Small interfering RNA: Difference between revisions
m Disambiguate Vector (biology) to Vector (molecular biology) using popups |
No edit summary |
||
| Line 82: | Line 82: | ||
*[http://www.human-siRNA-database.net HuSiDa: Human siRNA Database] |
*[http://www.human-siRNA-database.net HuSiDa: Human siRNA Database] |
||
*[http://siRNA.cgb.ki.se siRNAdb: a database of siRNA sequences] |
*[http://siRNA.cgb.ki.se siRNAdb: a database of siRNA sequences] |
||
siRNA focused Companies: |
|||
*[http://www.alnylam.com Alnylam Pharmaceuticals] |
|||
*[http://www.anadyspharma.com Anadys Pharmaceuticals] |
|||
*[http://www.ambion.com Ambion (a division of Applied Biosystems, Inc.)] |
|||
*[http://www.armagen.com ArmaGen Technologies,Inc] |
|||
*[http://www.bdbiosciences.com BD Biosciences] |
|||
*[http://www.benitec.com Benitec] |
|||
*[http://www.bionomics.com.au Bionomics] |
|||
*[http://www.biospring.de BioSpring] |
|||
*[http://www.calandopharma.com Calando Pharmaceuticals] |
|||
*[http://www.celera.com Celera Genomics] |
|||
*[http://www.cenix-bioscience.com Cenix BioScience] |
|||
*[http://www.ceptyr.com Ceptyr] |
|||
*[http://www.dharmacon.com Dharmacon] |
|||
*[http://www.genlantis.com Genlantis] |
|||
*[http://www.ingenium-ag.com Ingenium Pharmaceuticals] |
|||
*[http://www.idtdna.com/Scitools/Applications/Predesign/ Integrated DNA Technologies, Inc.] |
|||
*[http://www.invitrogen.com Invitrogen] |
|||
*[http://www.invivogen.com InvivoGen] |
|||
*[http://www.isispharm.com Isis Pharmaceuticals] |
|||
*[http://www.nucleonicsinc.com Nucleonics Inc] |
|||
*[http://www.pfizer.com Pfizer Inc] |
|||
*[http://www.rnai.co.jp RNAi Co.Ltd] |
|||
*[http://www.roche-kulmbach.de/portal/eipf/de/RocheKulmbach/RocheKulmbach Roche Kulmbach GmbH] |
|||
*[http://www.sigmaaldrich.com Sigma-Aldrich] |
|||
*[http://www.sirna.com Sirna Therapeutics, Inc.] |
|||
*[http://www.sirion-biotech.com SIRION-Biotech] |
|||
*[http://www.thermo.com/sirna Thermo Fisher Scientific] |
|||
*[http://www.qiagen.com QIAGEN Sample & Assay Technologies] |
|||
esiRNA (Endoribonuclease-prepared siRNA) |
esiRNA (Endoribonuclease-prepared siRNA) |
||
*[http://www.sigma.com/esirna MISSION esiRNA, Sigma-Aldrich] |
*[http://www.sigma.com/esirna MISSION esiRNA, Sigma-Aldrich] |
||
{{natural antisense transcripts}} |
{{natural antisense transcripts}} |
||
Revision as of 01:59, 23 July 2009

Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA molecules, 20-25 nucleotides in length, that play a variety of roles in biology. Most notably, siRNA is involved in the RNA interference (RNAi) pathway, where it interferes with the expression of a specific gene. In addition to their role in the RNAi pathway, siRNAs also act in RNAi-related pathways, e.g., as an antiviral mechanism or in shaping the chromatin structure of a genome; the complexity of these pathways is only now being elucidated.
SiRNAs were first discovered by David Baulcombe's group at the Sainsbury Laboratory in Norwich, England, as part of post-transcriptional gene silencing (PTGS) in plants. The group published their findings in Science in a paper titled "A species of small antisense RNA in posttranscriptional gene silencing in plants".[1] Shortly thereafter, in 2001, synthetic siRNAs were shown to be able to induce RNAi in mammalian cells by Thomas Tuschl and colleagues in a paper published in Nature.[2] This discovery led to a surge in interest in harnessing RNAi for biomedical research and drug development.
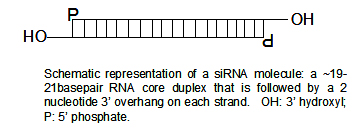
Structure
SiRNAs have a well-defined structure: a short (usually 21-nt) double strand of RNA (dsRNA) with 2-nt 3' overhangs on either end:
Each strand has a 5' phosphate group and a 3' hydroxyl (-OH) group. This structure is the result of processing by dicer, an enzyme that converts either long dsRNAs or small hairpin RNAs into siRNAs.[3] SiRNAs can also be exogenously (artificially) introduced into cells by various transfection methods to bring about the specific knockdown of a gene of interest. Essentially any gene of which the sequence is known can thus be targeted based on sequence complementarity with an appropriately tailored siRNA. This has made siRNAs an important tool for gene function and drug target validation studies in the post-genomic era.
RNAi induction using siRNAs or their biosynthetic precursors

Transfection of an exogenous siRNA can be problematic because the gene knockdown effect is only transient, particularly in rapidly dividing cells. One way of overcoming this challenge is to modify the siRNA in such a way as to allow it to be expressed by an appropriate vector, e.g., a plasmid. This is done by the introduction of a loop between the two strands, thus producing a single transcript, which can be processed into a functional siRNA. Such transcription cassettes typically use an RNA polymerase III promoter (e.g., U6 or H1), which usually directs the transcription of small nuclear RNAs (snRNAs) (U6 is involved in gene splicing; H1 is the RNase component of human RNase P). It is assumed (although not known for certain) that the resulting siRNA transcript is then processed by Dicer.
RNA activation
It has recently been found that dsRNA can also activate gene expression, a mechanism that has been termed "small RNA-induced gene activation" or RNAa. It has been shown that dsRNAs targeting gene promoters induce potent transcriptional activation of associated genes. RNAa was demonstrated in human cells using synthetic dsRNAs, termed "small activating RNAs" (saRNAs). It is currently not known whether RNAa is conserved in other organisms.[4]
Challenges: avoiding nonspecific effects
Because RNAi intersects with a number of other pathways, it is not surprising that on occasion nonspecific effects are triggered by the experimental introduction of an siRNA. When a mammalian cell encounters a double-stranded RNA such as an siRNA, it may mistake it as a viral by-product and mount an immune response. Furthermore, because structurally related microRNAs modulate gene expression largely via incomplete complementarity with a target mRNA, the introduction of an siRNA may cause unintended off-targeting.
Innate immunity
Introduction of too much siRNA can result in nonspecific events due to activation of innate immune responses. Most evidence to date suggests that this is probably due to activation of the dsRNA sensor PKR, although retinoic acid inducible gene I (RIG-I) may also be involved. The induction of cytokines via toll-like receptor 7 (TLR7) has also been described. One promising method of reducing the nonspecific effects is to convert the siRNA into a microRNA. MicroRNAs occur naturally, and by harnessing this endogenous pathway it should be possible to achieve similar gene knockdown at comparatively low concentrations of resulting siRNAs. This should minimize nonspecific effects.
Off-targeting
Off-targeting is another challenge to the use of siRNAs as a gene knockdown tool. Here, genes with incomplete complementarity are inadvertently downregulated by the siRNA (effectively, the siRNA acts as an miRNA), leading to problems in data interpretation and potential toxicity. This, however, can be partly addressed by designing appropriate control experiments, and siRNA design algorithms are currently being developed to produce siRNAs free from off-targeting. Genome-wide expression analysis, e.g., by microarray technology, can then be used to verify this and further refine the algorithms. A 2006 paper from the laboratory of Dr. Khvorova implicates 6- or 7-basepair-long stretches from position 2 onward in the siRNA matching with 3'UTR regions in off-targeted genes.[5]
Possible therapeutic applications and challenges
Given the ability to knock down essentially any gene of interest, RNAi via siRNAs has generated a great deal of interest in both basic[6] and applied biology. There are an increasing number of large-scale RNAi screens that are designed to identify the important genes in various biological pathways. Because disease processes also depend on the activity of multiple genes, it is expected that in some situations turning off the activity of a gene with an siRNA could produce a therapeutic benefit.
However, applying RNAi via siRNAs to living animals, especially humans, poses many challenges. Experimentally, siRNAs show different effectiveness in different cell types in a manner as yet poorly understood: some cells respond well to siRNAs and show a robust knockdown, whereas others show no such knockdown (even despite efficient transfection).
Phase I results of the first two therapeutic RNAi trials (indicated for age-related macular degeneration, aka AMD) reported at the end of 2005 demonstrated that siRNAs are well tolerated and have suitable pharmacokinetic properties.[7] siRNAs and related RNAi induction methods therefore stand to become an important new class of drugs in the foreseeable future.
In 2008, a team of researchers from Texas Tech University and Harvard University announced the development of a siRNA-based treatment that may ultimately counteract the Human Immunodeficiency Virus (HIV). Human cells infected with HIV, injected into rats, have been cured by the experimental treatment. Clinical trials on humans are expected to begin by 2010.[8][9][10]
SiRNAs in fiction
Ian McEwan's novel Saturday suggests an siRNA-based treatment for Huntington's disease.
See also
MicroRNAs (miRNA) are a related class of gene regulatory small RNAs, typically 21-23nt in length. They typically differ from siRNAs because they are processed from single-stranded RNA precursors and show only partial complementarity to mRNA targets. They have been implicated in a wide range of functions such as cell growth and apoptosis, development, neuronal plasticity and remodeling, and even insulin secretion. MiRNAs have also been implicated in disease: e.g., an overabundance of miRNA has been reported in cases of Fragile X Mental Retardation, and some cancers are associated with up- and downregulation of certain miRNA genes.
Initial studies have indicated that miRNAs regulate gene expression posttranscriptionally at the level of translational inhibition at P-bodies in the cytoplasm. However, miRNAs may also guide mRNA cleavage in a manner similar to siRNAs. This is often the case in plants where the target sites are typically highly complementary to the miRNA. Whereas target sites in plant mRNAs can be found in the 5'UTR, open-reading frames, and 3'UTR, in animals it is the 3' UTR that is the main target. This difference between plants and animals may be explained by their different modes of gene silencing.
miRNAs are first transcribed as part of a primary microRNA (pri-miRNA). This is then processed by the Drosha with the help of Pasha/DGCR8 (=Microprocessor complex) into pre-miRNAs. The ~75nt pre-miRNA is then exported to the cytoplasm by exportin-5,[11] where it is then diced into 21-23nt siRNA-like molecules by Dicer. In some cases, multiple miRNAs can be found on the same pri-miRNA.
References
- ^ Hamilton A, Baulcombe D (1999). "A species of small antisense RNA in posttranscriptional gene silencing in plants". Science. 286 (5441): 950–2. doi:10.1126/science.286.5441.950. PMID 10542148.
- ^ Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001). "Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells". Nature. 411 (6836): 494–8. doi:10.1038/35078107. PMID 11373684.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bernstein E, Caudy A, Hammond S, Hannon G (2001). "Role for a bidentate ribonuclease in the initiation step of RNA interference". Nature. 409 (6818): 363–6. doi:10.1038/35053110. PMID 11201747.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Li LC (2008). "Small RNA-Mediated Gene Activation". RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 978-1-904455-25-7.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Birmingham A, Anderson E, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, Marshall W, Khvorova A (2006). "3' UTR seed matches, but not overall identity, are associated with RNAi off-targets". Nat Methods. 3 (3): 199–204. doi:10.1038/nmeth854. PMID 16489337.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Alekseev OM, Richardson RT, Alekseev O, O'Rand MG. Analysis of gene expression profiles in HeLa cells in response to overexpression or siRNA-mediated depletion of NASP. Reprod Biol Endocrinol. 2009 May 13;7:45.PMID: 19439102
- ^ Tansey B (11 August 2006). "Macular degeneration treatment interferes with RNA messages". San Francisco Chronicle.
- ^ "Texas Tech Researchers May Have Found AIDS Cure". KCBD. 2008-08-07. Retrieved 2008-08-16.
{{cite news}}: Check date values in:|date=(help) - ^ Swaminathan, Nikhil (2008-08-07). "Researchers Silence HIV in Mice Engineered to Be Like Humans". Scientific American. Retrieved 2008-08-16.
{{cite news}}: Check date values in:|date=(help) - ^ "Researchers halt spread of HIV with RNAi". Harvard Medical School. Retrieved 2008-08-16.
- ^ Yi, R., Qin, Y., Macara, I.G., Cullen, B.R. (2003). "Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs". Genes & Development. 17 (24): 3011–3016. doi:10.1101/gad.1158803. PMID 14681208.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
General background
- Hannon G, Rossi J (2004). "Unlocking the potential of the human genome with RNA interference". Nature. 431 (7006): 371–8. doi:10.1038/nature02870. PMID 15372045.
- Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature. 391 (6669): 806–11. doi:10.1038/35888. PMID 9486653.
{{cite journal}}: CS1 maint: multiple names: authors list (link) The original paper by Fire et al. describing RNAi - Elbashir S, Lendeckel W, Tuschl T (2001). "RNA interference is mediated by 21- and 22-nucleotide RNAs". Genes Dev. 15 (2): 188–200. doi:10.1101/gad.862301. PMID 11157775.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Zamore P, Tuschl T, Sharp P, Bartel D (2000). "RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals". Cell. 101 (1): 25–33. doi:10.1016/S0092-8674(00)80620-0. PMID 10778853.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Hamilton A, Baulcombe D (1999). "A species of small antisense RNA in posttranscriptional gene silencing in plants". Science. 286 (5441): 950–2. doi:10.1126/science.286.5441.950. PMID 10542148. First description of siRNAs.
- Gartel, A.L. (2006). "RNA interference in cancer". Biomolecular Engineering. 23 (1): 17–34. doi:10.1016/j.bioeng.2006.01.002.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)
Non-specific effects
- Zhang Z, Weinschenk T, Guo K, Schluesener H (2006). "siRNA binding proteins of microglial cells: PKR is an unanticipated ligand". J Cell Biochem. 97 (6): 1217–29. doi:10.1002/jcb.20716. PMID 16315288.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Kim D, Behlke M, Rose S, Chang M, Choi S, Rossi J (2005). "Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy". Nat Biotechnol. 23 (2): 222–6. doi:10.1038/nbt1051. PMID 15619617.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Sledz C, Williams B (2004). "RNA interference and double-stranded-RNA-activated pathways". Biochem Soc Trans. 32 (Pt 6): 952–6. doi:10.1042/BST0320952. PMID 15506933.
- Pebernard S, Iggo R (2004). "Determinants of interferon-stimulated gene induction by RNAi vectors". Differentiation. 72 (2–3): 103–11. doi:10.1111/j.1432-0436.2004.07202001.x. PMID 15066190.
- Sledz C, Holko M, de Veer M, Silverman R, Williams B (2003). "Activation of the interferon system by short-interfering RNAs". Nat Cell Biol. 5 (9): 834–9. doi:10.1038/ncb1038. PMID 12942087.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
Tools for design and quality:
- siRNA at Whitehead: A web-based siRNA Selection Program
- Deqor: A web-based tool for the design and quality control of siRNAs
- siDirect: a web-based online software system for designing siRNA sequences
- siSearch: a siRNA design tool using a number of popular design methods
- SpecificityServer: a tool for checking the specificity of a given siRNA
- SEQ2SVM: software for designing siRNA sequences
- miRacle: tool for prediction of siRNA and microRNA targets using an algorithm which incorporates RNA secondary structure
- InvivoGen's siRNA Wizard: A web-based tool for designing siRNA and scrambled siRNA sequences
Databases:
siRNA focused Companies:
- Alnylam Pharmaceuticals
- Anadys Pharmaceuticals
- Ambion (a division of Applied Biosystems, Inc.)
- ArmaGen Technologies,Inc
- BD Biosciences
- Benitec
- Bionomics
- BioSpring
- Calando Pharmaceuticals
- Celera Genomics
- Cenix BioScience
- Ceptyr
- Dharmacon
- Genlantis
- Ingenium Pharmaceuticals
- Integrated DNA Technologies, Inc.
- Invitrogen
- InvivoGen
- Isis Pharmaceuticals
- Nucleonics Inc
- Pfizer Inc
- RNAi Co.Ltd
- Roche Kulmbach GmbH
- Sigma-Aldrich
- Sirna Therapeutics, Inc.
- SIRION-Biotech
- Thermo Fisher Scientific
- QIAGEN Sample & Assay Technologies
esiRNA (Endoribonuclease-prepared siRNA)