Philadelphia chromosome
| Philadelphia chromosome | |
|---|---|
| Specialty | Oncology |
Philadelphia chromosome or Philadelphia translocation is a specific chromosomal abnormality that is associated with chronic myelogenous leukemia (CML). It is the result of a reciprocal translocation between chromosome 9 and 22, and is specifically designated t(9;22)(q34;q11). The presence of this translocation is a highly sensitive test for CML, since 95% of people with CML have this abnormality (the remainder have either a cryptic translocation that is invisible on G-banded chromosome preparations, or a variant translocation involving another chromosome or chromosomes as well as the long arm of chromosomes 9 and 22). However, the presence of the Philadelphia (Ph) chromosome is not sufficiently specific to diagnose CML, since it is also found in acute lymphoblastic leukemia[1] (ALL, 25–30% in adult and 2–10% in pediatric cases) and occasionally in acute myelogenous leukemia (AML).
Molecular biology
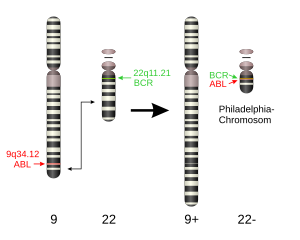
The exact chromosomal defect in Philadelphia chromosome is a translocation, in which parts of two chromosomes, 9 and 22, swap places. The result is that a fusion gene is created by juxtapositioning the Abl1 gene on chromosome 9 (region q34) to a part of the BCR ("breakpoint cluster region") gene on chromosome 22 (region q11). This is a reciprocal translocation, creating an elongated chromosome 9 (der 9), and a truncated chromosome 22 (the Philadelphia chromosome).[2][3] In agreement with the International System for Human Cytogenetic Nomenclature (ISCN), this chromosomal translocation is designated as t(9;22)(q34;q11). Abl stands for "Abelson", the name of a leukemia virus which carries a similar protein.
The result of the translocation is the oncogenic BCR-ABL gene fusion, located on the shorter derivative 22 chromosome. This gene encodes the Bcr-abl fusion protein. Depending on the precise location of the fusion the molecular weight of the protein can range from 185 to 210 kDa. For this reason bcr-abl is sometimes called p210 or p185. Three clinically important variants are the p190, p210 and p230 isoforms.[4] p190 is generally associated with acute lymphoblastic leukemia (ALL), while p210 is generally associated with chronic myeloid leukemia but can also be associated with ALL.[5] p230 is usually associated with chronic neutrophilic leukemia.[5] Additionally, the p190 isoform can also be expressed as a splice variant of p210.[6]
Because the Abl gene expresses a membrane-associated protein, a tyrosine kinase, the BCR-Abl transcript is also translated into a tyrosine kinase, adding a phosphate group to tyrosine. Although the BCR region also expresses serine/threonine kinases, the tyrosine kinase function is very relevant for drug therapy. Tyrosine kinase inhibitors (such as imatinib and sunitinib) are important drugs against a variety of cancers including CML, renal cell carcinoma (RCC) and gastrointestinal stromal tumors (GISTs).
The fused BCR-Abl protein interacts with the interleukin-3 receptor beta(c) subunit. The ABL tyrosine kinase activity of BCR-Abl is elevated relative to wild-type ABL[7] . Since ABL activates a number of cell cycle-controlling proteins and enzymes, the result of the BCR-Abl fusion is to speed up cell division. Moreover, it inhibits DNA repair, causing genomic instability and potentially causing the feared blast crisis in CML.
Nomenclature
Philadelphia chromosome is designated Ph (or Ph') chromosome and the translocation is termed t(9;22)(q34.1;q11.2).
Therapy
Tyrosine kinase inhibitors

In the late 1990s, STI-571 (imatinib, Gleevec/Glivec) was identified by the pharmaceutical company Novartis (then known as Ciba Geigy) in high-throughput screens for tyrosine kinase inhibitors. Subsequent clinical trials led by Dr. Brian J. Druker at Oregon Health & Science University in collaboration with Dr. Charles Sawyers and Dr. Moshe Talpaz demonstrated that STI-571 inhibits proliferation of BCR-ABL-expressing hematopoietic cells. Although it did not eradicate CML cells, it did greatly limit the growth of the tumor clone and decreased the risk of the feared "blast crisis".[citation needed] In 2000 John Kuriyan determined the mechanism by which STI-571 inhibits the Abl kinase domain.[8] It was marketed in 2001 by Novartis as imatinib mesylate (Gleevec in the US, Glivec in Europe). Other pharmacological inhibitors are being developed, which are more potent and/or are active against the emerging Gleevec/Glivec resistant BCR-abl clones in treated patients. The majority of these resistant clones are point-mutations in the kinase of BCR-abl. New inhibitors include dasatinib and nilotinib, which are significantly more potent than imatinib and may overcome resistance.
Treatment of pediatric Ph+ ALL with a combination of standard chemotherapy and RTK inhibitors may result in remission,[citation needed] but the curative potential is unknown.
Blood or marrow transplants
COG study AALL 0031, which examines the use of Gleevec with standard chemotherapeutic regimens and bone marrow transplant from HLA-matched related donors for high risk ALL (including Ph+ ALL), has concluded, and findings will be published in the near future.[citation needed]
A potentially curative, but risky option for pediatric Ph+ ALL or Ph+ CML includes bone marrow transplant or cord blood transplant, but chemotherapy is favored by some for achieving first remission (CR1). For some, bone marrow transplant from a matched sibling donor or a matched, unrelated donor may be favored when remission is obtained.
Cord blood transplant is favored by some when a 10/10 bone marrow match is not available, and cord blood transplant may have some advantages, including a reduced incidence of graft-vs-host disease (GVHD), which is a common and significant complication of transplant. However, transplant with cord blood sometimes requires longer periods of time for engraftment, which may increase the potential for complications due to infection. Regardless of the type of transplant, transplant-related mortality and relapse are possible, and the rates may change as treatment protocols improve. For second remission (CR2), if achieved, both chemotherapy and transplant options are possible, and many physicians prefer transplant.[citation needed]
History
The Philadelphia chromosome was first discovered and described in 1960 by Peter Nowell from University of Pennsylvania School of Medicine[9] and David Hungerford from the Fox Chase Cancer Center's[10] Institute for Cancer Research and was therefore named after the city in which both facilities are located.
In 1973, Janet D. Rowley at the University of Chicago identified the mechanism by which the Philadelphia chromosome arises as a translocation.[11][12]
See also
References
- ^ Talpaz M, Shah NP, Kantarjian H; et al. (2006). "Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias". N. Engl. J. Med. 354 (24): 2531–41. doi:10.1056/NEJMoa055229. PMID 16775234.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 12755554, please use {{cite journal}} with
|pmid=12755554instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 8656667, please use {{cite journal}} with
|pmid=8656667instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 12191565, please use {{cite journal}} with
|pmid=12191565instead. - ^ a b Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 18665825, please use {{cite journal}} with
|pmid=18665825instead. - ^ Attention: This template ({{cite pmid}}) is deprecated. To cite the publication identified by PMID 9858221, please use {{cite journal}} with
|pmid=9858221instead. - ^ http://www.ncbi.nlm.nih.gov/pubmed/11345193.
{{cite web}}: Missing or empty|title=(help) - ^ Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J (2000). "AStructural mechanism for STI-571 inhibition of abelson tyrosine kinase". Science. 289 (5486): 1857–9. doi:10.1126/science.289.5486.1938. PMID 10988075.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nowell P, Hungerford D (November 1960). "A minute chromosome in chronic granulocytic leukemia" (PDF). Science. 132 (3438): 1497. doi:10.1126/science.132.3438.1488.
- ^ Fox Chase Cancer Center. "50th Anniversary of the Discovery of the Philadelphia Chromsome".
{{cite journal}}: Cite journal requires|journal=(help) - ^ Rowley JD (1973). "Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining". Nature. 243 (5405): 290–3. doi:10.1038/243290a0. PMID 4126434.
- ^ Claudia Dreifus (2011-02-07). "The Matriarch of Modern Cancer Genetics". New York Times.
External links
- Philadelphia+chromosome at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- bcr-abl+Fusion+Proteins at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
