Vibrio cholerae: Difference between revisions
m Bot: Migrating 27 interwiki links, now provided by Wikidata on d:q160821 (Report Errors) |
→Genome: Wrong link |
||
| Line 19: | Line 19: | ||
==Genome== |
==Genome== |
||
''V. cholerae'' has two circular [[chromosomes]], together totalling 4 million [[base pairs]] of [[DNA]] sequence and 3,885 predicted [[genes]].<ref>{{cite journal |last1=Fraser |first1=Claire M. |last2=Heidelberg |first2=John F. |last3=Eisen |first3=Jonathan A. |last4=Nelson |first4=William C. |last5=Clayton |first5=Rebecca A. |last6=Gwinn |first6=Michelle L. |last7=Dodson |first7=Robert J. |last8=Haft |first8=Daniel H. |last9=Hickey |first9=Erin K. |title=DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae |journal=Nature |volume=406 |issue=6795 |pages=477–83 |year=2000 |pmid=10952301 |doi=10.1038/35020000}}</ref> The genes for cholera toxin are carried by CTXphi (CTXφ), a [[temperate]] [[bacteriophage]] inserted into the ''V. cholerae'' genome. CTXφ can transmit cholera toxin genes from one ''V. cholerae'' strain to another, one form of [[horizontal gene transfer]]. The genes for toxin coregulated pilus are coded by the VPI [[pathogenicity island]] (VPIφ). |
''V. cholerae'' has two circular [[chromosomes]], together totalling 4 million [[base pairs]] of [[DNA]] sequence and 3,885 predicted [[genes]].<ref>{{cite journal |last1=Fraser |first1=Claire M. |last2=Heidelberg |first2=John F. |last3=Eisen |first3=Jonathan A. |last4=Nelson |first4=William C. |last5=Clayton |first5=Rebecca A. |last6=Gwinn |first6=Michelle L. |last7=Dodson |first7=Robert J. |last8=Haft |first8=Daniel H. |last9=Hickey |first9=Erin K. |title=DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae |journal=Nature |volume=406 |issue=6795 |pages=477–83 |year=2000 |pmid=10952301 |doi=10.1038/35020000}}</ref> The genes for cholera toxin are carried by CTXphi (CTXφ), a [[temperateness (virology)|temperate]] [[bacteriophage]] inserted into the ''V. cholerae'' genome. CTXφ can transmit cholera toxin genes from one ''V. cholerae'' strain to another, one form of [[horizontal gene transfer]]. The genes for toxin coregulated pilus are coded by the VPI [[pathogenicity island]] (VPIφ). |
||
===Bacteriophage CTXφ=== |
===Bacteriophage CTXφ=== |
||
Revision as of 10:08, 28 February 2013
| Vibrio cholerae | |
|---|---|
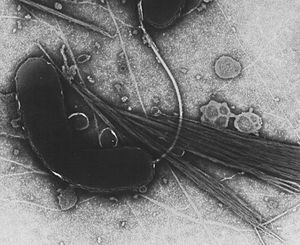
| |
| TEM image | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Class: | |
| Order: | Vibrionales
|
| Family: | |
| Genus: | |
| Species: | V. cholerae
|
| Binomial name | |
| Vibrio cholerae Pacini 1854
| |
Vibrio cholerae is a Gram-negative, comma-shaped bacterium. Some strains of V. cholerae cause the disease cholera. V. cholerae is facultatively anaerobic and has a flagellum at one cell pole. V. cholerae was first isolated as the cause of cholera by Italian anatomist Filippo Pacini in 1854, but his discovery was not widely known until Robert Koch, working independently 30 years later, publicized the knowledge and the means of fighting the disease.[1][2]
Pathogenesis
V. cholerae pathogenicity genes code for proteins directly or indirectly involved in the virulence of the bacteria. During infection, V. cholerae secretes cholera toxin, a protein that causes profuse, watery diarrhea. Colonization of the small intestine also requires the toxin coregulated pilus (TCP), a thin, flexible, filamentous appendage on the surface of bacterial cells.
Genome
V. cholerae has two circular chromosomes, together totalling 4 million base pairs of DNA sequence and 3,885 predicted genes.[3] The genes for cholera toxin are carried by CTXphi (CTXφ), a temperate bacteriophage inserted into the V. cholerae genome. CTXφ can transmit cholera toxin genes from one V. cholerae strain to another, one form of horizontal gene transfer. The genes for toxin coregulated pilus are coded by the VPI pathogenicity island (VPIφ).
Bacteriophage CTXφ
CTXφ (also called CTXphi) is a filamentous phage that contains the genes for cholera toxin. Infectious CTXφ particles are produced when V. cholerae infects humans. Phage particles are secreted from bacterial cells without lysis. When CTXφ infects V. cholerae cells, it integrates into specific sites on either chromosome. These sites often contain tandem arrays of integrated CTXφ prophage. In addition to the ctxA and ctxB genes encoding cholera toxin, CTXφ contains eight genes involved in phage reproduction, packaging, secretion, integration, and regulation. The CTXφ genome is 6.9 kb long.[4]
VPI pathogenicity island
The VPI pathogenicity island contains genes involved in the creation of toxin coregulated pilus (TCP). It is a large genetic element (~40 kb) flanked by two repetitive regions (att-like sites), resembling a transposon in structure. The VPI pathogenicity island is composed of two gene clusters: the TCP cluster and the ACF cluster. Among the genes of the VPI pathogenicity island, twenty genes have been identified, where some genes are outside of the clusters like tagA, tagB and aldA[clarification needed].[5] The acf cluster is composed of 4 genes: acfABC and tagE, encoding a putative accessory colonization factor activated by toxR. The tcp cluster is composed of 15 genes: tcpABCDEFHIJPQRST and regulatory gene toxT.
Ecology and epidemiology
The main reservoirs of V. cholerae are people and aquatic sources such as brackish water and estuaries, often in association with copepods or other zooplankton, shellfish, and aquatic plants.
Cholera infections are most commonly acquired from drinking water in which V. cholerae is found naturally or into which it has been introduced from the feces of an infected person. Other common vehicles include contaminated fish and shellfish, produce, or leftover cooked grains that have not been properly reheated. Transmission from person to person, even to health care workers during epidemics, is rarely documented.
Diversity and evolution
Two serogroups of V. cholerae, O1 and O139, cause outbreaks of cholera. O1 causes the majority of outbreaks, while O139 – first identified in Bangladesh in 1992 – is confined to South-East Asia. Many other serogroups of V. cholerae, with or without the cholera toxin gene (including the nontoxigenic strains of the O1 and O139 serogroups), can cause a cholera-like illness. Only toxigenic strains of serogroups O1 and O139 have caused widespread epidemics.
V. cholerae O1 has 2 biotypes, classical and El Tor, and each biotype has 2 distinct serotypes, Inaba and Ogawa. The symptoms of infection are indistinguishable, although more people infected with the El Tor biotype remain asymptomatic or have only a mild illness. In recent years, infections with the classical biotype of V. cholerae O1 have become rare and are limited to parts of Bangladesh and India.[6] Recently, new variant strains have been detected in several parts of Asia and Africa. Observations suggest that these strains cause more severe cholera with higher case fatality rates.
Gallery
-
Vibrio cholera bacteria
-
Diagram of the bacterium, V. cholerae
See also
References
Notes
- ^ Bentivoglio, M; Pacini, P (1995). "Filippo Pacini: A determined observer". Brain Research Bulletin. 38 (2): 161–5. doi:10.1016/0361-9230(95)00083-Q. PMID 7583342.
- ^ Howard-Jones, N (1984). "Robert Koch and the cholera vibrio: a centenary". BMJ. 288 (6414): 379–81. doi:10.1136/bmj.288.6414.379. PMC 1444283. PMID 6419937.
- ^ Fraser, Claire M.; Heidelberg, John F.; Eisen, Jonathan A.; Nelson, William C.; Clayton, Rebecca A.; Gwinn, Michelle L.; Dodson, Robert J.; Haft, Daniel H.; Hickey, Erin K. (2000). "DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae". Nature. 406 (6795): 477–83. doi:10.1038/35020000. PMID 10952301.
- ^ McLeod, S. M.; Kimsey, H. H.; Davis, B. M.; Waldor, M. K. (2005). "CTXφ and Vibrio cholerae: exploring a newly recognized type of phage-host cell relationship". Molecular Microbiology. 57: 347–356. doi:10.1111/j.1365-2958.04676.x. PMID 15978069.
- ^ Karaolis, David K. R.; Somara, Sita; Maneval, David R.; Johnson, Judith A.; Kaper, James B. (1999). "A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria". Nature. 399 (6734): 375–9. doi:10.1038/20715. PMID 10360577.
- ^ Siddique, A.K.; Baqui, A.H.; Eusof, A.; Haider, K.; Hossain, M.A.; Bashir, I.; Zaman, K. (1991). "Survival of classic cholera in Bangladesh". The Lancet. 337 (8750): 1125–1127. doi:10.1016/0140-6736(91)92789-5.


