MT-ATP6: Difference between revisions
m Updated protein structure link |
m fixed research reference |
||
| Line 3: | Line 3: | ||
[[File:Map of the human mitochondrial genome.svg|thumb|300px|Location of the ''MT-ATP6'' gene on the L strand of the human mitochondrial genome. ''MT-ATP6'' is one of the two ATP synthase mitochondrial genes (red boxes).]] |
[[File:Map of the human mitochondrial genome.svg|thumb|300px|Location of the ''MT-ATP6'' gene on the L strand of the human mitochondrial genome. ''MT-ATP6'' is one of the two ATP synthase mitochondrial genes (red boxes).]] |
||
'''''MT-ATP6''''' (or '''''ATP6''''') is a [[mitochondrial gene]] encoding the '''ATP synthase F<sub>o</sub> subunit 6''' (or '''subunit/chain A)'''. This subunit belongs to the F<sub>o</sub> complex of transmembrane F-type [[ATP synthase]].<ref name="pmid7219534">{{cite journal | vauthors = Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG | title = Sequence and organization of the human mitochondrial genome | journal = Nature | volume = 290 | issue = 5806 | pages = 457–65 | date = April 1981 | pmid = 7219534 | doi = 10.1038/290457a0 }}</ref> The MT-ATP6 protein forms one part of a large enzyme called ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation. Specifically, one segment of ATP synthase allows positively charged particles, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Mutations in the ''MT-ATP6'' gene have been found in approximately 10 to 20 percent of people with [[Leigh syndrome]]. |
'''''MT-ATP6''''' (or '''''ATP6''''') is a [[mitochondrial gene]] encoding the '''ATP synthase F<sub>o</sub> subunit 6''' (or '''subunit/chain A)'''. This subunit belongs to the F<sub>o</sub> complex of transmembrane F-type [[ATP synthase]].<ref name="pmid7219534">{{cite journal | vauthors = Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG | title = Sequence and organization of the human mitochondrial genome | journal = Nature | volume = 290 | issue = 5806 | pages = 457–65 | date = April 1981 | pmid = 7219534 | doi = 10.1038/290457a0 }}</ref> The MT-ATP6 protein forms one part of a large enzyme called ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of [[oxidative phosphorylation]]. Specifically, one segment of ATP synthase allows positively charged particles, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Mutations in the ''MT-ATP6'' gene have been found in approximately 10 to 20 percent of people with [[Leigh syndrome]]. |
||
==Gene== |
==Gene== |
||
| Line 46: | Line 46: | ||
== Research == |
== Research == |
||
The [[SENS Research Foundation]] have published a paper detailing the successful allotopic expression of replacement DNA for the MT-ATP6 gene in the cell nuclear DNA.<ref> |
The [[SENS Research Foundation]] have published a paper detailing the successful allotopic expression of replacement DNA for the MT-ATP6 gene in the cell nuclear DNA.<ref>{{cite journal |last1=Boominathan |first1=Amutha |last2=Vanhoozer |first2=Shon |last3=Basisty |first3=Nathan |last4=Powers |first4=Kathleen |last5=Crampton |first5=Alexandra L. |last6=Wang |first6=Xiaobin |last7=Friedricks |first7=Natalie |last8=Schilling |first8=Birgit |last9=Brand |first9=Martin D. |last10=O'Connor |first10=Matthew S. |title=Stable nuclear expression of and genes rescues a mtDNA Complex V mutant |journal=Nucleic Acids Research |date=4 September 2016 |volume=44 |issue=19 |pages=9342–9357 |doi=https://doi.org/10.1093/nar/gkw756}}</ref> |
||
== References == |
== References == |
||
Revision as of 00:02, 31 July 2018
| ATP6 | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ATP6, ATPase6, MTATP synthase Fo subunit 6 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 516060; MGI: 99927; HomoloGene: 5012; GeneCards: ATP6; OMA:ATP6 - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
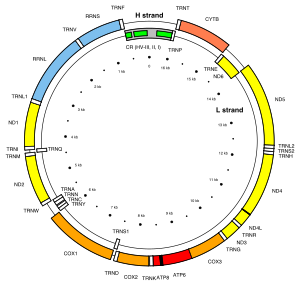
MT-ATP6 (or ATP6) is a mitochondrial gene encoding the ATP synthase Fo subunit 6 (or subunit/chain A). This subunit belongs to the Fo complex of transmembrane F-type ATP synthase.[5] The MT-ATP6 protein forms one part of a large enzyme called ATP synthase. This enzyme, which is also known as complex V, is responsible for the final step of oxidative phosphorylation. Specifically, one segment of ATP synthase allows positively charged particles, called protons, to flow across a specialized membrane inside mitochondria. Another segment of the enzyme uses the energy created by this proton flow to convert a molecule called adenosine diphosphate (ADP) to ATP. Mutations in the MT-ATP6 gene have been found in approximately 10 to 20 percent of people with Leigh syndrome.
Gene
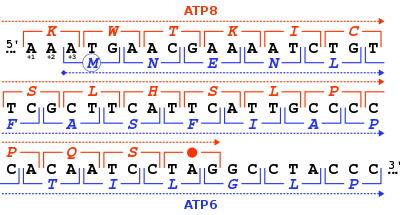
The MT-ATP6 gene provides information for making a protein that is essential for normal mitochondrial function. The human MT-ATP6 gene, located in mitochondrial DNA, is 681 base pairs in length.[6]
An unusual feature of MT-ATP6 is the 46-nucleotide gene overlap of its first codons with the end of the MT-ATP8 gene. With respect to the MT-ATP6 reading frame (+3), the MT-ATP8 gene ends in the +1 reading frame with a TAG stop codon.
Protein structure
| ATP synthase A chain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | ATP-synt_A | ||||||||||
| Pfam | PF00119 | ||||||||||
| InterPro | IPR000568 | ||||||||||
| PROSITE | PDOC00420 | ||||||||||
| SCOP2 | 1c17 / SCOPe / SUPFAM | ||||||||||
| OPM superfamily | 5 | ||||||||||
| OPM protein | 1c17 | ||||||||||
| |||||||||||
The MT-ATP6 protein weighs 24.8 kDa and is composed of 226 amino acids.[7][8] The protein is a subunit of the F1Fo ATPase, also known as Complex V, which consists of 14 nuclear- and 2 mitochondrial-encoded subunits. As an A subunit, MT-ATP6 is contained within the non-catalytic, transmembrane Fo portion of the complex.[6] The nomenclature of the enzyme has a long history. The F1 fraction derives its name from the term "Fraction 1" and Fo (written as a subscript letter "o", not "zero") derives its name from being the binding fraction for oligomycin, a type of naturally-derived antibiotic that is able to inhibit the Fo unit of ATP synthase.[9][10] The Fo region of ATP synthase is a proton pore that is embedded in the mitochondrial membrane. It consists of three main subunits A, B, and C, and (in humans) six additional subunits, d, e, f, g, F6, and 8 (or A6L). 3D structure of E. coli homologue of this subunit was modeled based on electron microscopy data (chain M of PDB: 1c17). It forms a transmembrane 4-α-bundle.
Function
This subunit is a key component of the proton channel, and may play a direct role in the translocation of protons across the membrane. Catalysis in the F1 complex depends upon the rotation of the central stalk and Fo c-ring, which in turn is driven by the flux of protons through the membrane via the interface between the F0 c-ring and subunit A. The peripheral stalk links subunit A to the external surface of the F1 domain, and is thought to act as a stator to counter the tendency of subunit A and the F1alpha3 beta3 catalytic portion to rotate with the central rotary element.[11]
Clinical significance
Pathogenic variants of the mitochondrial gene MT-ATP6 are known to cause mtDNA-associated Leigh syndrome, a progressive brain disorder that usually appears in infancy or early childhood. Affected children may experience delayed development, muscle weakness, problems with movement, or difficulty breathing.[12] Other variants known to cause mtDNA-associated Leigh syndrome involve MT-TL1, MT-TK, MT-TW, MT-TV, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND5, MT-ND6 and MT-CO3. Abnormalities in mitochondrial energy generation result in neurodegenerative disorders like Leigh syndrome, which is characterized by an onset of symptoms between 12 months and three years of age. The symptoms frequently present themselves following a viral infection and include movement disorders and peripheral neuropathy, as well as hypotonia, spasticity and cerebellar ataxia. Roughly half of affected patients die of respiratory or cardiac failure by the age of three. Leigh syndrome is a maternally inherited disorder and its diagnosis is established through genetic testing of the aforementioned mitochondrial genes, including MT-ATP6.[13] MT-ATP6 gene mutations associated with Leigh syndrome change one DNA building block (nucleotide) in the MT-ATP6 gene. The most common genetic change replaces the nucleotide thymine with the nucleotide guanine at position 8993 (written as T8993G). The mutations that cause Leigh syndrome impair the function or stability of the ATP synthase complex, inhibiting ATP production and impairing oxidative phosphorylation. Although the exact mechanism is unclear, researchers believe that impaired oxidative phosphorylation can lead to cell death because of decreased energy available in the cell. Certain tissues that require large amounts of energy, such as the brain, muscles, and heart, seem especially sensitive to decreases in cellular energy. Cell death in the brain likely causes the characteristic changes in the brain seen in Leigh syndrome, which contribute to the signs and symptoms of the condition. Cell death in other sensitive tissues may also contribute to the features of Leigh syndrome. A heteroplasmic T→C MT-ATP6 mutation at position 9185 results in the substitution of a highly conserved leucine to proline at codon 220 and a heteroplasmic T→C missense mutation at position 9191 converted a highly conserved leucine to a proline at position 222 of the polypeptide, leading to a Leigh-type phenotype. The T9185C mutation resulted in a mild and reversible phenotype, with 97% of the patient's muscle and blood samples reflecting the mutation. The T9191C mutation presented a much more severe phenotype that resulted in the death of the patient at 2 years of age.[14] Mutations to these oxidative phosphorylation genes have been associated with a variety of neurodegenerative disorders, including Leber's hereditary optic neuropathy (LHON), mitochondrial encephalomyopathy with stroke-like episodes (MELAS) and the previously mentioned Leigh syndrome.
Some of the mutations of the ATP6 gene that cause Leigh syndrome are also responsible for a similar, but less severe, condition called neuropathy, ataxia, and retinitis pigmentosa (NARP).[15] A small number of mutations in the MT-ATP6 gene have been identified in people with NARP. Each of these mutations changes one nucleotide in the MT-ATP6 gene. As in Leigh syndrome, the most common genetic change associated with NARP replaces the nucleotide thymine with the nucleotide guanine at position 8993 (written as T8993G). The mutations that cause NARP alter the structure or function of ATP synthase, reducing the ability of mitochondria to produce ATP. Although the precise effects of these mutations are unclear, researchers continue to investigate how changes in the MT-ATP6 gene interfere with ATP production and lead to muscle weakness, vision loss, and the other features of NARP.[12]
Most of the body's cells contain thousands of mitochondria, each with one or more copies of mitochondrial DNA. The severity of some mitochondrial disorders is associated with the percentage of mitochondria in each cell that has a particular genetic change. People with Leigh syndrome due to a MT-ATP6 gene mutation tend to have a very high percentage of mitochondria with the mutation (from more than 90 percent to 95 percent). The less-severe features of NARP result from a lower percentage of mitochondria with the mutation, typically 70 percent to 90 percent. Because these two conditions result from the same genetic changes and can occur in different members of a single family, researchers believe that they may represent a spectrum of overlapping features instead of two distinct syndromes.[12]
A condition called familial bilateral striatal necrosis, which is similar to Leigh syndrome, can also result from changes in the MT-ATP6 gene. In the few reported cases with these mutations, affected children have had delayed development, problems with movement and coordination, weak muscle tone (hypotonia), and an unusually small head size (microcephaly). Researchers have not determined why MT-ATP6 mutations result in this combination of signs and symptoms in children with bilateral striatal necrosis.[12]
Research
The SENS Research Foundation have published a paper detailing the successful allotopic expression of replacement DNA for the MT-ATP6 gene in the cell nuclear DNA.[16]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000198899 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000064357 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (April 1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–65. doi:10.1038/290457a0. PMID 7219534.
- ^ a b "Entrez Gene: MT-ATP6 mitochondrially encoded ATP synthase 6".
- ^ Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, Deng N, Kim AK, Choi JH, Zelaya I, Liem D, Meyer D, Odeberg J, Fang C, Lu HJ, Xu T, Weiss J, Duan H, Uhlen M, Yates JR, Apweiler R, Ge J, Hermjakob H, Ping P (Oct 2013). "Integration of cardiac proteome biology and medicine by a specialized knowledgebase". Circulation Research. 113 (9): 1043–53. doi:10.1161/CIRCRESAHA.113.301151. PMC 4076475. PMID 23965338.
- ^ "ATP synthase subunit A, mitochondrial". Cardiac Organellar Protein Atlas Knowledgebase (COPaKB).
- ^ Kagawa Y, Racker E (May 1966). "Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase". The Journal of Biological Chemistry. 241 (10): 2461–6. PMID 4223640.
- ^ Mccarty RE (November 1992). "A plant biochemist's view of H+
-ATPases and ATP synthases". The Journal of Experimental Biology. 172 (Pt 1): 431–441. PMID 9874753. - ^ Carbajo RJ, Kellas FA, Runswick MJ, Montgomery MG, Walker JE, Neuhaus D (August 2005). "Structure of the F1-binding domain of the stator of bovine F1Fo-ATPase and how it binds an alpha-subunit". Journal of Molecular Biology. 351 (4): 824–38. doi:10.1016/j.jmb.2005.06.012. PMID 16045926.
- ^ a b c d "MT-ATP6". Genetics Home Resource. NCBI.
- ^ Thorburn DR, Rahman S (1993–2015). "Mitochondrial DNA-Associated Leigh Syndrome and NARP". In Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, Bird TD, Dolan CR, Fong CT, Smith RJ, Stephens K (eds.). GeneReviews [Internet]. Seattle (WA): University of Washington, Seattle.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ Moslemi AR, Darin N, Tulinius M, Oldfors A, Holme E (October 2005). "Two new mutations in the MTATP6 gene associated with Leigh syndrome". Neuropediatrics. 36 (5): 314–8. doi:10.1055/s-2005-872845. PMID 16217706.
- ^ Baracca A, Sgarbi G, Mattiazzi M, Casalena G, Pagnotta E, Valentino ML, Moggio M, Lenaz G, Carelli V, Solaini G (July 2007). "Biochemical phenotypes associated with the mitochondrial ATP6 gene mutations at nt8993". Biochimica et Biophysica Acta. 1767 (7): 913–9. doi:10.1016/j.bbabio.2007.05.005. PMID 17568559.
- ^ Boominathan, Amutha; Vanhoozer, Shon; Basisty, Nathan; Powers, Kathleen; Crampton, Alexandra L.; Wang, Xiaobin; Friedricks, Natalie; Schilling, Birgit; Brand, Martin D.; O'Connor, Matthew S. (4 September 2016). "Stable nuclear expression of and genes rescues a mtDNA Complex V mutant". Nucleic Acids Research. 44 (19): 9342–9357. doi:https://doi.org/10.1093/nar/gkw756.
{{cite journal}}: Check|doi=value (help); External link in|doi=
Further reading
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ (June 2006). "Harvesting the fruit of the human mtDNA tree". Trends in Genetics. 22 (6): 339–45. doi:10.1016/j.tig.2006.04.001. PMID 16678300.
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (December 2000). "Mitochondrial genome variation and the origin of modern humans". Nature. 408 (6813): 708–13. doi:10.1038/35047064. PMID 11130070.
- Manfredi G, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA (April 2002). "Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus". Nature Genetics. 30 (4): 394–9. doi:10.1038/ng851. PMID 11925565.
- Torigoe T, Izumi H, Ishiguchi H, Uramoto H, Murakami T, Ise T, Yoshida Y, Tanabe M, Nomoto M, Itoh H, Kohno K (September 2002). "Enhanced expression of the human vacuolar H+-ATPase c subunit gene (ATP6L) in response to anticancer agents". The Journal of Biological Chemistry. 277 (39): 36534–43. doi:10.1074/jbc.M202605200. PMID 12133827.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - Mishmar D, Ruiz-Pesini E, Golik P, Macaulay V, Clark AG, Hosseini S, Brandon M, Easley K, Chen E, Brown MD, Sukernik RI, Olckers A, Wallace DC (January 2003). "Natural selection shaped regional mtDNA variation in humans". Proceedings of the National Academy of Sciences of the United States of America. 100 (1): 171–6. doi:10.1073/pnas.0136972100. PMC 140917. PMID 12509511.
- Ingman M, Gyllensten U (July 2003). "Mitochondrial genome variation and evolutionary history of Australian and New Guinean aborigines". Genome Research. 13 (7): 1600–6. doi:10.1101/gr.686603. PMC 403733. PMID 12840039.
- Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP (September 2003). "Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences". American Journal of Human Genetics. 73 (3): 671–6. doi:10.1086/377718. PMC 1180693. PMID 12870132.
- Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZM (September 2003). "Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria". Human Molecular Genetics. 12 (18): 2341–8. doi:10.1093/hmg/ddg238. PMID 12915481.
- Reuter TY, Medhurst AL, Waisfisz Q, Zhi Y, Herterich S, Hoehn H, Gross HJ, Joenje H, Hoatlin ME, Mathew CG, Huber PA (October 2003). "Yeast two-hybrid screens imply involvement of Fanconi anemia proteins in transcription regulation, cell signaling, oxidative metabolism, and cellular transport". Experimental Cell Research. 289 (2): 211–21. doi:10.1016/S0014-4827(03)00261-1. PMID 14499622.
- Dubot A, Godinot C, Dumur V, Sablonnière B, Stojkovic T, Cuisset JM, Vojtiskova A, Pecina P, Jesina P, Houstek J (January 2004). "GUG is an efficient initiation codon to translate the human mitochondrial ATP6 gene". Biochemical and Biophysical Research Communications. 313 (3): 687–93. doi:10.1016/j.bbrc.2003.12.013. PMID 14697245.
- Coble MD, Just RS, O'Callaghan JE, Letmanyi IH, Peterson CT, Irwin JA, Parsons TJ (June 2004). "Single nucleotide polymorphisms over the entire mtDNA genome that increase the power of forensic testing in Caucasians". International Journal of Legal Medicine. 118 (3): 137–46. doi:10.1007/s00414-004-0427-6. PMID 14760490.
- Carrozzo R, Rizza T, Stringaro A, Pierini R, Mormone E, Santorelli FM, Malorni W, Matarrese P (July 2004). "Maternally-inherited Leigh syndrome-related mutations bolster mitochondrial-mediated apoptosis". Journal of Neurochemistry. 90 (2): 490–501. doi:10.1111/j.1471-4159.2004.02505.x. PMID 15228605.
External links
- GeneReviews/NCBI/NIH/UW entry on Mitochondrial DNA-Associated Leigh Syndrome and NARP
- MT-ATP6+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.