DNA nanotechnology

DNA nanotechnology is the study of the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. This use is enabled by the strict base pairing rules of nucleic acids, which cause only those portions of nucleic acid strands with complementary base sequences to bind together to form strong, rigid nucleic acid double helix molecules. This allows for the rational design of base sequences that will selectively assemble to form a desired target structure, leading to a unique ability to form designed, complex structures with precise control over nanoscale features. DNA is the dominant material used, but structures incorporating other nucleic acids such as RNA and peptide nucleic acid (PNA) have also been constructed, leading to the occasional use of the alternate name nucleic acid nanotechnology to describe the field.
The conceptual foundation for DNA nanotechnology was first laid out by Nadrian Seeman in the early 1980s, and the field began to attract widespread interest in the mid 2000s. Researchers in the field have created both static structures such as two- and three-dimensional crystal lattices, nanotubes, polyhedra, and arbitrary shapes made by the DNA origami method; and functional structures such as molecular machines and DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in crystallography and spectroscopy for protein structure determination. Potential real-world applications in molecular scale electronics and nanomedicine are also being investigated.
Fundamental concepts


Properties of nucleic acids
The goal of nanotechnology is to construct materials and devices with features on a scale less than 100 nanometers. DNA nanotechnology is an example of molecular self-assembly, a bottom-up approach to nanotechnology where pre-existing molecular components spontaneously form a larger, organized structure due to their physical and chemical properties.[3] Nucleic acids such as DNA are well-suited to nanoscale construction, as a nucleic acid double helix has a diameter of 2 nm and a helical repeat length of 3.5 nm. The key property which makes nucleic acids more useful for constructing structures than other materials is that the binding between two nucleic acid strands depends on simple base pairing rules which are well understood, and form a specific structure upon binding, making the assembly of nucleic acid structures easy to control through nucleic acid design. This property is absent in other materials used in nanotechnology, including proteins, for which protein design is very difficult, and nanoparticles, which lack the capability for specific assembly on their own.[4]
The structure of a nucleic acid molecule consists of a sequence of nucleotides distinguished by which nucleobase they contain. In DNA, the four bases present are adenine (A), cytosine (C), guanine (G), and thymine (T). Nucleic acids have the property that two molecules will only bind to each other to form a double helix if the two sequences are complementary, meaning that they form matching sequences of base pairs, with A only binding to T, and C only to G.[4][5] Because the formation of correctly matched base pairs is energetically favorable, nucleic acid strands are expected in most cases to bind to each other in the conformation that maximizes the number of correctly paired bases. The sequences of bases in a system of strands thus determine the pattern of binding and the overall structure in an easily controllable way. In DNA nanotechnology, the base sequences of strands are rationally designed by researchers so that the base pairing interactions cause the strands to assemble in the desired conformation.[4][6]
Subfields
DNA nanotechnology is sometimes divided into two overlapping subfields: structural DNA nanotechnology and dynamic DNA nanotechnology. Structural DNA nanotechnology, sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials that assemble into a static, equilibrium end state. On the other hand, dynamic DNA nanotechnology focuses on complexes with useful non-equilibrium behavior such as the ability to reconfigure based on a chemical or physical stimulus. Some complexes, such as nucleic acid nanomechanical devices, combine features of both the structural and dynamic subfields.[7][8]
The complexes constructed in structural DNA nanotechnology use branched nucleic acid structures containing junctions. (In contrast, most biological DNA exists as an unbranched double helix.) One of the simplest branched structures is a four-arm junction that consists of four individual DNA strands, portions of which are complementary in a specific pattern. Unlike in natural Holliday junctions, each arm in the artificial immobile four-arm junction has a different base sequence, causing the junction point to be fixed at a certain position. Multiple junctions can also be combined in the same molecule, such as in the widely-used double-crossover (DX) motif, which contains two parallel double helical domains with individual strands crossing between the domains at two crossover points. Each crossover point is itself topologically a four-arm junction, but is constrained to a single orientation, as opposed to the flexible single four-arm junction, providing a rigidity that makes the DX motif suitable as a structural building block for larger DNA complexes.[4][6]
Dynamic DNA nanotechnology uses a mechanism called toehold-mediated strand displacement to allow the nucleic acid complexes to reconfigure in response to the addition of a new nucleic acid strand. In this reaction, the incoming strand binds to a single-stranded toehold region of a double-stranded complex, and then displaces one of the strands bound in the original complex through a branch migration process. The overall effect is that one of the strands in the complex is replaced with another one.[7] In addition, reconfigurable structures and devices can be made using functional nucleic acids such as deoxyribozymes and ribozymes, which are capable of performing chemical reactions, and aptamers, which can bind to specific proteins or small molecules.[9]
Design
DNA nanostructures must be rationally designed so that the individual nucleic acid strands will assemble into the desired structures. This process usually begins with the specification of a desired target structure or functionality. Then, the overall secondary structure of the target molecule is determined, specifying the arrangement of nucleic acid strands within the structure, and which portions of those strands should be bound to each other. The last step is the primary structure design, which is the specification of the actual base sequences of each nucleic acid strand.[10][11]
Structural design
The first step in designing a nucleic acid nanostructure is to decide how a given structure should be represented by a specific arrangement of nucleic acid strands. This design step determines the secondary structure, or the positions of the base pairs that hold the individual strands together in the desired shape.[10] Several approaches have been demonstrated:
- Tile-based structures. This approach breaks the target structure into smaller units with strong binding between the strands contained in each unit, and weaker interactions between the units. It is often used to make periodic lattices, but can also be used to implement algorithmic self-assembly, making them a platform for DNA computing. This was the dominant design strategy used from the mid 1990s until the mid 2000s, when the DNA origami methodology was developed.[10][12]
- Folding structures. An alternative to the tile-based approach, folding approaches make the nanostructure from a single long strand. This long strand can either have a designed sequence that folds due to its interactions with itself, or it can be folded into the desired shape by using shorter, "staple" strands. This latter method is called DNA origami, which allows the creation of nanoscale two- and three-dimensional shapes (see Discrete structures below).[13][14]
- Dynamic assembly. This approach directly controls the kinetics of DNA self-assembly, specifying all of the intermediate steps in the reaction mechanism in addition to the final product. This is done using a nucleic acid hairpin structure as the starting material, which assembles in a cascade reaction in a specific order. This class of methods has the advantage of proceeding isothermally, and thus does not involve the thermal annealing step that is required by solely thermodynamic approaches.[13][15]
Sequence design
After any of the above approaches are used to design the secondary structure of a target molecule, an actual sequence of nucleotides that will form into the desired structure must be devised. Nucleic acid design is the process of assigning a specific nucleic acid base sequence to each of a structure's constituent strands so that they will associate into a desired conformation. Most methods have the goal of designing sequences so that the target structure has the lowest energy, and is thus the most thermodynamically favorable, while misassembled structures have higher energies and are thus disfavored. This is done either through simple, faster heuristic methods such as sequence symmetry minimization, or by using a full nearest-neighbor thermodynamic model, which is more accurate but slower and more computationally intensive. Geometric models are used to examine tertiary structure of the nanostructures and to ensure that the complexes are not overly strained.[11][16]
Nucleic acid design has similar goals to protein design: in both, the sequence of monomers is designed to favor the desired assembled structure and to disfavor alternate structures. Nucleic acid design has the advantage of being much computationally simpler than protein design, since the simplicity of Watson-Crick base pairing rules leads to simple heuristic methods that yield experimentally robust designs. However, nucleic acid structures are less versatile than proteins in their functionality.[16]
Structural DNA nanotechnology
Structural DNA nanotechnology, sometimes abbreviated as SDN, focuses on synthesizing and characterizing nucleic acid complexes and materials where the assembly has a static, equilibrium endpoint. Structural DNA nanotechnology is based on the fact that the nucleic acid double helix has a robust, defined three-dimensional geometry that makes it possible to predict and design the structures of more complex nucleic acid molecules. Many such structures have been created, including two- and three-dimensional structures; and periodic, aperiodic, and discrete structures.[8]
Extended lattices
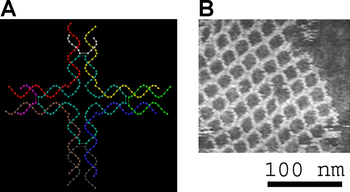
Small nucleic acid complexes can be equipped with sticky ends in order to combine them into larger two-dimensional periodic lattices containing a specific tessellated pattern of the individual molecular tiles.[8] The earliest example of this used DX, or double-crossover, molecules as the basic tiles, each containing four sticky ends designed with sequences that caused the DX units to combine into periodic two-dimensional flat sheets that are essentially rigid two-dimensional crystals of DNA.[19][20] Two-dimensional arrays have been made from other motifs as well, including the Holliday junction rhombus lattice,[21] and various DX-based arrays making use of a double-cohesion scheme.[22][23]
Two-dimensional arrays can also be made to exhibit aperiodic structures whose assembly implements a specific algorithm, making them one form of DNA computing.[24] The DX tiles can have their sticky end sequences chosen so that they act as Wang tiles, allowing them to perform computation. A DX array whose assembly encodes an XOR operation has been demonstrated; this allows the DNA array to implement a cellular automaton that generates a fractal called the Sierpinski gasket.[18] Another system has the function of a binary counter, displaying a representation of increasing binary numbers as it grows. These results show that computation can be incorporated into the assembly of DNA arrays, increasing its scope beyond simple periodic arrays.[25]
DX arrays have been made to form hollow 4–20 nm-diameter nanotubes, which are essentially two-dimensional lattices which curve back upon themselves.[10] These DNA nanotubes are somewhat similar in size and shape to carbon nanotubes, and while they lack the electrical conduction ability of carbon nanotubes, DNA nanotubes are more easily modified and connected to other structures. There have been multiple schemes for constructing DNA nanotubes, one of which uses the inherent curvature of DX tiles to form a DX lattice to curl around itself and close into a tube.[26] An alternative design uses single-stranded tiles for which the rigidity of the tube is an emergent property. This method has the benefit of being able to specify the circumference of the nanotube in a simple, modular fashion.[27]
The creation of three-dimensional lattices out of DNA was the earliest goal of DNA nanotechnology, but this proved to be one of the most difficult to realize. Success in constructing three-dimensional DNA lattices using a motif based on the concept of tensegrity, a balance between tension and compression forces, was finally reported in 2009.[24][28]
Discrete structures
Researchers have synthesized a number of three-dimensional DNA molecules that each have the connectivity of a polyhedron, such as a cube or octahedron, meaning that the DNA duplexes trace the edges of a polyhedron with a DNA junction at each vertex.[13] The earliest demonstrations of DNA polyhedra were very work-intensive, requiring multiple ligations and solid-phase synthesis steps to create catenated polyhedra.[29] Subsequent work yielded polyhedra whose synthesis was much easier. These include a DNA octahedron made from a long single strand designed to fold into the correct conformation,[30] and a tetrahedron that can be produced from four DNA strands in a single step, pictured at the top of this article.[1]
Nanostructures of arbitrary, non-regular shapes are usually made using the DNA origami method. These structures consist of a long, natural virus strand as a "scaffold", which is made to fold into the desired shape by computationally designed short "staple" strands. This method has the advantages of being easy to design, as the base sequence is predetermined by the scaffold strand sequence, and not requiring high strand purity and accurate stoichiometry, as most other DNA nanotechnology methods do. DNA origami was first demonstrated for two-dimensional shapes, such as a smiley face and a coarse map of North America.[13][14] Solid three-dimensional structures can be made by using parallel DNA helices arranged in a honeycomb pattern,[31] and structures with two-dimensional faces can be made to fold into a hollow overall three-dimensional shape, akin to a cardboard box. These can be programmed to open and reveal or release a molecular cargo in response to a stimulus, making them potentially useful as programmable molecular cages.[32][33]
Templated assembly
Nucleic acid structures can be made to incorporate or molecules other than nucleic acids, sometimes called heteroelements, including molecules such as proteins, metallic nanoparticles, quantum dots, and fullerenes. This allows the construction of materials and devices with a range of functionalities much greater than is possible with nucleic acids alone. The goal is to use the self-assembly of the nucleic acid structures to template the assembly of the nanoparticles hosted on them, controlling their position and in some cases orientation.[13][34] Many of these schemes use a covalent attachment scheme, using oligonucleotides with amide or thiol functional groups as a chemical handle to bind the heteroelements. This covalent binding scheme has been used to arrange gold nanoparticles on a DX-based array,[35] and also to arrange streptavidin protein molecules into specific patterns on a DX array.[36] A non-covalent hosting scheme using Dervan polyamides on a DX array was used to arrange streptavidin proteins in a specific pattern on a DX array.[37] Carbon nanotubes have been hosted on DNA arrays in a pattern allowing the assembly to act as a molecular electronic device, a carbon nanotube field-effect transistor.[38] In addition, there are nucleic acid metallization methods, in which the nucleic acid is replaced by a metal which assumes the general shape of the original nucleic acid structure,[39] and schemes for using nucleic acid nanostructures as lithography masks, transferring their pattern into a solid surface.[40]
Dynamic DNA nanotechnology
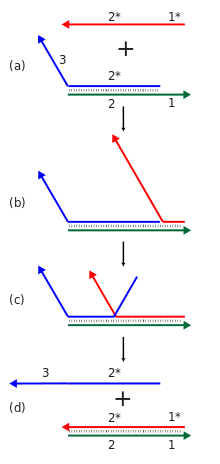
Dynamic DNA nanotechnology focuses on creating molecules with designed dynamic functionalities related to their overall structures, such as computation and mechanical motion. There is some overlap between structural and dynamic DNA nanotechnology, as structures can be formed through annealing and then reconfigured dynamically, or can be made to form dynamically in the first place.[13][41]
Nanomechanical devices
DNA complexes have been made that change their conformation upon some stimulus, making them one form of nanorobotics. These structures are initially formed in the same way as the static structures made in structural DNA nanotechnology, but are designed so that dynamic reconfiguration is possible after the initial assembly.[7][41] The earliest such device made use of the transition between the B-DNA and Z-DNA forms to respond to a change in buffer conditions by undergoing a twisting motion.[42] This reliance on buffer conditions, however, caused all devices to change state at the same time. Subsequent systems could change states based upon the presence of control strands, allowing multiple devices to be independently operated in solution. Some examples of such systems are a "molecular tweezers" design that has an open and a closed state,[43] a device that could switch from a paranemic-crossover (PX) conformation to a double-junction (JX2) conformation, undergoing rotational motion in the process,[44] and a two-dimensional array that could dynamically expand and contract in response to control strands.[45] Structures have also been made which dynamically open and/or close, potentially acting as a molecular cage to release or reveal a functional cargo upon opening.[32][46][47]
DNA walkers are a class of nucleic acid nanomachines that exhibit directional motion along a linear track. A large number of schemes have been demonstrated.[41] One strategy is to control the motion of the walker along the track using control strands that need to be manually added in sequence.[48][49] Another approach is to make use of restriction enzymes or deoxyribozymes to cleave the strands and cause the walker to move forward, which has the advantage of running autonomously.[50][51] A later system could walk upon a two-dimensional surface rather than a linear track, and demonstrated the ability to selectively pick up and move molecular cargo.[52] Additionally, a linear walker has been demonstrated that performs DNA-templated synthesis as the walker advances along the track, allowing autonomous multistep chemical synthesis directed by the walker.[53]
Strand displacement cascades
Cascades of strand displacement reactions can be used for either computational or structural purposes. An individual strand displacement reaction involves revealing a new sequence in response to the presence of some initiator strand. Many such reactions can be linked into a cascade where the newly-revealed output sequence of one reaction can initiate another strand displacement reaction elsewhere. This in turn allows for the construction of chemical reaction networks with many components, exhibiting complex computational and information processing abilities. These cascades are made energetically favorable through the formation of new base pairs and/or the entropy gain resulting from disassembly reactions. Strand displacement cascades allow for isothermal operation of the assembly or computational process, as opposed to traditional nucleic acid assembly's requirement for a thermal annealing step, where the temperature is raised and then slowly lowered to ensure proper formation of the desired structure. Strand displacement cascades can also support catalytic functionality of the initiator species, where less than one equivalent of the initiator can cause the reaction to go to completion.[7][15]
Strand displacement complexes can be used to make molecular logic gates capable of complex computation. Unlike traditional electronic computers, which use electric current as inputs and outputs, molecular computers use the concentrations of specific chemical species as signals. In the case of nucleic acid strand displacement circuits, the signal is the presence of nucleic acid strands that are released or consumed by binding and unbinding events to other strands in displacement complexes. This approach has been used to make logic gates such as AND, OR, and NOT gates.[54] More recently, a four-bit circuit was demonstrated that can compute the square root of the integers 0–15, using a system of gates containing 130 DNA strands.[55]
Another use of strand displacement cascades is to make dynamically assembled structures. These use a hairpin structure for the reactants, so that when the input strand binds, the newly revealed sequence is on the same molecule rather than disassembling. This allows new opened hairpins to be added to a growing complex. This approach has been used to make simple structures such as three- and four-arm junctions and dendrimers.[15]
Applications
DNA nanotechnology provides one of the only ways to form designed, complex structures with precise control over nanoscale features. The field is beginning to see application to solve basic science problems in structural biology and biophysics. One such application, the earliest envisaged for the field, is in crystallography, where molecules that are difficult to crystallize in isolation could be arranged and oriented within a three-dimensional nucleic acid lattice, thus allowing determination of their structure. DNA origami rods have also been used to replace liquid crystals in residual dipolar coupling experiments in protein NMR spectroscopy; using DNA origami is advantageous because, unlike liquid crystals, they are tolerant of the detergents needed to suspend membrane proteins in solution. It has been demonstrated that DNA walkers can be used as nanoscale assembly lines to move nanoparticles and direct chemical synthesis. Furthermore, DNA origami structures have aided in the biophysical studies of enzyme function and protein folding.[8][56]
DNA nanotechnology is also moving towards potential real-world applications. It has been suggested that the ability of nucleic acid arrays to arrange other molecules has potential applications in molecular scale electronics. The assembly of a nucleic acid structure could be used to template the assembly of a molecular electronic elements such as molecular wires, providing a method for nanometer-scale control of the placement and overall architecture of the device analogous to a molecular breadboard.[8][13] DNA nanotechnology has been called a form of programmable matter because of the coupling of computation to its material properties.[57]
There are potential applications for DNA nanotechnology in nanomedicine, making use of its ability to perform computation in a biocompatible format to make "smart drugs" for targeted drug delivery. One such system being investigated uses a hollow DNA box containing proteins that induce apoptosis, or cell death, that will only open when in proximity to a cancer cell.[56][58] There has additionally been interest in expressing these artificial structures in engineered living bacterial cells, most likely using the transcribed RNA for the assembly, although it is unknown whether these complex structures are able to efficiently fold or assemble in the cell's cytoplasm. If successful, this could enable directed evolution of nucleic acid nanostructures.[13]
Materials and methods
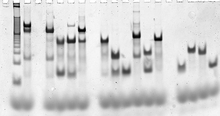
The sequences of the individual DNA strands that make up the target structure are designed computationally, using molecular modeling and thermodynamic modeling software.[11][16] Once the sequences have been designed, the nucleic acid molecules themselves are synthesized using standard oligonucleotide synthesis methods. This process is usually automated by using a machine called an oligonucleotide synthesizer, and nucleic acids of custom sequence are commercially available from many vendors.[59] For methods that require pure strands of known concentration, the nucleic acid strands can be purified using denaturing gel electrophoresis,[60] and concentrations are determined using one of several nucleic acid quantitation methods using ultraviolet absorbance spectroscopy.[61]
The fully formed target structures are usually characterized by native gel electrophoresis, which provides information about the size and shape of DNA molecules. An electrophoretic mobility shift assay can indicate whether a structure incorporates all the individual desired strands.[62] Fluorescent labeling and Förster resonance energy transfer (FRET) are sometimes used to characterize the structure of the molecules.[63]
Nucleic acid structures can be directly imaged by atomic force microscopy, which is well suited to extended two-dimensional structures, but is less useful for discrete three-dimensional structures due to the interaction of the microscope tip with the fragile nucleic acid structure. For imaging three-dimensional structures, transmission electron microscopy and cryo-electron microscopy are important methods. Extended three-dimensional lattices are analyzed by X-ray crystallography.[64][65]
History

The conceptual foundation for DNA nanotechnology was first laid out by Nadrian Seeman in the early 1980s.[66] Seeman's original motivation was to create a three-dimensional DNA lattice for orienting other large molecules, which would simplify their crystallographic study by eliminating the difficult process of obtaining pure crystals. This idea had reportedly come to him in late 1980, after realizing the similarity between the woodcut Depth by M. C. Escher and an array of DNA six-arm junctions.[6][67] A number of natural branched DNA molecules were known at the time, including the DNA replication fork and the mobile Holliday junction, but Seeman's insight was that immobile nucleic acid junctions could be created by properly designing the strand sequences to remove symmetry in the assembled molecule, and that these immobile junctions could in principle be combined into rigid crystalline lattices. The first theoretical paper proposing this scheme was published in 1982, and the first experimental demonstration of an immobile DNA junction was published the following year.[13][4]
In 1991, Seeman's laboratory published the synthesis of a cube made of DNA, the first synthetic three-dimensional nucleic acid nanostructure, for which he received the 1995 Feynman Prize in Nanotechnology. This was followed by a DNA truncated octahedron. However, it soon became clear that these molecules, polygonal shapes with flexible junctions as their vertices, were not rigid enough to form extended three-dimensional lattices. Seeman developed the more rigid double-crossover (DX) motif, and in 1998, in collaboration with Erik Winfree, published the creation of two-dimensional lattices of DX tiles.[6][66][68] These tile-based structures had the advantage that they provided the capability to implement DNA computing, which was demonstrated by Winfree and Paul Rothemund in their 2004 paper on the algorithmic self-assembly of a Sierpinski gasket structure, and for which they shared the 2006 Feynman Prize in Nanotechnology. Winfree's key insight was that the DX tiles could be used as Wang tiles, meaning that their assembly was capable of performing computation.[66] The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to achieve it.[56]
New capabilities continued to be discovered for designed DNA structures throughout the 2000s. The first DNA nanomachine—a motif that changes its structure in response to an input—was demonstrated in 1999 by Seeman. An improved system, which was the first nucleic acid device to make use of toehold-mediated strand displacement, was demonstrated by Bernard Yurke the following year. The next advance was to translate this into mechanical motion, and in 2004 and 2005, a number of DNA walker systems were demonstrated by the groups of Seeman, Niles Pierce, Andrew Turberfield, and Chengde Mao.[41] The idea of using DNA arrays to template the assembly of other molecules such as nanoparticles and proteins, first suggested by Bruche Robinson and Seeman in 1987,[69] was demonstrated in 2006 and 2007 by the groups of Hao Yan, Peter Dervan, and Thomas LaBean.[4][34]
In 2006, Rothemund first demonstrated the DNA origami technique for easily and robustly creating folded DNA molecules of any shape. Rothemund had conceived of this method as being conceptually intermediate between Seeman's DX lattices, which used many short strands, and William Shih's DNA octahedron, which consisted mostly of one very long strand. Rothemund's DNA origami contains a long strand whose folding is assisted by a number of short strands. This method allowed the creation of much larger structures than were previously possible, and which are less technically demanding to design and synthesize.[68] DNA origami was the cover story of Nature on March 15, 2006.[14] Rothemund's research demonstrating two-dimensional DNA origami structures was followed by the demonstration of solid three-dimensional DNA origami by Douglas et al. in 2009,[31] while the labs of Jørgen Kjems and Yan demonstrated hollow three-dimensional structures made out of two-dimensional faces.[56]
DNA nanotechnology was initially met with some skepticism due to the unusual non-biological use of nucleic acids as materials for building structures and doing computation, and the preponderance of proof of principle experiments that extended the capabilities of the field but were far from actual applications. Seeman's 1991 paper on the synthesis of the DNA cube was rejected by the journal Science after one reviewer praised its originality while another criticized it for its lack of biological relevance. By the early 2010s, however, the field was considered to have increased its capabilities to the point that applications for basic science research were beginning to be realized, and practical applications in medicine and other fields were beginning to be considered feasible.[56][70] The field had grown from very few active laboratories in 2001 to at least 60 in 2010, which increased the talent pool and thus the number of scientific advances in the field during that decade.[24]
See also
- Nucleic acid structure
- Molecular models of DNA
- International Society for Nanoscale Science, Computation, and Engineering
- List of nucleic acid simulation software
References
- ^ a b DNA polyhedra: Goodman, Russel P. (9 December 2005). "Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication". Science. 310 (5754): 1661–1665. Bibcode:2005Sci...310.1661G. doi:10.1126/science.1120367. PMID 16339440.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Overview: Mao, Chengde (2004). "The emergence of complexity: lessons from DNA". PLoS Biology. 2 (12): 2036–2038. doi:10.1371/journal.pbio.0020431. PMC 535573. PMID 15597116.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Background: Pelesko, John A. (2007). Self-assembly: the science of things that put themselves together. New York: Chapman & Hall/CRC. pp. 5, 7. ISBN 978-1-58488-687-7.
- ^ a b c d e f Overview: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1146/annurev-biochem-060308-102244, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1146/annurev-biochem-060308-102244instead. - ^ Background: Long, Eric C. (1996). "Fundamentals of nucleic acids". In Hecht, Sidney M (ed.). Bioorganic chemistry: nucleic acids. New York: Oxford University Press. pp. 4–10. ISBN 0-19-508467-5.
- ^ a b c d Overview: Seeman, Nadrian C. (2004). "Nanotechnology and the double helix". Scientific American. 290 (6): 64–75. doi:10.1038/scientificamerican0604-64. PMID 15195395.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d Dynamic DNA nanotechnology: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nchem.957, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nchem.957instead. - ^ a b c d e Structural DNA nanotechnology: Seeman, Nadrian C. (2007). "An overview of structural DNA nanotechnology". Molecular Biotechnology. 37 (3): 246–257. doi:10.1007/s12033-007-0059-4. PMID 17952671.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Dynamic DNA nanotechnology: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/j.copbio.2006.10.004, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/j.copbio.2006.10.004instead. - ^ a b c d Design: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/anie.200502358, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/anie.200502358instead. - ^ a b c Design: Brenneman, Arwen; Condon, Anne (25 September 2002). "Strand design for biomolecular computation". Theoretical Computer Science. 287: 39–58. doi:10.1016/S0304-3975(02)00135-4.
- ^ Overview: Lin, Chenxiang; Liu, Yan; Rinker, Sherri; Yan, Hao (11 August 2006). "DNA tile based self-assembly: building complex nanoarchitectures". ChemPhysChem. 7 (8): 1641–1647. doi:10.1002/cphc.200600260. PMID 16832805.
- ^ a b c d e f g h i Overview: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nnano.2011.187, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nnano.2011.187instead. - ^ a b c DNA origami: Rothemund, Paul W. K. (16 March 2006). "Folding DNA to create nanoscale shapes and patterns". Nature. 440 (7082): 297–302. Bibcode:2006Natur.440..297R. doi:10.1038/nature04586. PMID 16541064.
- ^ a b c Kinetic assembly: Yin, Peng; Choi, Harry M. T.; Calvert, Colby R.; Pierce, Niles A. (17 January 2008). "Programming biomolecular self-assembly pathways". Nature. 451 (7176): 318–322. Bibcode:2008Natur.451..318Y. doi:10.1038/nature06451. PMID 18202654.
- ^ a b c Design: Dirks, Robert M. (15 February 2004). "Paradigms for computational nucleic acid design". Nucleic Acids Research. 32 (4): 1392–1403. doi:10.1093/nar/gkh291. PMC 390280. PMID 14990744.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Other arrays: Strong, Michael (2004). "Protein Nanomachines". PLoS Biology. 2 (3): e73. doi:10.1371/journal.pbio.0020073. PMC 368168. PMID 15024422.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b Algorithmic self-assembly: Rothemund, Paul W. K. (2004). "Algorithmic self-assembly of DNA Sierpinski triangles". PLoS Biology. 2 (12): 2041–2053. doi:10.1371/journal.pbio.0020424. PMC 534809. PMID 15583715.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ DX arrays: Winfree, Erik (6 August 1998). "Design and self-assembly of two-dimensional DNA crystals". Nature. 394 (6693): 529–544. Bibcode:1998Natur.394..539W. doi:10.1038/28998. PMID 9707114.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DX arrays: Liu, Furong (10 February 1999). "Modifying the surface features of two-dimensional DNA crystals". Journal of the American Chemical Society. 121 (5): 917–922. doi:10.1021/ja982824a.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Other arrays: Mao, Chengde (16 June 1999). "Designed two-dimensional DNA Holliday junction arrays visualized by atomic force microscopy". Journal of the American Chemical Society. 121 (23): 5437–5443. doi:10.1021/ja9900398.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Other arrays: Constantinou, Pamela E. (21 September 2006). "Double cohesion in structural DNA nanotechnology". Organic and Biomolecular Chemistry. 4 (18): 3414–3419. doi:10.1039/b605212f. PMID 17036134.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Other arrays: Mathieu, Frederick (2005). "Six-helix bundles designed from DNA". Nano Letters. 5 (4): 661–665. Bibcode:2005NanoL...5..661M. doi:10.1021/nl050084f. PMID 15826105.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c History: Seeman, Nadrian (9 June 2010). "Structural DNA nanotechnology: growing along with Nano Letters". Nano Letters. 10 (6): 1971–1978. Bibcode:2010NanoL..10.1971S. doi:10.1021/nl101262u. PMC 2901229. PMID 20486672.
- ^ Algorithmic self-assembly: Barish, Robert D.; Rothemund, Paul W. K.; Winfree, Erik (2005). "Two computational primitives for algorithmic self-assembly: copying and counting". Nano Letters. 5 (12): 2586–2592. Bibcode:2005NanoL...5.2586B. doi:10.1021/nl052038l. PMID 16351220.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ DNA nanotubes: Rothemund, Paul W. K. (22 December 2004). "Design and Characterization of Programmable DNA Nanotubes". Journal of the American Chemical Society. 126 (50): 16344–16352. doi:10.1021/ja044319l. PMID 15600335.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA nanotubes: Yin, P.; Hariadi, R. F.; Sahu, S.; Choi, H. M. T.; Park, S. H.; Labean, T. H.; Reif, J. H. (8 August 2008). "Programming DNA Tube Circumferences". Science. 321 (5890): 824–826. Bibcode:2008Sci...321..824Y. doi:10.1126/science.1157312. PMID 18687961.
- ^ Three-dimensional arrays: Zheng, Jianping; Birktoft, Jens J.; Chen, Yi; Wang, Tong; Sha, Ruojie; Constantinou, Pamela E.; Ginell, Stephan L.; Mao, Chengde (3 September 2009). "From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal". Nature. 461 (7260): 74–77. Bibcode:2009Natur.461...74Z. doi:10.1038/nature08274. PMC 2764300. PMID 19727196.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA polyhedra: Zhang, Yuwen (1 March 1994). "Construction of a DNA-truncated octahedron". Journal of the American Chemical Society. 116 (5): 1661–1669. doi:10.1021/ja00084a006.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA polyhedra: Shih, William M. (12 February 2004). "A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron". Nature. 427 (6975): 618–621. doi:10.1038/nature02307. PMID 14961116.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b DNA origami: Douglas, Shawn M.; Dietz, Hendrik; Liedl, Tim; Högberg, Björn; Graf, Franziska; Shih, William M. (21 May 2009). "Self-assembly of DNA into nanoscale three-dimensional shapes". Nature. 459 (7245): 414–418. Bibcode:2009Natur.459..414D. doi:10.1038/nature08016. PMC 2688462. PMID 19458720.
- ^ a b DNA boxes: Andersen, Ebbe S.; Dong, Mingdong; Nielsen, Morten M.; Jahn, Kasper; Subramani, Ramesh; Mamdouh, Wael; Golas, Monika M.; Sander, Bjoern (7 May 2009). "Self-assembly of a nanoscale DNA box with a controllable lid". Nature. 459 (7243): 73–76. Bibcode:2009Natur.459...73A. doi:10.1038/nature07971. PMID 19424153.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA boxes: Ke, Yonggang; Sharma, Jaswinder; Liu, Minghui; Jahn, Kasper; Liu, Yan; Yan, Hao (10 June 2009). "Scaffolded DNA origami of a DNA tetrahedron molecular container". Nano Letters. 9 (6): 2445–2447. Bibcode:2009NanoL...9.2445K. doi:10.1021/nl901165f. PMID 19419184.
- ^ a b Overview: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/cbic.200900286, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/cbic.200900286instead. - ^ Nanoarchitecture: Zheng, Jiwen (2006). "2D Nanoparticle Arrays Show the Organizational Power of Robust DNA Motifs". Nano Letters. 6 (7): 1502–1504. Bibcode:2006NanoL...6.1502Z. doi:10.1021/nl060994c. PMID 16834438.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Nanoarchitecture: Park, Sung Ha (2006). "Finite-size, fully addressable DNA tile lattices formed by hierarchical assembly procedures". Angewandte Chemie. 118 (40): 749–753. doi:10.1002/ange.200690141.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Nanoarchitecture: Cohen, Justin D. (22 October 2007). "Addressing single molecules on DNA nanostructures". Angewandte Chemie International Edition. 46 (42): 7956–7959. doi:10.1002/anie.200702767. PMID 17763481.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Nanoarchitecture: Maune, Hareem T.; Han, Si-Ping; Barish, Robert D.; Bockrath, Marc; Goddard, III, William A.; Rothemund, Paul W. K.; Winfree, Erik (2009). "Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates". Nature Nanotechnology. 5 (1): 61–66. Bibcode:2010NatNa...5...61M. doi:10.1038/nnano.2009.311. PMID 19898497.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Nanoarchitecture: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1021/nn1035075, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1021/nn1035075instead. - ^ Nanoarchitecture: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/anie.200460257, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/anie.200460257instead. - ^ a b c d DNA machines: Bath, Jonathan; Turberfield, Andrew J. (2007). "DNA nanomachines". Nature Nanotechnology. 2 (5): 275–284. Bibcode:2007NatNa...2..275B. doi:10.1038/nnano.2007.104. PMID 18654284.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ DNA machines: Mao, Chengde (14 January 1999). "A DNA nanomechanical device based on the B-Z transition". Nature. 397 (6715): 144–146. Bibcode:1999Natur.397..144M. doi:10.1038/16437. PMID 9923675.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA machines: Yurke, Bernard (10 August 2000). "A DNA-fuelled molecular machine made of DNA". Nature. 406 (6796): 605–609. Bibcode:2000Natur.406..605Y. doi:10.1038/35020524. PMID 10949296.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA machines: Yan, Hao (3 January 2002). "A robust DNA mechanical device controlled by hybridization topology". Nature. 415 (6867): 62–65. Bibcode:2002Natur.415...62Y. doi:10.1038/415062a. PMID 11780115.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ DNA machines: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/ange.200351818, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/ange.200351818instead. - ^ DNA machines: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1038/nnano.2008.3, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1038/nnano.2008.3instead. - ^ Applications: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1126/science.1214081, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1126/science.1214081instead. - ^ DNA walkers: Shin, Jong-Shik; Pierce, Niles A. (8 September 2004). "A synthetic DNA walker for molecular transport". Journal of the American Chemical Society. 126 (35): 10834–10835. doi:10.1021/ja047543j. PMID 15339155.
- ^ DNA walkers: Sherman, William B.; Seeman, Nadrian C. (2004). "A precisely controlled DNA biped walking device". Nano Letters. 4 (7): 1203–1207. Bibcode:2004NanoL...4.1203S. doi:10.1021/nl049527q.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ DNA walkers: Tian, Ye; He, Yu; Chen, Yi; Yin, Peng; Mao, Chengde (11 July 2005). "A DNAzyme that walks processively and autonomously along a one-dimensional track". Angewandte Chemie. 117 (28): 4429–4432. doi:10.1002/ange.200500703.
- ^ DNA walkers: Bath, Jonathan; Green, Simon J.; Turberfield, Andrew J. (11 July 2005). "A free-running DNA motor powered by a nicking enzyme". Angewandte Chemie International Edition. 44 (28): 4358–4361. doi:10.1002/anie.200501262.
- ^ Functional DNA walkers: Lund, Kyle; Manzo, Anthony J.; Dabby, Nadine; Michelotti, Nicole; Johnson-Buck, Alexander; Nangreave, Jeanette; Taylor, Steven; Pei, Renjun (13 May 2010). "Molecular robots guided by prescriptive landscapes". Nature. 465 (7295): 206–210. Bibcode:2010Natur.465..206L. doi:10.1038/nature09012. PMC 2907518. PMID 20463735.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Functional DNA walkers: He, Yu; Liu, David R. (2010). "Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker". Nature Nanotechnology. 5 (11): 778–782. Bibcode:2010NatNa...5..778H. doi:10.1038/nnano.2010.190. PMC 2974042. PMID 20935654.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Strand displacement cascades: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1126/science.1132493, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1126/science.1132493instead. - ^ Strand displacement cascades: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1126/science.1200520, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1126/science.1200520instead. - ^ a b c d e History/applications: Service, Robert F. (3 June 2011). "DNA nanotechnology grows up". Science. 332 (6034): 1140–1143. doi:10.1126/science.332.6034.1140.
- ^ Applications: Rietman, Edward A. (2001). Molecular engineering of nanosystems. Springer. pp. 209–212. ISBN 978-0-387-98988-4. Retrieved 17 April 2011.
- ^ Applications: Jungmann, Ralf; Renner, Stephan; Simmel, Friedrich C. (2008). "From DNA nanotechnology to synthetic biology". HFSP journal. 2 (2): 99–109. doi:10.2976/1.2896331. PMC 2645571. PMID 19404476.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/0471142727.mb0211s42, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/0471142727.mb0211s42instead. - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/0471142727.mb0212s42 , please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/0471142727.mb0212s42instead. - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/9780470089941.et0202s5, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/9780470089941.et0202s5instead. - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/0471142727.mb0207s47, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/0471142727.mb0207s47instead. - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1002/0471142700.nc1110s11, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1002/0471142700.nc1110s11instead. - ^ Methods: Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1007/978-1-61779-142-0_1, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1007/978-1-61779-142-0_1instead. - ^ Methods: Bloomfield, Victor A. (2000). Nucleic acids: structures, properties, and functions. Sausalito, Calif: University Science Books. pp. 84–86, 396–407. ISBN 0-935702-49-0.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c History: Pelesko, John A. (2007). Self-assembly: the science of things that put themselves together. New York: Chapman & Hall/CRC. pp. 201, 242, 259. ISBN 978-1-58488-687-7.
- ^ History: See "Current crystallization protocol". Nadrian Seeman Lab. for a statement of the problem, and "DNA cages containing oriented guests". Nadiran Seeman Laboratory. for the proposed solution.
- ^ a b DNA origami: Rothemund, Paul W. K. (2006). "Scaffolded DNA origami: from generalized multicrossovers to polygonal networks". In Chen, Junghuei; Jonoska, Natasha; Rozenberg, Grzegorz (eds.). Nanotechnology: science and computation. Natural Computing Series. New York: Springer. pp. 3–21. doi:10.1007/3-540-30296-4_1. ISBN 978-3-540-30295-7.
- ^ Nanoarchitecture: Robinson, Bruche H. (1987). "The design of a biochip: a self-assembling molecular-scale memory device". Protein Engineering. 1 (4): 295–300. doi:10.1093/protein/1.4.295. PMID 3508280.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ History: Hopkin, Karen (2011). "Profile: 3-D seer". The Scientist. Retrieved 8 August 2011.
{{cite journal}}: Unknown parameter|month=ignored (help)
Further reading
General:
- Seeman, Nadrian C. (2004). "Nanotechnology and the double helix". Scientific American. 290 (6): 64–75. doi:10.1038/scientificamerican0604-64. PMID 15195395.
{{cite journal}}: Unknown parameter|month=ignored (help)—An article written for laypeople by the founder of the field - Seeman, Nadrian C. (9 June 2010). "Structural DNA nanotechnology: growing along with Nano Letters". Nano Letters. 10 (6): 1971–1978. Bibcode:2010NanoL..10.1971S. doi:10.1021/nl101262u. PMC 2901229. PMID 20486672.—A review of results in the period 2001–2010
- Seeman, Nadrian C. (2010). "Nanomaterials based on DNA". Annual Review of Biochemistry. 79: 65–87. doi:10.1146/annurev-biochem-060308-102244. PMID 20222824.—A more comprehensive review including both old and new results in the field
- Service, Robert F. (3 June 2011). "DNA nanotechnology grows up". Science. 332 (6034): 1140–1143. doi:10.1126/science.332.6034.1140. and doi:10.1126/science.332.6034.1142.—A news article focusing on the history of the field and development of new applications
Specific subfields:
- Bath, Jonathan; Turberfield, Andrew J. (5 May 2007). "DNA nanomachines". Nature Nanotechnology. 2 (5): 275–284. Bibcode:2007NatNa...2..275B. doi:10.1038/nnano.2007.104. PMID 18654284.—A review of nucleic acid nanomechanical devices
- Feldkamp, Udo; Niemeyer, Christof M. (13 March 2006). "Rational design of DNA nanoarchitectures". Angewandte Chemie International Edition. 45 (12): 1856–76. doi:10.1002/anie.200502358. PMID 16470892.—A review coming from the viewpoint of secondary structure design
- Lin, Chenxiang; Liu, Yan; Rinker, Sherri; Yan, Hao (11 August 2006). "DNA tile based self-assembly: building complex nanoarchitectures". ChemPhysChem. 7 (8): 1641–1647. doi:10.1002/cphc.200600260. PMID 16832805.—A minireview specifically focusing on tile-based assembly
- Zhang, David Yu; Seelig, Georg (2011). "Dynamic DNA nanotechnology using strand-displacement reactions". Nature Chemistry. 3 (2): 103–113. Bibcode:2011NatCh...3..103Z. doi:10.1038/nchem.957. PMID 21258382.
{{cite journal}}: Unknown parameter|month=ignored (help)—A review of DNA systems making use of strand displacement mechanisms
External links
- International Society for Nanoscale Science, Computation and Engineering
- What is Bionanotechnology?—a video introduction to DNA nanotechnology
