Lipopolysaccharide

Lipopolysaccharides (LPS), also known as lipoglycans, are large molecules consisting of a lipid and a polysaccharide joined by a covalent bond; they are found in the outer membrane of Gram-negative bacteria, act as endotoxins and elicit strong immune responses in animals.
Functions
LPS is the major component of the outer membrane of Gram-negative bacteria, contributing greatly to the structural integrity of the bacteria, and protecting the membrane from certain kinds of chemical attack. LPS also increases the negative charge of the cell membrane and helps stabilize the overall membrane structure. It is of crucial importance to gram negative bacteria, whose death results if it is mutated or removed. LPS is an endotoxin, and induces a strong response from normal animal immune systems.
LPS acts as the prototypical endotoxin because it binds the CD14/TLR4/MD2 receptor complex, which promotes the secretion of pro-inflammatory cytokines in many cell types, but especially in macrophages. In Immunology, the term "LPS challenge" refers to the process of exposing a subject to an LPS which may act as a toxin.
LPS is also an exogenous pyrogen (external fever-inducing substance).
Being of crucial importance to gram negative bacteria, these molecules make candidate targets for new antimicrobial agents.
Some researchers doubt reports of generalized toxic effects attributed to all lipopolysaccharides, particularly for cyanobacteria.[1]
Composition
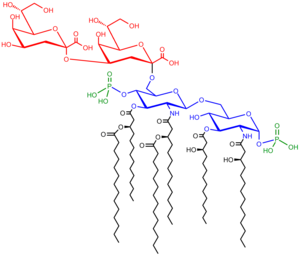
It comprises three parts:
O-antigen
When LPS contains a repetitive glycan polymer this is referred to as the O antigen, O polysaccharide, or O side chain of the bacteria. The O antigen is attached to the core oligosaccharide, and comprises the outermost domain of the LPS molecule. The composition of the O chain varies from strain to strain, for example there are over 160 different O antigen structures produced by different E. coli strains.[2] The presence or absence of O chains determine whether the LPS is considered rough or smooth. Full length O-chains would render the LPS smooth while the absence or reduction of O-chains would make the LPS rough.[3] Bacteria with rough LPS usually have more penetrable cell membranes to hydrophobic antibiotics since a rough LPS is more hydrophobic.[4] O antigen is exposed on the very outer surface of the bacterial cell, and as a consequence, is a target for recognition by host antibodies.
Core
The Core domain always contains an oligosaccharide component which attaches directly to lipid A and commonly contains sugars such as heptose and 3-deoxy-D-mannooctulosonic Acid (also known as KDO, keto-deoxyoctulosonate).[5] The LPS Cores of many bacteria also contain non-carbohydrate components, such as phosphate, amino acids, and ethanolamine substitutents.
Lipid A
Lipid A is normally a phosphorylated glucosamine disaccharide decorated with multiple fatty acids. These hydrophobic fatty acid chains anchor the LPS into the bacterial membrane and the rest of the LPS projects from the cell surface. The lipid A domain is responsible for much of the toxicity of Gram-negative bacteria. When bacterial cells are lysed by the immune system, fragments of membrane containing lipid A are released into the circulation, causing fever, diarrhea, and possible fatal endotoxic shock (also called septic shock).
Biosynthesis and Transport
LPS modifications
The making of LPS can be modified in order to present a specific sugar structure. Those can be recognised by either other LPS (which enables to inhibit LPS toxins) or glycosyltransferases which use those sugar structure to add more specific sugars. It has recently been shown that a specific enzyme in the intestine (alkaline phosphatase) can detoxify LPS by removing the two phosphate groups found on LPS carbohydrates [8]. This may function as an adaptive mechanism to help the host manage potentially toxic effects of gram-negative bacteria normally found in the small intestine.
Variability and effect upon specificity
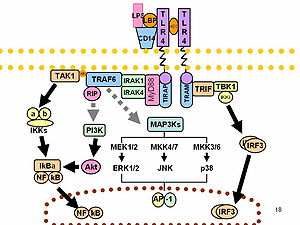
O-antigens (the outer carbohydrates) are the most variable portion of the LPS molecule, imparting the antigenic specificity. In contrast, lipid A is the most conserved part. However, —lipid A composition also may vary (e.g., in number and nature of acyl chains even within or between genera). Some of these variations may impart antagonistic properties to these LPS. For example Rhodobacter sphaeroides diphosphoryl lipid A (RsDPLA) is a potent antagonist of LPS in human cells, but is an agonist in hamster and equine cells.
It has been speculated that conical Lipid A (e.g. from E. coli) are more agonistic, less conical lipid A like those of Porphyromonas gingivalis may activate a different signal (TLR2 instead of TLR4), and completely cylindrical lipid A like that of Rhodobacter sphaeroides is antagonistic to TLRs.[9][10]
Lipopolysaccharide gene clusters are highly variable between different strains, subspecies, species of bacterial pathogens of plants and animals.[11][12]
Immune response
LPS function has been under experimental research for several years due to its role in activating many transcription factors. LPS challenge also produces many types of mediators involved in septic shock. Humans are much more sensitive to LPS than other animals (e.g., mice). A dose of 1µg/kg induces shock in humans, but mice will tolerate a dose up to a thousand times higher.[13] This may relate to differences in the level of circulating natural antibodies between the two species.[14][15]
See also
References
- ^ Stewart I, Schluter PJ, Shaw GR (2006). "Cyanobacterial lipopolysaccharides and human health - a review". Environ Health. 5: 7. doi:10.1186/1476-069X-5-7. PMC 1489932. PMID 16563160.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Christian Raetz and Chris Whitfield (2002) Lipopolysaccharide Endotoxins Annu. Rev. Biochem. 71:635-700
- ^ Rittig MG; et al. (2004). "Smooth and rough lipopolysaccharide phenotypes of Brucella induce different intracellular trafficking and cytokine/chemokine release in human monocytes". Journal of Leukocyte Biology. 5 (4): 196–200. doi:10.1189/jlb.0103015. PMID 12960272.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Tsujimoto H; et al. (2003). "Diffusion of macrolide antibiotics through the outer membrane of Moraxella catarrhalis". Journal of Infection and Chemotherapy. 74 (4): 1045–1055. doi:10.1007/s101569900025. PMID 11810516.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Hershberger C and Binkley SB (1968). "Chemistry and Metabolism of 3-Deoxy-d-mannooctulosonic Acid. I. STEREOCHEMICAL DETERMINATION". Journal of Biological Chemistry. 243 (7): 1578–1584. PMID 4296687.
- ^ Wang, Xiaoyuan and Quinn, Peter J. (2010). "Lipopolysaccharide:Biosynthetic pathway and structure modification". Progress in Lipid Research. 49: 97–107. doi:10.1016/j.plipres.2009.06.002. PMID 19815028.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ruiz, Natividad; Kahne, Daniel; Silhavy, Thomas J (2009). "Transport of lipopolysaccharide across the cell envelope: the long road of discovery". Nature Reviews Microbiology. 7 (9): 677–683. doi:10.1038/nrmicro2184. PMID 19633680.
- ^ Bates J.M.; et al. (2007). "Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota". Cell Host and Microbe. 2(6) (6): 371–382. doi:10.1016/j.chom.2007.10.010. PMC 2730374. PMID 18078689.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Netea M; et al. (2002). "Does the shape of lipid A determine the interaction of LPS with Toll-like receptors?". Trends Immunol. 23 (3): 135–9. doi:10.1016/S1471-4906(01)02169-X. PMID 11864841.
{{cite journal}}: Explicit use of et al. in:|author=(help) - ^ Seydel U, Oikawa M, Fukase K, Kusumoto S, Brandenburg K (2000). "Intrinsic conformation of lipid A is responsible for agonistic and antagonistic activity". Eur J Biochem. 267 (10): 3032–9. doi:10.1046/j.1432-1033.2000.01326.x. PMID 10806403.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Reeves P, Wang L (2002). "Genomic organization of LPS-specific loci". Curr Top Microbiol Immunol. 264 (1): 109–35. PMID 12014174.
- ^ Patil P, Sonti R (2004). "Variation suggestive of horizontal gene transfer at a lipopolysaccharide (lps) biosynthetic locus in Xanthomonas oryzae pv. oryzae, the bacterial leaf blight pathogen of rice". BMC Microbiol. 4 (1): 40. doi:10.1186/1471-2180-4-40. PMC 524487. PMID 15473911.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Resilience to bacterial infection: Difference between species could be due to proteins in serum". J Infect Dis. 201: 223–232. 2010. doi:10.1086/649557.
- ^ Reid RR, Prodeus AP, Khan W, Hsu T, Rosen FS, Carroll MC (1997). "Endotoxin shock in antibody-deficient mice: unraveling the role of natural antibody and complement in the clearance of lipopolysaccharide". J. Immunol. 159 (2): 970–5. PMID 9218618.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J (1998). "A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection". J. Exp. Med. 188 (12): 2381–6. PMC 2212438. PMID 9858525.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- Lipopolysaccharides at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

![LPS Final Assembly: O-antigen subunits are translocated across the inner membrane (by Wzx) where they are polymerized (by Wzy, chain length determined by Wzz) and ligated (by WaaL) on to complete Core-Lipid A molecules (which were translocated by MsbA).[6]](http://upload.wikimedia.org/wikipedia/commons/thumb/1/1a/LPS-Assembly.svg/120px-LPS-Assembly.svg.png)
![LPS Transport: Completed LPS molecules are transported across the periplasm and outer membrane by the proteins LptA, B, C, D, E, F, and G[7]](http://upload.wikimedia.org/wikipedia/commons/thumb/2/21/LPS-Transport.svg/120px-LPS-Transport.svg.png)