2011}}</ref> As shown in an animal study with diabetic mice, the topical application of a mixture of sugar (70%) and 3% povidone-iodine paste is an effective agent for the treatment of diabetic ulcers with MRSA infection.<ref>{{cite journal |author=Shi CM, Nakao H, Yamazaki M, Tsuboi R, Ogawa H |title=Mixture of sugar and povidone-iodine stimulates healing of MRSA-infected skin ulcers on db/db mice |journal=Arch. Dermatol. Res. |volume=299 |issue=9 |pages=449–56 |year=2007 |month=November |pmid=17680256 |doi=10.1007/s00403-007-0776-3 }}</ref> |
||
| Line 180: | Line 180: | ||
== Treatment == |
== Treatment == |
||
Both CA-MRSA and HA-MRSA are resistant to traditional anti-staphylococcal [[beta-lactam antibiotic]]s, such as [[cephalexin]]. CA-MRSA has a greater spectrum of antimicrobial susceptibility, including to [[Sulfonamide (medicine)|sulfa drugs]] (like [[co-trimoxazole]]/trimethoprim-sulfamethoxazole), [[Tetracycline antibiotics|tetracyclines]] (like [[doxycycline]] and [[minocycline]]) and [[clindamycin]], but the drug of choice for treating CA-MRSA is now believed to be [[vancomycin]], according to a Henry Ford Hospital Study. HA-MRSA is resistant even to these antibiotics and often is susceptible only to vancomycin. Newer drugs, such as [[linezolid]] (belonging to the newer [[oxazolidinones]] class) and [[daptomycin]], are effective against both CA-MRSA and HA-MRSA. Linezolid is now felt to be the best drug for treating patients with MRSA pneumonia.<ref>{{cite journal |author=Wunderink RG, Rello J, Cammarata SK, Croos-Dabrera RV, Kollef MH |title=Linezolid vs vancomycin: analysis of two double-blind studies of patients with methicillin-resistant ''Staphylococcus aureus'' nosocomial pneumonia |journal=Chest |volume=124 |issue=5 |pages=1789–97 |year=2003 |month=November |pmid=14605050 }}</ref>{{dubious|reason=No RCTs have shown this and virtually no hospitals use this in their MRSA protocol for pneumonia|date=January 2013}} Ceftaroline and ceftabiparole, new fifth generation cephalosporins, are the first beta-lactam antibiotics approved in the US to treat MRSA infections (skin and soft tissue only).{{Citation needed|date=February 2012}} |
Both CA-MRSA and HA-MRSA are resistant to traditional anti-staphylococcal [[beta-lactam antibiotic]]s, such as [[cephalexin]]. CA-MRSA has a greater spectrum of antimicrobial susceptibility, including to [[Sulfonamide (medicine)|sulfa drugs]] (like [[co-trimoxazole]]/trimethoprim-sulfamethoxazole), [[Tetracycline antibiotics|tetracyclines]] (like [[doxycycline]] and [[minocycline]]) and [[clindamycin]], but the drug of choice for treating CA-MRSA is now believed to be [[vancomycin]], according to a Henry Ford Hospital Study. HA-MRSA is resistant even to these antibiotics and often is susceptible only to vancomycin. Newer drugs, such as [[linezolid]] (belonging to the newer [[oxazolidinones]] class) and [[daptomycin]], are effective against both CA-MRSA and HA-MRSA. Linezolid is now felt to be the best drug for treating patients with MRSA pneumonia.<ref>{{cite journal |author=Wunderink RG, Rello J, Cammarata SK, Croos-Dabrera RV, Kollef MH |title=Linezolid vs vancomycin: analysis of two double-blind studies of patients with methicillin-resistant ''Staphylococcus aureus'' nosocomial pneumonia |journal=Chest |volume=124 |issue=5 |pages=1789–97 |year=2003 |month=November |pmid=14605050 |doi=10.1378/chest.124.5.1789 }}</ref>{{dubious|reason=No RCTs have shown this and virtually no hospitals use this in their MRSA protocol for pneumonia|date=January 2013}} Ceftaroline and ceftabiparole, new fifth generation cephalosporins, are the first beta-lactam antibiotics approved in the US to treat MRSA infections (skin and soft tissue only).{{Citation needed|date=February 2012}} |
||
Vancomycin and [[teicoplanin]] are [[glycopeptide antibiotics]] used to treat MRSA infections.<ref>{{cite journal |author=Schentag JJ, Hyatt JM, Carr JR, Paladino JA, Birmingham MC, Zimmer GS, Cumbo TJ |title=Genesis of methicillin-resistant ''Staphylococcus aureus'' (MRSA), how treatment of MRSA infections has selected for vancomycin-resistant Enterococcus faecium, and the importance of antibiotic management and infection control |journal=Clin. Infect. Dis. |volume=26 |issue=5 |pages=1204–14 |year=1998 |pmid=9597254 |doi=10.1086/520287}}</ref> |
Vancomycin and [[teicoplanin]] are [[glycopeptide antibiotics]] used to treat MRSA infections.<ref>{{cite journal |author=Schentag JJ, Hyatt JM, Carr JR, Paladino JA, Birmingham MC, Zimmer GS, Cumbo TJ |title=Genesis of methicillin-resistant ''Staphylococcus aureus'' (MRSA), how treatment of MRSA infections has selected for vancomycin-resistant Enterococcus faecium, and the importance of antibiotic management and infection control |journal=Clin. Infect. Dis. |volume=26 |issue=5 |pages=1204–14 |year=1998 |pmid=9597254 |doi=10.1086/520287}}</ref> |
||
[[Teicoplanin]] is a structural [[Congener (chemistry)|congener]] of vancomycin that has a similar activity spectrum but a longer [[Exponential decay#Natural sciences|half-life]].<ref>{{cite journal |author=Rybak MJ, Lerner SA, Levine DP, Albrecht LM, McNeil PL, Thompson GA, Kenny MT, Yuh L |title=Teicoplanin pharmacokinetics in intravenous drug abusers being treated for bacterial endocarditis |journal=Antimicrob. Agents Chemother. |volume=35 |issue=4 |pages=696–700 |year=1991 |pmid=1829880 |pmc=245081}}</ref> |
[[Teicoplanin]] is a structural [[Congener (chemistry)|congener]] of vancomycin that has a similar activity spectrum but a longer [[Exponential decay#Natural sciences|half-life]].<ref>{{cite journal |author=Rybak MJ, Lerner SA, Levine DP, Albrecht LM, McNeil PL, Thompson GA, Kenny MT, Yuh L |title=Teicoplanin pharmacokinetics in intravenous drug abusers being treated for bacterial endocarditis |journal=Antimicrob. Agents Chemother. |volume=35 |issue=4 |pages=696–700 |year=1991 |pmid=1829880 |pmc=245081 |doi=10.1128/AAC.35.4.696}}</ref> |
||
Because the oral absorption of vancomycin and [[teicoplanin]] is very low, these agents must be administered intravenously to control systemic infections.<ref>{{cite journal |author=Janknegt R |title=The treatment of staphylococcal infections with special reference to pharmacokinetic, pharmacodynamic, and pharmacoeconomic considerations |journal=Pharmacy world & science : PWS |volume=19 |issue=3 |pages=133–41 |year=1997 |pmid=9259029 |doi=10.1023/A:1008609718457}}</ref> Treatment of MRSA infection with vancomycin can be complicated, due to its inconvenient route of administration. Moreover, many clinicians believe that the efficacy of vancomycin against MRSA is inferior to that of anti-staphylococcal [[beta-lactam antibiotic]]s against methicillin-susceptible ''Staphylococcus aureus'' (MSSA).<ref>{{cite journal | journal=Medicine (Baltimore) | year=2003 | volume=82 | issue=5 | pages=333–9 | title=''Staphylococcus aureus'' bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study | author=Chang FY, Peacock JE Jr, Musher DM, ''et al.'' | pmid = 14530782 | doi=10.1097/01.md.0000091184.93122.09}}</ref><ref>{{cite journal | journal=Scand J Infect Dis | year=2005 | volume=37 | issue=8 | pages=572–8 | title=The role of vancomycin in the persistence or recurrence of ''Staphylococcus aureus'' bacteraemia | author=Siegman-Igra Y, Reich P, Orni-Wasserlauf R, Schwartz D, Giladi M. | pmid = 16138425 | doi=10.1080/00365540510038488}}</ref> |
Because the oral absorption of vancomycin and [[teicoplanin]] is very low, these agents must be administered intravenously to control systemic infections.<ref>{{cite journal |author=Janknegt R |title=The treatment of staphylococcal infections with special reference to pharmacokinetic, pharmacodynamic, and pharmacoeconomic considerations |journal=Pharmacy world & science : PWS |volume=19 |issue=3 |pages=133–41 |year=1997 |pmid=9259029 |doi=10.1023/A:1008609718457}}</ref> Treatment of MRSA infection with vancomycin can be complicated, due to its inconvenient route of administration. Moreover, many clinicians believe that the efficacy of vancomycin against MRSA is inferior to that of anti-staphylococcal [[beta-lactam antibiotic]]s against methicillin-susceptible ''Staphylococcus aureus'' (MSSA).<ref>{{cite journal | journal=Medicine (Baltimore) | year=2003 | volume=82 | issue=5 | pages=333–9 | title=''Staphylococcus aureus'' bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study | author=Chang FY, Peacock JE Jr, Musher DM, ''et al.'' | pmid = 14530782 | doi=10.1097/01.md.0000091184.93122.09}}</ref><ref>{{cite journal | journal=Scand J Infect Dis | year=2005 | volume=37 | issue=8 | pages=572–8 | title=The role of vancomycin in the persistence or recurrence of ''Staphylococcus aureus'' bacteraemia | author=Siegman-Igra Y, Reich P, Orni-Wasserlauf R, Schwartz D, Giladi M. | pmid = 16138425 | doi=10.1080/00365540510038488}}</ref> |
||
Several newly discovered strains of MRSA show [[antibiotic resistance]] even to vancomycin and [[teicoplanin]]. These new evolutions of the MRSA bacterium have been dubbed [[Vancomycin-resistant Staphylococcus aureus|Vancomycin intermediate-resistant ''Staphylococcus aureus'' (VISA)]].<ref>{{cite journal |author=Sieradzki K, Tomasz A |title=Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of ''Staphylococcus aureus'' |journal=J. Bacteriol. |volume=179 |issue=8 |pages=2557–66 |year=1997 |pmid=9098053 |pmc=179004}}</ref> |
Several newly discovered strains of MRSA show [[antibiotic resistance]] even to vancomycin and [[teicoplanin]]. These new evolutions of the MRSA bacterium have been dubbed [[Vancomycin-resistant Staphylococcus aureus|Vancomycin intermediate-resistant ''Staphylococcus aureus'' (VISA)]].<ref>{{cite journal |author=Sieradzki K, Tomasz A |title=Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of ''Staphylococcus aureus'' |journal=J. Bacteriol. |volume=179 |issue=8 |pages=2557–66 |year=1997 |pmid=9098053 |pmc=179004}}</ref> |
||
<ref>{{cite journal | author=Schito GC | title=The importance of the development of antibiotic resistance in ''Staphylococcus aureus'' | journal=Clin Microbiol Infect | year=2006 | pages=3–8 | volume=12 |issue=Suppl 1 | pmid = 16445718 | doi=10.1111/j.1469-0691.2006.01343.x}}</ref> [[Linezolid]], [[quinupristin/dalfopristin]], [[daptomycin]], [[ceftaroline]], and [[tigecycline]] are used to treat more severe infections that do not respond to glycopeptides such as vancomycin.<ref>{{cite journal |author=Mongkolrattanothai K, Boyle S, Kahana MD, Daum RS |title=Severe ''Staphylococcus aureus'' infections caused by clonally related community-associated methicillin-susceptible and methicillin-resistant isolates |journal=Clin. Infect. Dis. |volume=37 |issue=8 |pages=1050–8 |year=2003 |pmid=14523769 |doi=10.1086/378277}}</ref> Current guidelines recommend [[daptomycin]] for VISA bloodstream infections and endocarditis. <ref>{{cite journal |author=Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, J Rybak M, Talan DA, Chambers HF. |title=Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary |
<ref>{{cite journal | author=Schito GC | title=The importance of the development of antibiotic resistance in ''Staphylococcus aureus'' | journal=Clin Microbiol Infect | year=2006 | pages=3–8 | volume=12 |issue=Suppl 1 | pmid = 16445718 | doi=10.1111/j.1469-0691.2006.01343.x}}</ref> [[Linezolid]], [[quinupristin/dalfopristin]], [[daptomycin]], [[ceftaroline]], and [[tigecycline]] are used to treat more severe infections that do not respond to glycopeptides such as vancomycin.<ref>{{cite journal |author=Mongkolrattanothai K, Boyle S, Kahana MD, Daum RS |title=Severe ''Staphylococcus aureus'' infections caused by clonally related community-associated methicillin-susceptible and methicillin-resistant isolates |journal=Clin. Infect. Dis. |volume=37 |issue=8 |pages=1050–8 |year=2003 |pmid=14523769 |doi=10.1086/378277}}</ref> Current guidelines recommend [[daptomycin]] for VISA bloodstream infections and endocarditis. <ref>{{cite journal |author=Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, J Rybak M, Talan DA, Chambers HF. |title=Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary |journal=Clin Infect Dis. |date=2011 Feb 1 |volume=52 |issue=3 |page=285–92 |doi=10.1093/cid/cir034 |pmid=21217178 |pages=285–92}}</ref> |
||
There have been claims that [[bacteriophage]] can be used to cure MRSA.<ref>[http://www.phageinternational.com/ptc.htm Phage International - Subsidiaries - Phage Therapy Center<!-- Bot generated title -->]</ref><ref>{{cite news| url=http://www.wired.com/wired/archive/11.10/phages.html | work=Wired | title=Wired 11.10: How Ravenous Soviet Viruses Will Save the World}}</ref> |
There have been claims that [[bacteriophage]] can be used to cure MRSA.<ref>[http://www.phageinternational.com/ptc.htm Phage International - Subsidiaries - Phage Therapy Center<!-- Bot generated title -->]</ref><ref>{{cite news| url=http://www.wired.com/wired/archive/11.10/phages.html | work=Wired | title=Wired 11.10: How Ravenous Soviet Viruses Will Save the World}}</ref> |
||
Revision as of 07:38, 16 August 2013
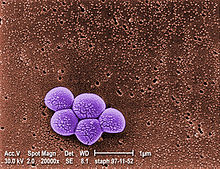
Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterium responsible for several difficult-to-treat infections in humans. It is also called oxacillin-resistant Staphylococcus aureus (ORSA). MRSA is any strain of Staphylococcus aureus that has developed, through the process of natural selection, resistance to beta-lactam antibiotics, which include the penicillins (methicillin, dicloxacillin, nafcillin, oxacillin, etc.) and the cephalosporins. Strains unable to resist these antibiotics are classified as methicillin-sensitive Staphylococcus aureus, or MSSA. The evolution of such resistance does not cause the organism to be more intrinsically virulent than strains of Staphylococcus aureus that have no antibiotic resistance, but resistance does make MRSA infection more difficult to treat with standard types of antibiotics and thus more dangerous.
MRSA is especially troublesome in hospitals, prisons and nursing homes, where patients with open wounds, invasive devices, and weakened immune systems are at greater risk of infection than the general public.
Signs and symptoms

S. aureus most commonly colonizes the anterior nares (the nostrils). The rest of the respiratory tract, open wounds, intravenous catheters, and the urinary tract are also potential sites for infection. Healthy individuals may carry MRSA asymptomatically for periods ranging from a few weeks to many years. Patients with compromised immune systems are at a significantly greater risk of symptomatic secondary infection.
In most patients, MRSA can be detected by swabbing the nostrils and isolating the bacteria found inside. Combined with extra sanitary measures for those in contact with infected patients, screening patients admitted to hospitals has been found to be effective in minimizing the spread of MRSA in hospitals in the United States,[1] Denmark, Finland, and the Netherlands.[2]
MRSA may progress substantially within 24–48 hours of initial topical symptoms. After 72 hours, MRSA can take hold in human tissues and eventually become resistant to treatment. The initial presentation of MRSA is small red bumps that resemble pimples, spider bites, or boils; they may be accompanied by fever and, occasionally, rashes. Within a few days, the bumps become larger and more painful; they eventually open into deep, pus-filled boils.[3] About 75 percent of community-associated (CA-) MRSA infections are localized to skin and soft tissue and usually can be treated effectively.[citation needed] But some CA-MRSA strains display enhanced virulence, spreading more rapidly and causing illness much more severe than traditional healthcare-associated (HA-) MRSA infections, and they can affect vital organs and lead to widespread infection (sepsis), toxic shock syndrome, and necrotizing ("flesh-eating") pneumonia. This is thought to be due to toxins carried by CA-MRSA strains, such as PVL and PSM, though PVL was recently found not to be a factor in a study by the National Institute of Allergy and Infectious Diseases (NIAID) at the NIH. It is not known why some healthy people develop CA-MRSA skin infections that are treatable while others infected with the same strain develop severe infections or die.[4]
People are very commonly colonized with CA-MRSA and completely asymptomatic. The most common manifestations of CA-MRSA are simple skin infections, such as impetigo, boils, abscesses, folliculitis, and cellulitis. Rarer, but more serious manifestations can occur, such as necrotizing fasciitis and pyomyositis (most commonly found in the tropics), necrotizing pneumonia, infective endocarditis (which affects the valves of the heart), and bone and joint infections.[5] CA-MRSA often results in abscess formation that requires incision and drainage. Before the spread of MRSA into the community, abscesses were not considered contagious, because it was assumed that infection required violation of skin integrity and the introduction of staphylococci from normal skin colonization. However, newly emerging CA-MRSA is transmissible (similar, but with very important differences) from Hospital-Associated MRSA. CA-MRSA is less likely than other forms of MRSA to cause cellulitis.
Risk factors
Some of the populations at risk:
- People with weak immune systems (HIV/AIDS, lupus, or cancer sufferers; transplant recipients, severe asthmatics, etc.)
- Diabetics[6]
- Intravenous drug users [7]
- Users of quinolone antibiotics[8]
- Young children[citation needed]
- The elderly[citation needed]
- College students living in dormitories[7]
- People staying or working in a health care facility for an extended period of time[7]
- People who spend time in coastal waters where MRSA is present, such as some beaches in Florida and the west coast of the United States[9][10]
- People who spend time in confined spaces with other people, including occupants of homeless shelters and warming centers, prison inmates, military recruits in basic training,[11] and individuals who spend considerable time in changerooms or gyms.[citation needed]
- Urban under-served[12]
- Indigenous populations, including Native Americans, Native Alaskans, and Australian Aborigines[12]
- Veterinarians, Livestock handlers, and Pet owners[12]
Hospital patients
Many MRSA infections occur in hospitals and healthcare facilities. When infections occur in this manner it is known as healthcare acquired MRSA or HA-MRSA. These Rates of MRSA infection are also increased in hospitalized patients who are treated with quinolones. Healthcare provider-to-patient transfer is common, especially when healthcare providers move from patient to patient without performing necessary hand-washing techniques between patients.[8][13]
Prison inmates, military recruits, and the homeless
Prisons, military barracks, and homeless shelters can be crowded and confined, and poor hygiene practices may proliferate, thus putting inhabitants at increased risk of contracting MRSA.[12] Cases of MRSA in such populations were first reported in the United States, and then in Canada. The earliest reports were made by the Center for Disease Control (CDC) in US state prisons. Subsequently reports of a massive rise in skin and soft tissue infections were reported by the CDC in the Los Angeles County Jail system in 2001, and this has continued. Pan et al. reported on the changing epidemiology of MRSA skin infection in the San Francisco County Jail, noting the MRSA accounted for more than 70% of S. aureus infection in the jail by 2002. Lowy and colleagues reported on frequent MRSA skin infections in New York State Prisons. Two reports on inmates in Maryland have demonstrated frequent colonization with MRSA.
In the news media hundreds of reports of MRSA outbreaks in prisons appeared between 2000 and 2008. For example, in February 2008, The Tulsa County Jail in the U.S. State of Oklahoma started treating an average of twelve Staphylococcus cases per month.[14] A report on skin and soft tissue infections in the Cook County Jail in Chicago in 2004–05 demonstrated that MRSA was the most common cause of these infections among cultured lesions and furthermore that few risk factors were more strongly associated with MRSA infections than infections caused by methicillin-susceptible S. aureus. In response to these and many other reports on MRSA infections among incarcerated and recently incarcerated persons, the Federal Bureau of Prisons has released guidelines for the management and control of the infections although few studies provide an evidence base for these guidelines.
People in contact with live food-producing animals
Cases of MRSA have increased in livestock animals. CC398 is a new variant of MRSA that has emerged in animals and is found in intensively reared production animals (primarily pigs, but also cattle and poultry), where it can be transmitted to humans. Though dangerous to humans, CC398 is often asymptomatic in food-producing animals.[15]
A 2011 study reported 47% of the meat and poultry sold in surveyed U.S. grocery stores was contaminated with S. aureus and, of those, 52%—or 24.4% of the total—were resistant to at least three classes of antibiotics. "Now we need to determine what this means in terms of risk to the consumer," said Dr. Keim, a co-author of the paper.[16] Some samples of commercially sold meat products in Japan were also found to harbor MRSA strains.[17]
Athletes
In the United States, there have been increasing numbers of reports of outbreaks of MRSA colonization and infection through skin contact in locker rooms and gyms, even among healthy populations.[citation needed] A study published in the New England Journal of Medicine linked MRSA to the abrasions caused by artificial turf.[18] Three studies by the Texas State Department of Health found that the infection rate among football players was 16 times the national average. In October 2006, a high school football player was temporarily paralyzed from MRSA-infected turf burns. His infection returned in January 2007 and required three surgeries to remove infected tissue, as well as three weeks of hospital stay.[19]
Children
MRSA is also becoming a problem in pediatric settings,[20] including hospital nurseries.[21] A 2007 study found that 4.6% of patients in U.S. health care facilities were infected or colonized with MRSA.[22] MRSA is becoming a major health concern in children because they are more likely to exhibit minor scrapes, cuts, bruises, and bug bites than adults. Children as well as adults are at higher risk of getting MRSA who come in contact with day care centers, playgrounds, locker rooms, camps, dormitories, classrooms and other school settings, and gyms and workout facilities. Parents should be especially cautious of children who participate in activities where there is shared sports equipment such as football helmets and uniforms.[23]
Diagnosis

Diagnostic microbiology laboratories and reference laboratories are key for identifying outbreaks of MRSA. New rapid techniques for the identification and characterization of MRSA have been developed. This notwithstanding, the bacterium generally must be cultured via blood, urine, sputum, or other body fluid cultures, and cultured in the lab in sufficient quantities to perform these confirmatory tests first. Consequently, there is no quick and easy method to diagnose a MRSA infection. Therefore, initial treatment is often based upon 'strong suspicion' by the treating physician, since any delay in treating this type of infection can have fatal consequences. These techniques include quantitative PCR and are increasingly being employed in clinical laboratories for the rapid detection and identification of MRSA strains.[24][25]
Another common laboratory test is a rapid latex agglutination test that detects the PBP2a protein. PBP2a is a variant penicillin-binding protein that imparts the ability of S. aureus to be resistant to oxacillin.[26]
Genetics
Antimicrobial resistance is genetically based; resistance is mediated by the acquisition of extrachromosomal genetic elements containing resistance genes. Exemplary are plasmids, transposable genetic elements, and genomic islands, which are transferred between bacteria via horizontal gene transfer.[27] A defining characteristic of MRSA is its ability to thrive in the presence of penicillin-like antibiotics, which normally prevent bacterial growth by inhibiting synthesis of cell wall material. This is due to a resistance gene, mecA, which stops β-lactam antibiotics from inactivating the enzymes (transpeptidases) that are critical for cell wall synthesis.
SCCmec
Staphylococcal cassette chromosome mec (SCCmec) is a genomic island of unknown origin containing the antibiotic resistance gene mecA.[28][29] SCCmec contains additional genes beyond mecA, including the cytolysin gene psm-mec, which may suppress virulence in hospital-acquired MRSA strains.[30] SCCmec also contains ccrA and ccrB; both genes encode recombinases that mediate the site-specific integration and excision of the SCCmec element from the S. aureus chromosome.[28][29] Currently, six unique SCCmec types ranging in size from 21–67 kb have been identified;[28] they are designated types I-VI and are distinguished by variation in mec and ccr gene complexes.[27] Owing to the size of the SCCmec element and the constraints of horizontal gene transfer, a limited number of clones is thought to be responsible for the spread of MRSA infections.[28]
Different SCCmec genotypes confer different microbiological characteristics, such as different antimicrobial resistance rates.[31] Different genotypes are also associated with different types of infections. Types I-III SCCmec are large elements that typically contain additional resistance genes and are characteristically isolated from HA-MRSA strains.[29][31] Conversely, CA-MRSA is associated with types IV and V, which are smaller and lack resistance genes other than mecA.[29][31]
mecA
mecA is responsible for resistance to methicillin and other β-lactam antibiotics. After acquisition of mecA, the gene must be integrated and localized in the S. aureus chromosome.[28] mecA encodes penicillin-binding protein 2a (PBP2a), which differs from other penicillin-binding proteins as its active site does not bind methicillin or other β-lactam antibiotics.[28] As such, PBP2a can continue to catalyze the transpeptidation reaction required for peptidoglycan cross-linking, enabling cell wall synthesis in the presence of antibiotics. As a consequence of the inability of PBP2a to interact with β-lactam moieties, acquisition of mecA confers resistance to all β-lactam antibiotics in addition to methicillin.[28][32]
mecA is under the control of two regulatory genes, mecI and mecR1. MecI is usually bound to the mecA promoter and functions as a repressor.[27][29] In the presence of a β-lactam antibiotic, MecR1 initiates a signal transduction cascade that leads to transcriptional activation of mecA.[27][29] This is achieved by MecR1-mediated cleavage of MecI, which alleviates MecI repression.[27] mecA is further controlled by two co-repressors, BlaI and BlaR1. blaI and blaR1 are homologous to mecI and mecR1, respectively, and normally function as regulators of blaZ, which is responsible for penicillin resistance.[28][33] The DNA sequences bound by MecI and BlaI are identical;[28] therefore, BlaI can also bind the mecA operator to repress transcription of mecA.[33]
Strains
Acquisition of SCCmec in methicillin-sensitive staphylococcus aureus (MSSA) gives rise to a number of genetically different MRSA lineages. These genetic variations within different MRSA strains possibly explain the variability in virulence and associated MRSA infections.[34] The first MRSA strain, ST250 MRSA-1 originated from SCCmec and ST250-MSSA integration.[34] Historically, major MRSA clones: ST2470-MRSA-I, ST239-MRSA-III, ST5-MRSA-II, and ST5-MRSA-IV were responsible for causing hospital-acquired MRSA (HA-MRSA) infections.[34] ST239-MRSA-III, known as the Brazilian clone, was highly transmissible compared to others and distributed in Argentina, Czech Republic, and Portugal.[34]
In the UK, where MRSA is commonly called "Golden Staph", the most common strains of MRSA are EMRSA15 and EMRSA16.[35] EMRSA16 is the best described epidemiologically: it originated in Kettering, England, and the full genomic sequence of this strain has been published.[36] EMRSA16 has been found to be identical to the ST36:USA200 strain, which circulates in the United States, and to carry the SCCmec type II, enterotoxin A and toxic shock syndrome toxin 1 genes.[37] Under the new international typing system, this strain is now called MRSA252. EMRSA 15 is also found to be one of the common MRSA strains in Asia. Other common strains include ST5:USA100 and EMRSA 1.[38] These strains are genetic characteristics of HA-MRSA.[39]
It is not entirely certain why some strains are highly transmissible and persistent in healthcare facilities.[34] One explanation is the characteristic pattern of antibiotic susceptibility. Both the EMRSA15 and EMRSA16 strains are resistant to erythromycin and ciprofloxacin. It is known that Staphylococcus aureus can survive intracellularly,[40] for example in the nasal mucosa [41] and in the tonsil tissue, .[42] Erythromycin and Ciprofloxacin are precisely the antibiotics that best penetrate intracellularly; it may be that these strains of S. aureus are therefore able to exploit an intracellular niche.
Community-acquired MRSA (CA-MRSA) strains emerged in late 1990 to 2000, infecting healthy people who had not been in contact with health care facilities.[39]A later study that analyzed data from more than 300 microbiology labs associated with hospitals all over the United States have found a seven-fold increase, jump from 3.6% of all MRSA infections to 28.2%, in the proportion of community-associated strains of MRSA between 1999 and 2006.[43] Researchers suggests that CA-MRSA did not evolve from the HA-MRSA.[39]This is further proven by molecular typing of CA-MRSA strains[44] and genome comparison between CA-MRSA and HA-MRSA, which indicate that novel MRSA strains integrated SCCmec into MSSA separately on its own.[39] By mid 2000, CA-MRSA was introduced into the health care systems and distinguishing CA-MRSA from HA-MRSA became a difficult process.[39]Community-acquired MRSA (CA-MRSA) is more easily treated and more virulent than hospital-acquired MRSA (HA-MRSA).[39] The genetic mechanism for the enhanced virulence in CA-MRSA remains an active area of research. Especially the Panton-Valentine leukocidin (PVL) genes are of interest because they are a unique feature of CA-MRSA.[34]
In the United States, most cases of CA-MRSA are caused by a CC8 strain designated ST8:USA300, which carries SCCmec type IV, Panton-Valentine leukocidin, PSM-alpha and enterotoxins Q and K,[37] and ST1:USA400.[45] The ST8:USA300 strain results in skin infections, necrotizing fasciitis and toxic shock syndrome, whereas the ST1:USA400 strain results in necrotizing pneumonia and pulmonary sepsis.[34] Other community-acquired strains of MRSA are ST8:USA500 and ST59:USA1000. In many nations of the world, MRSA strains with different predominant genetic background types have come to predominate among CA-MRSA strains; USA300 easily tops the list in the U. S. and is becoming more common in Canada after its first appearance there in 2004. For example, in Australia ST93 strains are common, while in continental Europe ST80 strains (Tristan et al., Emerging Infectious Diseases, 2006), which carry SCCmec type IV, predominate.[46] In Taiwan, ST59 strains, some of which are resistant to many non-beta-lactam antibiotics, have arisen as common causes of skin and soft tissue infections in the community. In a remote region of Alaska, unlike most of the continental U. S., USA300 was found rarely in a study of MRSA strains from outbreaks in 1996 and 2000 as well as in surveillance from 2004–06 (David et al., Emerg Infect Dis 2008).
In June 2011, the discovery of a new strain of MRSA was announced by two separate teams of researchers in the UK. Its genetic makeup was reportedly more similar to strains found in animals, and testing kits designed to detect MRSA were unable to identify it.[47] This MRSA strain, Clonal Complex 398 (CC398), is responsible for Livestock-associated MRSA (LA-MRSA) infections.[38] Although it is known to be more persistent in colonizing pigs and calves, there have been cases of LA-MRSA carriers with pneumonia, endocarditis, and necrotising fasciitis.[48]
Prevention
Screening programs
Patient screening upon hospital admission, with nasal cultures, prevents the cohabitation of MRSA carriers with non-carriers, and exposure to infected surfaces. The test used (whether a rapid molecular method or traditional culture) is not as important as the implementation of active screening.[49] In the United States and Canada, the Centers for Disease Control and Prevention issued guidelines on October 19, 2006, citing the need for additional research, but declined to recommend such screening.[50][51]
In some UK hospitals screening for MRSA is performed in every patient[52] and all NHS surgical patients, except for minor surgeries, are previously checked for MRSA.[53] There is no community screening in the UK; however, screening of individuals is offered by some private companies.[54]
In a US cohort of 1300 healthy children, 2.4% carried MRSA in their nose.[55]
Surface sanitizing

Alcohol has been proven to be an effective surface sanitizer against MRSA. Quaternary ammonium can be used in conjunction with alcohol to extend the longevity of the sanitizing action.[citation needed] The prevention of nosocomial infections involves routine and terminal cleaning. Non-flammable Alcohol Vapor in Carbon Dioxide systems (NAV-CO2) do not corrode metals or plastics used in medical environments and do not contribute to antibacterial resistance.
In healthcare environments, MRSA can survive on surfaces and fabrics, including privacy curtains or garments worn by care providers. Complete surface sanitation is necessary to eliminate MRSA in areas where patients are recovering from invasive procedures. Testing patients for MRSA upon admission, isolating MRSA-positive patients, decolonization of MRSA-positive patients, and terminal cleaning of patients' rooms and all other clinical areas they occupy is the current best practice protocol for nosocomial MRSA.
Studies published from 2004-2007 reported hydrogen peroxide vapor could be used to decontaminate busy hospital rooms, despite taking significantly longer than traditional cleaning. One study noted rapid recontamination by MRSA following the hydrogen peroxide application[56][57][58][59][60]
Also tested, in 2006, was a new type of surface cleaner, incorporating accelerated hydrogen peroxide, which was pronounced "a potential candidate" for use against the targeted microorganisms.[61]
Research on copper alloys
In 2008, after evaluating a wide body of research mandated specifically by the United States Environmental Protection Agency (EPA), registration approvals were granted by EPA in 2008 granting that copper alloys kill more than 99.9% of MRSA within two hours.
Subsequent research conducted at the University of Southampton (UK) compared the antimicrobial efficacies of copper and several non-copper proprietary coating products to kill MRSA.[62][63] At 20 °C, the drop-off in MRSA organisms on copper alloy C11000 is dramatic and almost complete (over 99.9% kill rate) within 75 minutes. However, neither a triclosan-based product nor two silver-containing based antimicrobial treatments (Ag-A and Ag-B) exhibited any meaningful efficacy against MRSA. Stainless steel S30400 did not exhibit any antimicrobial efficacy.
In 2004, the University of Southampton research team was the first to clearly demonstrate that copper inhibits MRSA.[64] On copper alloys — C19700 (99% copper), C24000 (80% copper), and C77000 (55% copper) — significant reductions in viability were achieved at room temperatures after 1.5 hours, 3.0 hours and 4.5 hours, respectively. Faster antimicrobial efficacies were associated with higher copper alloy content. Stainless steel did not exhibit any bactericidal benefits.
Hand washing
In September 2004,[65] after a successful pilot scheme to tackle MRSA, the UK National Health Service announced its Clean Your Hands campaign. Wards were required to ensure that alcohol-based hand rubs are placed near all beds so that staff can hand wash more regularly. It is thought that even if this cuts infection by no more than 1%, the plan will pay for itself many times over.[citation needed]
As with some other bacteria, MRSA is acquiring more resistance to some disinfectants and antiseptics. Although alcohol-based rubs remain somewhat effective, a more effective strategy is to wash hands with running water and an anti-microbial cleanser with persistent killing action, such as Chlorhexidine.[66] In another study chlorohexidine (Hibiclens), p-chloro-m-xylenol (Acute-Kare), hexachlorophene (Phisohex), and povidone-iodine (Betadine) were evaluated for their effectiveness. Of the four most commonly used antiseptics, povidone-iodine, when diluted 1:100, was the most rapidly bactericidal against both MRSA and methicillin-susceptible S. aureus.[67]
A June 2008 report, centered on a survey by the Association for Professionals in Infection Control and Epidemiology, concluded that poor hygiene habits remain the principal barrier to significant reductions in the spread of MRSA.
Use of surgical respirator
The US Food and Drug Administration (FDA) announced on 8 April 2011 that it had cleared a novel type of N95 Surgical Respirator, the SpectraShield 9500, that kills methicillin-resistant Staphylococcus aureus, Streptococcus pyogenes and Haemophilus influenzae. This mask is manufactured by Nexera Medical Ltd. of Richmond, British Columbia The mask blocks at least 95% of small particles in a standardized test. The FDA clearance also included evaluation by the National Institute of Occupational Health and Safety [68]
Proper disposal of hospital gowns
Used paper hospital gowns are associated with MRSA hospital infections, which could be avoided by proper disposal.[69]
Isolation
Excluding medical facilities, current US guidance does not require workers with MRSA infections to be routinely excluded from the general workplace.[70] Therefore, unless directed by a health care provider, exclusion from work should be reserved for those with wound drainage that cannot be covered and contained with a clean, dry bandage and for those who cannot maintain good hygiene practices.[70] Workers with active infections should be excluded from activities where skin-to-skin contact is likely to occur until their infections are healed. Health care workers should follow the Centers for Disease Control and Prevention's Guidelines for Infection Control in Health Care Personnel.[71]
To prevent the spread of staph or MRSA in the workplace, employers should ensure the availability of adequate facilities and supplies that encourage workers to practice good hygiene; that surface sanitizing in the workplace is followed; and that contaminated equipment are sanitized with Environmental Protection Agency (EPA)-registered disinfectants.[70]
Restricting antibiotic use
Glycopeptides, cephalosporins and in particular quinolones are associated with an increased risk of colonisation of MRSA. Reducing use of antibiotic classes that promote MRSA colonisation, especially fluoroquinolones, is recommended in current guidelines.[8][13]
Public health considerations
Mathematical models describe one way in which a loss of infection control can occur after measures for screening and isolation seem to be effective for years, as happened in the UK. In the "search and destroy" strategy that was employed by all UK hospitals until the mid-1990s, all patients with MRSA were immediately isolated, and all staff were screened for MRSA and were prevented from working until they had completed a course of eradication therapy that was proven to work. Loss of control occurs because colonised patients are discharged back into the community and then readmitted; when the number of colonised patients in the community reaches a certain threshold, the "search and destroy" strategy is overwhelmed.[72] One of the few countries not to have been overwhelmed by MRSA is the Netherlands: An important part of the success of the Dutch strategy may have been to attempt eradication of carriage upon discharge from hospital.[73]
The Centers for Disease Control and Prevention (CDC) estimated that about 1.7 million nosocomial infections occurred in the United States in 2002, with 99,000 associated deaths.[74] The estimated incidence is 4.5 nosocomial infections per 100 admissions, with direct costs (at 2004 prices) ranging from $10,500 (£5300, €8000 at 2006 rates) per case (for bloodstream, urinary tract, or respiratory infections in immunocompetent patients) to $111,000 (£57,000, €85,000) per case for antibiotic-resistant infections in the bloodstream in patients with transplants. With these numbers, conservative estimates of the total direct costs of nosocomial infections are above $17 billion. The reduction of such infections forms an important component of efforts to improve healthcare safety. (BMJ 2007)[citation needed] MRSA alone was associated with 8% of nosocomial infections reported to the CDC National Healthcare Safety Network from January 2006 to October 2007.[75]
This problem is not unique to one country; the British National Audit Office estimated that the incidence of nosocomial infections in Europe ranges from 4% to 10% of all hospital admissions. As of early 2005, the number of deaths in the United Kingdom attributed to MRSA has been estimated by various sources to lie in the area of 3,000 per year.[76] Staphylococcus bacteria account for almost half of all UK hospital infections. The issue of MRSA infections in hospitals has recently been a major political issue in the UK, playing a significant role in the debates over health policy in the United Kingdom general election held in 2005.
On January 6, 2008, half of 64 non-Chinese cases of MRSA infections in Hong Kong in 2007 were Filipino domestic helpers. Ho Pak-leung, professor of microbiology at the University of Hong Kong, traced the cause to high use of antibiotics. In 2007, there were 166 community cases in Hong Kong compared with 8,000 hospital-acquired MRSA case (155 recorded cases—91 involved Chinese locals, 33 Filipinos, 5 each for Americans and Indians, and 2 each from Nepal, Australia, Denmark and England).[77]
Worldwide, an estimated 2 billion people carry some form of S. aureus; of these, up to 53 million (2.7% of carriers) are thought to carry MRSA.[78] In the United States, 95 million carry S. aureus in their noses; of these, 2.5 million (2.6% of carriers) carry MRSA.[79] A population review conducted in three U.S. communities showed the annual incidence of CA-MRSA during 2001–2002 to be 18–25.7/100,000; most CA-MRSA isolates were associated with clinically relevant infections, and 23% of patients required hospitalization.[80]
One possible contribution to the increased spread of MRSA infections comes from the use of antibiotics in intensive pig farming. A 2008 study in Canada found MRSA in 10% of tested pork chops and ground pork; a U.S. study in the same year found MRSA in the noses of 70% of the tested farm pigs and in 45% of the tested pig farm workers.[81] There have also been anecdotal reports of increased MRSA infection rates in rural communities with pig farms.[82]
Healthcare facilities with high bed occupancy rates, high levels of temporary nursing staff, or low cleanliness scores no longer have significantly higher MRSA rates. Simple tabular evidence helps provide a clear picture of these changes, showing, for instance, that hospitals with occupancy over 90% had, in 2006–2007, MRSA rates little above those in hospitals with occupancy below 85%, in contrast to the period 2001–2004. In one sense, the disappearance of these relationships is puzzling. Reporters now blame IV cannula and catheters for spreading MRSA in hospitals. (Hospital organisation and speciality mix, 2008)[citation needed]
Decolonization
Care should be taken when trying to drain boils, as disruption of surrounding tissue can lead to larger infections, or even infection of the blood stream (often with fatal consequences).[83] Any drainage should be disposed of very carefully. After the drainage of boils or other treatment for MRSA, patients can shower at home using chlorhexidine (Hibiclens) or hexachlorophene (Phisohex) antiseptic soap (available over-the-counter at many pharmacies) from head to toe. Alternatively, a dilute bleach bath can be taken at a concentration of 1/2 cup bleach per 1/4-full bathtub of water.[84] Care should be taken to use a clean towel, and to ensure that nasal discharge doesn't infect the towel (see below).
All infectious lesions should be kept covered with a dressing.[83] Mupirocin (Bactroban) 2% ointment can be effective at reducing the size of lesions. A secondary covering of clothing is preferred.[85] As shown in an animal study with diabetic mice, the topical application of a mixture of sugar (70%) and 3% povidone-iodine paste is an effective agent for the treatment of diabetic ulcers with MRSA infection.[86]
The nose is a common refuge for MRSA, and a test swab can be taken of the nose to indicate whether MRSA is present.[87] If MRSA is detected via nasal culture, Mupirocin (Bactroban) 2% ointment can be applied inside each nostril twice daily for 7 days, using a cotton-tipped swab. However, care should be taken so that the swab doesn't penetrate into the sinus. Household members are recommended to follow the same decolonization protocol. After treatment, the nose should be swabbed again to ensure that the treatment was effective. If not, the process should be repeated.
Toilet seats are a common vector for infection, and wiping seats clean before and/or after use can help to prevent the spread of MRSA. Door handles, faucets, light switches (with care!), etc. can be disinfected regularly with disinfectant wipes.[85] Spray disinfectants can be used on upholstery. Carpets can be washed with disinfectant, and hardwood floors can be scrubbed with diluted tea tree oil (e.g. Melaleuca). Laundry soap containing tea tree oil may be effective at decontaminating clothing and bedding, especially if hot water and heavy soil cycles are used, however tea tree oil may cause a rash which MRSA can re-colonize. Alcohol-based sanitizers can be placed near bedsides, near sitting areas, in vehicles etc. to encourage their use.
Doctors may also prescribe antibiotics such as clindamycin, doxycycline or trimethoprim/sulfamethoxazole.
Community settings
The CDC offers suggestions for preventing the contraction and spread MRSA infection which are applicable to those in community settings, including incarcerated populations, childcare center employees, and athletes. To prevent MRSA infection, individuals should regularly wash hands using soap and water or an alcohol-based sanitizer, keep wounds clean and covered, avoid contact with other people's wounds, avoid sharing personal items such as razors or towels, shower after exercising at athletic facilities (including gyms, weight rooms, and school facilities), shower before using swimming pools or whirlpools, and maintain a clean environment.[88]
It may be difficult for people to maintain the necessary cleanliness if they do not have access to facilities such as public toilets with handwashing facilities. In the United Kingdom, the Workplace (Health, Safety and Welfare) Regulations 1992 requires businesses to provide toilets for their employees, along with washing facilities including soap or other suitable means of cleaning. Guidance on how many toilets to provide and what sort of washing facilities should be provided alongside them is given in the Workplace (Health, Safety and Welfare) Approved Code of Practice and Guidance L24, available from Health and Safety Executive Books. But there is no legal obligation on local authorities in the United Kingdom to provide public toilets, and although in 2008 the House of Commons Communities and Local Government Committee called for a duty on local authorities to develop a public toilet strategy [1] this was rejected by the Government [2].
Treatment
Both CA-MRSA and HA-MRSA are resistant to traditional anti-staphylococcal beta-lactam antibiotics, such as cephalexin. CA-MRSA has a greater spectrum of antimicrobial susceptibility, including to sulfa drugs (like co-trimoxazole/trimethoprim-sulfamethoxazole), tetracyclines (like doxycycline and minocycline) and clindamycin, but the drug of choice for treating CA-MRSA is now believed to be vancomycin, according to a Henry Ford Hospital Study. HA-MRSA is resistant even to these antibiotics and often is susceptible only to vancomycin. Newer drugs, such as linezolid (belonging to the newer oxazolidinones class) and daptomycin, are effective against both CA-MRSA and HA-MRSA. Linezolid is now felt to be the best drug for treating patients with MRSA pneumonia.[89][dubious – discuss] Ceftaroline and ceftabiparole, new fifth generation cephalosporins, are the first beta-lactam antibiotics approved in the US to treat MRSA infections (skin and soft tissue only).[citation needed]
Vancomycin and teicoplanin are glycopeptide antibiotics used to treat MRSA infections.[90] Teicoplanin is a structural congener of vancomycin that has a similar activity spectrum but a longer half-life.[91] Because the oral absorption of vancomycin and teicoplanin is very low, these agents must be administered intravenously to control systemic infections.[92] Treatment of MRSA infection with vancomycin can be complicated, due to its inconvenient route of administration. Moreover, many clinicians believe that the efficacy of vancomycin against MRSA is inferior to that of anti-staphylococcal beta-lactam antibiotics against methicillin-susceptible Staphylococcus aureus (MSSA).[93][94]
Several newly discovered strains of MRSA show antibiotic resistance even to vancomycin and teicoplanin. These new evolutions of the MRSA bacterium have been dubbed Vancomycin intermediate-resistant Staphylococcus aureus (VISA).[95] [96] Linezolid, quinupristin/dalfopristin, daptomycin, ceftaroline, and tigecycline are used to treat more severe infections that do not respond to glycopeptides such as vancomycin.[97] Current guidelines recommend daptomycin for VISA bloodstream infections and endocarditis. [98]
There have been claims that bacteriophage can be used to cure MRSA.[99][100]
The psychedelic mushroom Psilocybe semilanceata has been shown to strongly inhibit the growth of Staphylococcus aureus.[101]
Initial studies at the University of East London have demonstrated that allicin (a compound found in garlic) exhibits a strong antimicrobial response to the bacteria, indicating that it may one day lead to more effective treatments.[102]
A report released in 2010 details the efficacy of the active ingredients of a new composite dressing (hydrogen peroxide, tobramycin, chlorhexidine digluconate, chlorhexidine gluconate, levofloxacin, and silver) against MRSA.[103]
A 1990 study tested MRSA isolates obtained from veterans and found they could be killed by several substances, including bacitracin, nitrofurantoin, hydrogen peroxide, novobiocin, netilmicin and vancomycin. The study went on to conclude that netilmicin might be useful as an alternative to intravenous vancomycin, and suggested that topical applications of hydrogen peroxide may be useful to reduce MRSA on skin and some mucous membranes.[104]
History
The examples and perspective in this article may not represent a worldwide view of the subject. (December 2010) |
US and UK

In 1959 methicillin was licensed in England to treat penicillin-resistant S. aureus infections. Just as bacterial evolution had allowed microbes to develop resistance to penicillin, strains of S. aureus evolved to become resistant to methicillin. In 1961 the first MRSA isolates were reported in a British study, and between 1961-1967 there were infrequent hospital outbreaks in Western Europe and Australia.[105] The first United States hospital outbreak of MRSA occurred at the Boston City Hospital in 1968. Between 1968-mid-1990s the percent of S. aureus infections that were caused by MRSA increased steadily, and MRSA became recognized as an endemic pathogen. In 1974 2% of hospital-acquired S. aureus infections could be attributed to MRSA.[106] The rate had increased to 22% by 1995, and by 1997 the percent of hospital S. aureus infections attributable to MRSA had reached 50%.
The first report of CA-MRSA occurred in 1981, and in 1982 there was a large outbreak of CA-MRSA among intravenous drug users in Detroit, Michigan.[105] Additional outbreaks of CA-MRSA were reported through the 1980s and 1990s, including outbreaks among Australian Aboriginal populations that had never been exposed to hospitals. In the mid-1990s there were scattered reports of CA-MRSA outbreaks among US children. While HA-MRSA rates stabilized between 1998–2008, CA-MRSA rates continued to rise. A report released by The University of Chicago Children's Hospital comparing two time periods (1993–1995 and 1995–1997) found a 25-fold increase in the rate of hospitalizations due to MRSA among children in the United States.[107] In 1999 The University of Chicago reported the first deaths from invasive MRSA among otherwise healthy children in the United States.[105] By 2004 MRSA accounted for 64% of hospital-acquired S. aureus infections in the United States.
The Office for National Statistics reported 1,629 MRSA-related deaths in England and Wales during 2005, indicating a MRSA-related mortality rate half the rate of that in the United States for 2005, even though the figures from the British source were explained to be high because of "improved levels of reporting, possibly brought about by the continued high public profile of the disease"[108] during the time of the 2005 United Kingdom General Election. MRSA is thought to have caused 1,652 deaths in 2006 in UK up from 51 in 1993.[109]
It has been argued that the observed increased mortality among MRSA-infected patients may be the result of the increased underlying morbidity of these patients. Several studies, however, including one by Blot and colleagues, that have adjusted for underlying disease still found MRSA bacteremia to have a higher attributable mortality than methicillin-susceptible S. aureus (MSSA) bacteremia.[110]
A population-based study of the incidence of MRSA infections in San Francisco during 2004–05 demonstrated that nearly 1 in 300 residents suffered from such an infection in the course of a year and that greater than 85% of these infections occurred outside of the healthcare setting.[111] A 2004 study showed that patients in the United States with S. aureus infection had, on average, three times the length of hospital stay (14.3 vs. 4.5 days), incurred three times the total cost ($48,824 vs $14,141), and experienced five times the risk of in-hospital death (11.2% vs 2.3%) than patients without this infection.[112] In a meta-analysis of 31 studies, Cosgrove et al.,[113] concluded that MRSA bacteremia is associated with increased mortality as compared with MSSA bacteremia (odds ratio = 1.93; 95% CI = 1.93±0.39).[114] In addition, Wyllie et al. report a death rate of 34% within 30 days among patients infected with MRSA, a rate similar to the death rate of 27% seen among MSSA-infected patients.[115]
According to the CDC, the most recent estimates of the incidence of healthcare-associated infections that are attributable to MRSA in the United States indicate a decline in such infection rates. Incidence of MRSA central line-associated blood stream infections as reported by hundreds of intensive care units decreased 50-70% from 2001-2007.[106] A separate system tracking all hospital MRSA bloodstream infections found an overall 34% decrease between 2005-2008.[106]
MRSA is sometimes sub-categorised as community-acquired MRSA (CA-MRSA) or healthcare-associated MRSA (HA-MRSA), although the distinction is complex. Some researchers have defined CA-MRSA by the characteristics of patients whom it infects, while others define it by the genetic characteristics of the bacteria themselves. By 2005, identified CA-MRSA risk factors included athletes, military recruits, incarcerated people, emergency room patients, urban children, HIV-positive individuals, men who have sex with men, and indigenous populations.[105]
Worldwide
The first reported cases of CA-MRSA began to appear in the mid-1990s in Australia, New Zealand, the United States, the United Kingdom, France, Finland, Canada and Samoa, and were notable because they involved people who had not been exposed to a healthcare setting.[5]
Because measurement and reporting varies, it is difficult to compare rates of MRSA in different countries. An international comparison of 2004 MRSA-attributable S. aureus rates in middle and high income countries released by the Center For Disease Dynamics, Economics, and Policy in showed that Iceland had the lowest rate of infection, and Romania had the highest at over 70%.[116]
Research
Clinical
It has been reported that maggot therapy to clean out necrotic tissue of MRSA infection has been successful. Studies in diabetic patients reported significantly shorter treatment times than those achieved with standard treatments.[117][118][119]
Many antibiotics against MRSA are in phase II and phase III clinical trials. e.g.:
- Phase III : ceftobiprole, Ceftaroline, Dalbavancin, Telavancin, Aurograb, torezolid, iclaprim etc.
- Phase II : nemonoxacin.[120]
Pre-clinical
An entirely different and promising approach is phage therapy (e.g., at the Eliava Institute in Georgia[121]), which in mice had a reported efficacy against up to 95% of tested Staphylococcus isolates.[122]
On May 18, 2006, a report in Nature identified a new antibiotic, called platensimycin, that had demonstrated successful use against MRSA.[123][124]
A 2010 study noted significant antimicrobial action of Ulmo 90 and manuka UMF 25+ honey against several microorganisms, including MRSA. The investigators noted the superior antimicrobial action of Ulmo 90 honey, and suggested it be investigated further.[125] A separate 2010 study examined the use of medical-grade honey against several antibiotic-resistant strains of bacteria, including MRSA. The study concluded that the antimicrobial action of the honey studied was due to the activity of hydrogen peroxide, methylglyoxal, and a novel compound named bee defensin-1.[126]
Ocean-dwelling living sponges produce compounds that may make MRSA more susceptible to antibiotics.[127]
Some semi-toxic fungi/mushrooms excrete broad spectrum antibiotics, not all of which have been fully identified.[128]
Cannabinoids (components of Cannabis sativa), including cannabidiol (CBD), cannabinol (CBN), cannabichromene (CBC), tetrahydrocannabinol (THC) and cannabigerol (CBG), show activity against a variety of MRSA strains.[129]
See also
- Carbapenem resistant enterobacteriaceae
- Necrotizing fasciitis
- Staphylococcus aureus
- Toxic shock syndrome
Further reading
There are several web-based resources available for further research and treatment options.
- The Center for Disease Control maintains MRSA pages that provide information surrounding the bacteria, including prevention, statistics, at risk groups, causes, educational resources, and environmental factors.
- The National Institute for Occupational Safety and Health maintains a webpage providing information on the bacteria, with respect to the workplace setting, and steps towards mitigating risk from MRSA infection.
- The National Institutes of Health maintains Medline Plus, a site for patients that provides information surrounding diseases, their causes, and treatments. Their MRSA pages provide research and information surrounding causes and treatment options.
- The Mayo Clinic maintains an online site, updated by Mayo Clinic staff, that covers the basic definition, symptoms, and patient education for MRSA.
- The online medical resource site, WebMD, maintains on MRSA that provide basic information about the bacteria, as well as some multimedia resources for patient education.
- MRSA MD is an online medical site devoted solely to MRSA education, treatment, and prevention. Maintained by Dr. Kirk Bortel, MRSA MD provides treatment information and prevention education.
References
- ^ Study at the Veterans Affairs hospital in Pittsburgh: "Science Daily". [dead link]
- ^ McCaughey B. "Unnecessary Deaths: The Human and Financial Costs of Hospital Infections" (PDF) (2nd. ed.). Archived from the original (PDF) on July 11, 2007. Retrieved 2007-08-05.
- ^ "Symptoms". Mayo Clinic.
- ^ "MRSA Toxin Acquitted: Study Clears Suspected Key to Severe Bacterial Illness". NIH news release. National Institute of Health. 2006-11-06.
- ^ a b Raygada JL and Levine DP (March 30, 2009). "Managing CA-MRSA Infections: Current and Emerging Options". Infections in Medicine. 26 (2).
- ^ Lipsky BA, Tabak YP, Johannes RS, Vo L, Hyde L, Weigelt JA (2010). "Skin and soft tissue infections in hospitalised patients with diabetes: culture isolates and risk factors associated with mortality, length of stay and cost". Diabetologia. 53 (5): 914–23. doi:10.1007/s00125-010-1672-5. PMID 20146051.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c "MRSA Infections: People at Risk of Acquiring MRSA Infections". Centers for Disease Control and Prevention. 9 August 2010. Retrieved 13 May 2012.
- ^ a b c Tacconelli, E.; De Angelis, G.; Cataldo, MA.; Pozzi, E.; Cauda, R. (2008). "Does antibiotic exposure increase the risk of methicillin-resistant Staphylococcus aureus (MRSA) isolation? A systematic review and meta-analysis". J Antimicrob Chemother. 61 (1): 26–38. doi:10.1093/jac/dkm416. PMID 17986491.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Reuters (2009-02-16). "Study: Beachgoers More Likely to Catch MRSA". FoxNews.com.
{{cite news}}:|author=has generic name (help) - ^ Marilynn Marchione (2009-09-12). "Dangerous staph germs found at West Coast beaches". AP.
- ^ Zinderman, C.; Conner, B.; Malakooti, M.; LaMar, J.; Armstrong, A.; Bohnker, A. (2004). "Community-Acquired Methicillin-Resistant Staphylococcus aureus Among Military Recruits". Emerging Infectious Diseases. 10 (5): 941–4. doi:10.3201/eid1005.030604. PMC 3323224. PMID 15200838.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d David, Micheal Z. and Daum, Robert S. (2010). "Community-Associated Methicillin-Resistant Staphylococcus aureus: Epidemiology and Clinical Consequences of an Emerging Epidemic". Clinical microbiology reviews. 23 (6). American Society for Microbiology: 616–687. doi:10.1128/CMR.00081-09. PMC 2901661. PMID 20610826.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Muto, CA.; Jernigan, JA.; Ostrowsky, BE.; Richet, HM.; Jarvis, WR.; Boyce, JM.; Farr, BM. (2003). "SHEA guideline for preventing nosocomial transmission of multidrug-resistant strains of Staphylococcus aureus and enterococcus". Infect Control Hosp Epidemiol. 24 (5): 362–86. doi:10.1086/502213. PMID 12785411.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Staph (MRSA) Infection Eradicated For 14 Months
- ^ "Joint scientific report of ECDC, EFSA and EMEA on meticillin resistant Staphylococcus aureus (MRSA) in livestock, companion animals and food" (PDF). 2009-06-16. Retrieved 2009-09-19.
- ^ US meat and poultry is widely contaminated with drug-resistant Staph bacteria, study finds
- ^ Ogata, K (2012-02-03). "Commercially distributed meat as a potential vehicle for community-acquired methicillin-resistant Staphylococcus aureus". Applied and environmental microbiology. 78 (8): 2797–802. doi:10.1128/AEM.07470-11. PMC 3318828. PMID 22307310.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kazakova, SV (2005-02-03). "A clone of methicillin-resistant Staphylococcus aureus among professional football players". The New England Journal of Medicine. 352 (5): 468–75. doi:10.1056/NEJMoa042859. PMID 15689585.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Epstein, Victor (21 December 2007). "Texas Football Succumbs to Virulent Staph Infection From Turf". Bloomberg. Retrieved 10 June 2010.
- ^ Gray JW (2004). "MRSA: the problem reaches paediatrics". Arch. Dis. Child. 89 (4): 297–8. doi:10.1136/adc.2003.045534. PMC 1719885. PMID 15033832.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Bratu S, Eramo A, Kopec R; et al. (2005). "Community-associated methicillin-resistant Staphylococcus aureus in hospital nursery and maternity units". Emerging Infect. Dis. 11 (6): 808–13. doi:10.3201/eid1106.040885. PMC 3367583. PMID 15963273.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^
Association for Professionals in Infection Control & Epidemiology (June 25, 2007). "National Prevalence Study of Methicillin-Resistant Staphylococcus aureus (MRSA) in U.S. Healthcare Facilities". Archived from the original on September 7, 2007. Retrieved 2007-07-14.
{{cite web}}: External link in|author= - ^ Web MD
- ^ Francois P and Schrenzel J (2008). "Rapid Diagnosis and Typing of Staphylococcus aureus". Staphylococcus: Molecular Genetics. Caister Academic Press. ISBN 978-1-904455-29-5.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Mackay I M (editor). (2007). Real-Time PCR in Microbiology: From Diagnosis to Characterization. Caister Academic Press. ISBN 978-1-904455-18-9.
{{cite book}}:|author=has generic name (help) - ^ Seiken, Denka. "MRSA latex test for PBP2".
- ^ a b c d e Jensen SO, Lyon BR (2009). "Genetics of antimicrobial resistance in Staphylococcus aureus". Future Microbiol. 4 (5): 565–82. doi:10.2217/fmb.09.30. PMID 19492967.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d e f g h i Lowy FD (2003). "Antimicrobial resistance: the example of Staphylococcus aureus". J. Clin. Invest. 111 (9): 1265–73. doi:10.1172/JCI18535. PMC 154455. PMID 12727914.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d e f Pantosti A, Sanchini A, Monaco M (2007). "Mechanisms of antibiotic resistance in Staphylococcus aureus". Future Microbiol. 2 (3): 323–34. doi:10.2217/17460913.2.3.323. PMID 17661706.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kaito C, Saito Y, Nagano G; et al. (2011). Cheung, Ambrose (ed.). "Transcription and translation products of the cytolysin gene psm-mec on the mobile genetic element SCCmec regulate Staphylococcus aureus virulence". PLoS Pathog. 7 (2): e1001267. doi:10.1371/journal.ppat.1001267. PMC 3033363. PMID 21304931.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c Kuo SC, Chiang MC, Lee WS; et al. (2012). "Comparison of microbiological and clinical characteristics based on SCCmec typing in patients with community-onset meticillin-resistant Staphylococcus aureus (MRSA) bacteraemia". Int. J. Antimicrob. Agents. 39 (1): 22–6. doi:10.1016/j.ijantimicag.2011.08.014. PMID 21982834.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Sahebnasagh R, Saderi H, Owlia P. Detection of methicillin-resistant Staphylococcus aureus strains from clinical samples in Tehran by detection of the mecA and nuc genes. The First Iranian International Congress of Medical Bacteriology; 4-7 September; Tabriz, Iran. 2011. 195 pp.
- ^ a b Berger-Bächi B (1999). "Genetic basis of methicillin resistance in Staphylococcus aureus". Cell. Mol. Life Sci. 56 (9–10): 764–70. doi:10.1007/s000180050023. PMID 11212336.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d e f g Gordon RJ, Lowy FD (2008). "Pathogenesis of methicillin-resistant Staphylococcus aureus infection". Clin. Infect. Dis. 46 (Suppl 5): S350–9. doi:10.1086/533591. PMC 2474459. PMID 18462090.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Johnson AP, Aucken HM, Cavendish S; et al. (2001). "Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS)". J Antimicrob Chemother. 48 (1): 143–4. doi:10.1093/jac/48.1.143. PMID 11418528.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Holden MTG, Feil EJ, Lindsay JA; et al. (2004). "Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance". Proc Natl Acad Sci USA. 101 (26): 9786–91. doi:10.1073/pnas.0402521101. PMC 470752. PMID 15213324.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b Diep B, Carleton H, Chang R, Sensabaugh G, Perdreau-Remington F (2006). "Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus". J Infect Dis. 193 (11): 1495–503. doi:10.1086/503777. PMID 16652276.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Stefani, Stefania; Chung, Doo Ryeon; Lindsay, Jodi A.; Friedrich, Alex W.; Kearns, Angela M.; Westh, Henrik; MacKenzie, Fiona M. (2012). "Meticillin-resistant Staphylococcus aureus (MRSA): global epidemiology and harmonisation of typing methods". International Journal of Antimicrobial Agents. 39 (4): 273–82. doi:10.1016/j.ijantimicag.2011.09.030. ISSN 0924-8579. PMID 22230333.
- ^ a b c d e f Calfee DP (2011). "The epidemiology, treatment, and prevention of transmission of methicillin-resistant Staphylococcus aureus". J Infus Nurs. 34 (6): 359–64. doi:10.1097/NAN.0b013e31823061d6. PMID 22101629.
- ^ von Eiff C, Becker K, Metze D; et al. (2001). "Intracellular persistence of Staphylococcus aureus small-colony variants within keratinocytes: a cause for antibiotic treatment failure in a patient with Darier's disease". Clin Infect Dis. 32 (11): 1643–7. doi:10.1086/320519. PMID 11340539.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Clement S, Vaudaux P, François P; et al. (2005). "Evidence of an intracellular reservoir in the nasal mucosa of patients with recurrent Staphylococcus aureus rhinosinusitis". J Infect Dis. 192 (6): 1023–8. doi:10.1086/432735. PMID 16107955.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Zautner AE, Krause M, Stropahl G; et al. (2010). Bereswill, Stefan (ed.). "Intracellular persisting Staphylococcus aureus is the major pathogen in recurrent tonsillitis". PloS One. 5 (3): e9452. doi:10.1371/journal.pone.0009452. PMC 2830486. PMID 20209109.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ "New Study finds MRSA on the rise in hospital outpatients". 2009.
- ^ Daum, Robert S. (2007). "Skin and Soft-Tissue Infections Caused by Methicillin-Resistant Staphylococcus aureus". New England Journal of Medicine. 357 (4): 380–390. doi:10.1056/NEJMcp070747. PMID 17652653.
- ^ Wang R, Braughton KR, Kretschmer D; et al. (2007). "Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA". Nat. Med. 13 (12): 1510–4. doi:10.1038/nm1656. PMID 17994102.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Gould IM, David MZ, Esposito S; et al. (2012). "New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance". Int. J. Antimicrob. Agents. 39 (2): 96–104. doi:10.1016/j.ijantimicag.2011.09.028. PMID 22196394.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ahlstrom, Dick (2011-06-03). "New strain of MRSA superbug discovered in Dublin hospitals". The Irish Times.
- ^ Graveland H, Duim B, van Duijkeren E, Heederik D, Wagenaar JA (2011). "Livestock-associated methicillin-resistant Staphylococcus aureus in animals and humans". Int. J. Med. Microbiol. 301 (8): 630–4. doi:10.1016/j.ijmm.2011.09.004. PMID 21983338.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Tacconelli E, De Angelis G, de Waure C, Cataldo MA, La Torre G, Cauda R (2009). "Rapid screening tests for meticillin-resistant Staphylococcus aureus at hospital admission: systematic review and meta-analysis". Lancet Infect Dis. 9 (9): 546–54. doi:10.1016/S1473-3099(09)70150-1. PMID 19695491.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "To Catch a Deadly Germ," New York Times opinion
- ^ CDC Guideline "Management of Multidrug-Resistant Organisms in Healthcare Settings, 2006"
- ^ http://www.halifaxcourier.co.uk/latest-york-and-humberside-news/New-checks-in-hospitals-to.5123093.jp
- ^ "MRSA test for surgical patients". BBC News. 2009-03-31. Retrieved 2010-04-05.
- ^ Home Mrsa Test - Home
- ^ Fritz SA, Garbutt J, Elward A; et al. (2008). "Prevalence of and risk factors for community-acquired methicillin-resistant and methicillin-sensitive Staphylococcus aureus colonization in children seen in a practice-based research network". Pediatrics. 121 (6): 1090–8. doi:10.1542/peds.2007-2104. PMID 18519477.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Otter JA, Puchowicz M, Ryan D; et al. (2009). "Feasibility of routinely using hydrogen peroxide vapor to decontaminate rooms in a busy United States hospital". Infect Control Hosp Epidemiol. 30 (6): 574–7. doi:10.1086/597544. PMID 19415969.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bartels MD, Kristoffersen K, Slotsbjerg T, Rohde SM, Lundgren B, Westh H (2008). "Environmental meticillin-resistant Staphylococcus aureus (MRSA) disinfection using dry-mist-generated hydrogen peroxide". J. Hosp. Infect. 70 (1): 35–41. doi:10.1016/j.jhin.2008.05.018. PMID 18621434.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ French GL, Otter JA, Shannon KP, Adams NM, Watling D, Parks MJ (2004). "Tackling contamination of the hospital environment by methicillin-resistant Staphylococcus aureus (MRSA): a comparison between conventional terminal cleaning and hydrogen peroxide vapour decontamination". J. Hosp. Infect. 57 (1): 31–7. doi:10.1016/j.jhin.2004.03.006. PMID 15142713.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Otter JA, Cummins M, Ahmad F, van Tonder C, Drabu YJ (2007). "Assessing the biological efficacy and rate of recontamination following hydrogen peroxide vapour decontamination". J. Hosp. Infect. 67 (2): 182–8. doi:10.1016/j.jhin.2007.07.019. PMID 17884250.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hardy KJ, Gossain S, Henderson N; et al. (2007). "Rapid recontamination with MRSA of the environment of an intensive care unit after decontamination with hydrogen peroxide vapour". J. Hosp. Infect. 66 (4): 360–8. doi:10.1016/j.jhin.2007.05.009. PMID 17655975.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Omidbakhsh N, Sattar SA (2006). "Broad-spectrum microbicidal activity, toxicologic assessment, and materials compatibility of a new generation of accelerated hydrogen peroxide-based environmental surface disinfectant". Am J Infect Control. 34 (5): 251–7. doi:10.1016/j.ajic.2005.06.002. PMID 16765201.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Michels, H.T.; Noyce, J.O.; Keevil, C.W. (2009). "Effects of temperature and humidity on the efficacy of methicillin-resistant Staphylococcus aureus challenged antimicrobial materials containing silver and copper" (PDF). Letters in Applied Microbiology. 49 (2): 191–5. doi:10.1111/j.1472-765X.2009.02637.x. PMC 2779462. PMID 19413757.
- ^ Keevil, C.W., Noyce, J.O. (2007), Antimicrobial Efficacies of Copper, Stainless Steel, Microban, BioCote and AgIon with MRSA at 20 °C, unpublished data
- ^ Noyce, J.O. and Keevil, C.W. (2004), The Antimicrobial Effects of Copper and Copper-Based Alloys on Methicillin-resistant Staphylococcus aureus, Copper Development Association Poster Q-193 from Proceedings of the Annual General Meeting of the American Society for Microbiology, 24–27 May 2004, New Orleans; presented at the American Society for Microbiology General Meeting, New Orleans, LA May 24th.
- ^ "NPSA - About us".
- ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1128/AAC.00430-07, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1128/AAC.00430-07instead. - ^ Haley CE, Marling-Cason M, Smith JW, Luby JP, Mackowiak PA (1985). "Bactericidal activity of antiseptics against methicillin-resistant Staphylococcus aureus". J. Clin. Microbiol. 21 (6): 991–2. PMC 271835. PMID 4008627.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Gever J.Senior Editor, MedPage Today. Germicidal Surgical Mask Approved Published: April 09, 2011 http://www.medpagetoday.com/HospitalBasedMedicine/InfectionControl/25819
- ^ "Simple techniques slash hospital infections: meeting". Reuters. 2009-03-21.
- ^ a b c "NIOSH MRSA and the Workplace". United States National Institute for Occupational Safety and Health. Retrieved 2007-10-29.
- ^ CDC (1998). "Guidelines for Infection Control in Health Care Personnel, 1998". Centers for Disease Control and Prevention. Retrieved December 18, 2007.
- ^ Cooper BS, Medley GF, Stone SP; et al. (2004). "Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes". Proceedings of the National Academy of Sciences. 101 (27): 10223–8. doi:10.1073/pnas.0401324101. PMC 454191. PMID 15220470.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|unused_data=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bootsma MC, Diekmann O, Bonten MJ (2006). "Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing". Proc Natl Acad Sci USA. 103 (14): 5620–5. doi:10.1073/pnas.0510077103. PMC 1459403. PMID 16565219.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Klevens RM, Edwards JR, Richards CL; et al. (2007). "Estimating health care-associated infections and deaths in U.S. hospitals, 2002". Public Health Rep. 122 (2): 160–6. PMC 1820440. PMID 17357358.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Hidron AI, Edwards JR, Patel J; et al. (2008). "NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007". Infect Control Hosp Epidemiol. 29 (11): 996–1011. doi:10.1086/591861. PMID 18947320.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Johnson AP, Pearson A, Duckworth G (2005). "Surveillance and epidemiology of MRSA bacteraemia in the UK". J Antimicrob Chemother. 56 (3): 455–62. doi:10.1093/jac/dki266. PMID 16046464.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Inquirer.net, Cases of RP maids with 'superbug' infection growing in HK
- ^ "MRSA Infections". Keep Kids Healthy.
- ^ Graham P, Lin S, Larson E (2006). "A U.S. population-based survey of Staphylococcus aureus colonization". Ann Intern Med. 144 (5): 318–25. doi:10.7326/0003-4819-144-5-200603070-00006. PMID 16520472.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jernigan JA, Arnold K, Heilpern K, Kainer M, Woods C, Hughes JM (2006-05-12). "Methicillin-resistant Staphylococcus aureus as community pathogen". Symposium on Community-Associated Methicillin-resistant Staphylococcus aureus (Atlanta, Georgia, U.S.). Cited in Emerg Infect Dis. Centers for Disease Control and Prevention. Retrieved 2007-01-27.
{{cite conference}}: Unknown parameter|booktitle=ignored (|book-title=suggested) (help)CS1 maint: multiple names: authors list (link) - ^ First study finds MRSA in U.S. pigs and farmers, seattlepi.com, 4 June 2008
- ^ Our Pigs, Our Food, Our Health, The New York Times, 12 March 2009
- ^ a b "PubMed Health". US National Institutes of Health. Retrieved 20 November 2011.
- ^ "Optimal Bleach Concentration Required to Kill MRSA in Bath Water". American Academy of Pediatrics.
{{cite web}}: Missing or empty|url=(help) - ^ a b "Living With MRSA" (PDF). Group Health Cooperative/Tacoma-Pierce County Health Dept./Washington State Dept. of Health. Retrieved 20 November 2011.
- ^ Shi CM, Nakao H, Yamazaki M, Tsuboi R, Ogawa H (2007). "Mixture of sugar and povidone-iodine stimulates healing of MRSA-infected skin ulcers on db/db mice". Arch. Dermatol. Res. 299 (9): 449–56. doi:10.1007/s00403-007-0776-3. PMID 17680256.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "THE NOSE – "GROUND ZERO" FOR MRSA COLONIZATION". Ondine Biomedical Inc. Retrieved 20 November 2011.
- ^ "Personal Prevention of MRSA Skin Infections". CDC. 9 August 2010.
- ^ Wunderink RG, Rello J, Cammarata SK, Croos-Dabrera RV, Kollef MH (2003). "Linezolid vs vancomycin: analysis of two double-blind studies of patients with methicillin-resistant Staphylococcus aureus nosocomial pneumonia". Chest. 124 (5): 1789–97. doi:10.1378/chest.124.5.1789. PMID 14605050.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Schentag JJ, Hyatt JM, Carr JR, Paladino JA, Birmingham MC, Zimmer GS, Cumbo TJ (1998). "Genesis of methicillin-resistant Staphylococcus aureus (MRSA), how treatment of MRSA infections has selected for vancomycin-resistant Enterococcus faecium, and the importance of antibiotic management and infection control". Clin. Infect. Dis. 26 (5): 1204–14. doi:10.1086/520287. PMID 9597254.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rybak MJ, Lerner SA, Levine DP, Albrecht LM, McNeil PL, Thompson GA, Kenny MT, Yuh L (1991). "Teicoplanin pharmacokinetics in intravenous drug abusers being treated for bacterial endocarditis". Antimicrob. Agents Chemother. 35 (4): 696–700. doi:10.1128/AAC.35.4.696. PMC 245081. PMID 1829880.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Janknegt R (1997). "The treatment of staphylococcal infections with special reference to pharmacokinetic, pharmacodynamic, and pharmacoeconomic considerations". Pharmacy world & science : PWS. 19 (3): 133–41. doi:10.1023/A:1008609718457. PMID 9259029.
- ^ Chang FY, Peacock JE Jr, Musher DM; et al. (2003). "Staphylococcus aureus bacteremia: recurrence and the impact of antibiotic treatment in a prospective multicenter study". Medicine (Baltimore). 82 (5): 333–9. doi:10.1097/01.md.0000091184.93122.09. PMID 14530782.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Siegman-Igra Y, Reich P, Orni-Wasserlauf R, Schwartz D, Giladi M. (2005). "The role of vancomycin in the persistence or recurrence of Staphylococcus aureus bacteraemia". Scand J Infect Dis. 37 (8): 572–8. doi:10.1080/00365540510038488. PMID 16138425.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sieradzki K, Tomasz A (1997). "Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus". J. Bacteriol. 179 (8): 2557–66. PMC 179004. PMID 9098053.
- ^ Schito GC (2006). "The importance of the development of antibiotic resistance in Staphylococcus aureus". Clin Microbiol Infect. 12 (Suppl 1): 3–8. doi:10.1111/j.1469-0691.2006.01343.x. PMID 16445718.
- ^ Mongkolrattanothai K, Boyle S, Kahana MD, Daum RS (2003). "Severe Staphylococcus aureus infections caused by clonally related community-associated methicillin-susceptible and methicillin-resistant isolates". Clin. Infect. Dis. 37 (8): 1050–8. doi:10.1086/378277. PMID 14523769.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, J Rybak M, Talan DA, Chambers HF. (2011 Feb 1). "Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary". Clin Infect Dis. 52 (3): 285–92. doi:10.1093/cid/cir034. PMID 21217178.
{{cite journal}}: Check date values in:|date=(help); More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) - ^ Phage International - Subsidiaries - Phage Therapy Center
- ^ "Wired 11.10: How Ravenous Soviet Viruses Will Save the World". Wired.
- ^ Suay I, Arenal F, Asensio FJ, Basilio A, Cabello MA, Díez MT, García JB, González del Val A, Gorrochategui J, Hernández P, Peláez F, Vicente MF. (2000). "Screening of basidiomycetes for antimicrobial activities" (PDF). Antonie van Leeuwenhoek. 78 (2): 129–39. doi:10.1023/A:1026552024021. PMID 11204765.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cutler R.R.; Wilson, P (2004). "Antibacterial activity of a new, stable, aqueous extract of allicin against methicillin-resistant Staphylococcus aureus". British journal of biomedical science. 61 (2): 71–4. PMID 15250668.
- ^ Echague CG, Hair PS, Cunnion KM (2010). "A comparison of antibacterial activity against Methicillin-Resistant Staphylococcus aureus and gram-negative organisms for antimicrobial compounds in a unique composite wound dressing". Adv Skin Wound Care. 23 (9): 406–13. doi:10.1097/01.ASW.0000383213.95911.bc. PMID 20729646.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Flournoy DJ, Robinson MC (1990). "In vitro antimicrobial susceptibilities of 349 methicillin-resistant Staphylococcus aureus isolates from veterans". Methods Find Exp Clin Pharmacol. 12 (8): 541–4. PMID 2093132.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d "MRSA History Timeline: The First Half-Century, 1959-2009". The University of Chicago Medical Center. 2010.
- ^ a b c "MRSA Surveillance". Centers for Disease Control and Prevention. April 8, 2011.
- ^ http://mrsa-research-center.bsd.uchicago.edu/JAMA_02_25_1998_issue.pdf
- ^ UK Office for National Statistics Online (February 22, 2007), "MRSA Deaths continue to rise in 2005"
- ^ Hospitals struck by new killer bug An article by Manchester free newspaper 'Metro', May 7, 2008
- ^ Blot S, Vandewoude K, Hoste E, Colardyn F (2002). "Outcome and attributable mortality in critically Ill patients with bacteremia involving methicillin-susceptible and methicillin-resistant Staphylococcus aureus". Arch Intern Med. 162 (19): 2229–35. doi:10.1001/archinte.162.19.2229. PMID 12390067.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Liu C, Graber CJ, Karr M; et al. (2008). "A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005". Clin. Infect. Dis. 46 (11): 1637–46. doi:10.1086/587893. PMID 18433335.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Noskin GA, Rubin RJ, Schentag JJ, Kluytmans J, Hedblom EC, Smulders M, Lapetina E, Gemmen E (2005). "The Burden of Staphylococcus aureus Infections on Hospitals in the United States: An Analysis of the 2000 and 2001 Nationwide Inpatient Sample Database". Arch Intern Med. 165 (15): 1756–1761. doi:10.1001/archinte.165.15.1756. PMID 16087824.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y (2005). "The impact of Methicillin Resistance in Staphylococcus aureus Bacteremia on Patient Outcomes: Mortality, Length of Stay, and Hospital Charges" (– Scholar search). Infection Control and Hospital Epidemiology. 26 (2): 166–174. doi:10.1086/502522. PMID 15756888.
{{cite journal}}: External link in|format= - ^ Hardy KJ, Hawkey PM, Gao F, Oppenheim BA (2004). "Methicillin resistant Staphylococcus aureus in the critically ill". British Journal of Anaesthesia. 92 (1): 121–30. doi:10.1093/bja/aeh008. PMID 14665563.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wyllie D, Crook D, Peto T (2006). "Mortality after Staphylococcus aureus bacteraemia in two hospitals in Oxfordshire, 1997–2003: cohort study". BMJ. 333 (7562): 281. doi:10.1136/bmj.38834.421713.2F. PMC 1526943. PMID 16798756.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ CDDEP "MRSA Infection Rates by Country" (2004)
- ^ Bowling FL, Salgami EV, Boulton AJ (2007). "Larval therapy: a novel treatment in eliminating methicillin-resistant Staphylococcus aureus from diabetic foot ulcers". Diabetes Care. 30 (2): 370–1. doi:10.2337/dc06-2348. PMID 17259512.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Maggots help cure MRSA patients". BBC News. 2007-05-02.
- ^ "Maggots rid patients of MRSA". EurekAlert!/AAAS. 2007-05-03.
- ^ Clinical trial number NCT00685698 for "Safety and Efficacy Study of TG-873870 (Nemonoxacin) in Diabetic Foot Infections" at ClinicalTrials.gov
- ^ Murphy, Clare (2007-08-13). "'Red Army' virus to combat MRSA". BBC News.
- ^ Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S (2003). "Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11". J. Infect. Dis. 187 (4): 613–24. doi:10.1086/374001. PMID 12599078.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bayston R, Ashraf W, Smith T (2007). "Triclosan resistance in methicillin-resistant Staphylococcus aureus expressed as small colony variants: a novel mode of evasion of susceptibility to antiseptics". J. Antimicrob. Chemother. 59 (5): 848–53. doi:10.1093/jac/dkm031. PMID 17337510.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang J; Soisson, SM; Young, K; Shoop, W; Kodali, S; Galgoci, A; Painter, R; Parthasarathy, G; Tang, YS (2006). "Platensimycin is a selective FabF inhibitor with potent antibiotic properties". Nature. 441 (7091): 358–361. doi:10.1038/nature04784. PMID 16710421.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Sherlock O, Dolan A, Athman R; et al. (2010). "Comparison of the antimicrobial activity of Ulmo honey from Chile and Manuka honey against methicillin-resistant Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa". BMC Complement Altern Med. 10: 47. doi:10.1186/1472-6882-10-47. PMC 2942791. PMID 20813024.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Kwakman PH, te Velde AA, de Boer L, Speijer D, Vandenbroucke-Grauls CM, Zaat SA (2010). "How honey kills bacteria". FASEB J. 24 (7): 2576–82. doi:10.1096/fj.09-150789. PMID 20228250.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Sponge's secret weapon restores antibiotics' power
- ^ Psilocybe_semilanceata#Ecology_and_habitat
- ^ Appendino G, Gibbons S, Giana A, Pagani A, Grassi G, Stavri M, Smith E, Rahman M (2008). "Antibacterial Cannabinoids from Cannabis sativa: A Structure-Activity Study". J. Nat. Prod. 71 (8): 1427–30. doi:10.1021/np8002673. PMID 18681481.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
