Computational chemistry: Difference between revisions
added new edits and removed the template |
No edit summary |
||
| Line 291: | Line 291: | ||
{{Main article|Ab initio quantum chemistry methods}} |
{{Main article|Ab initio quantum chemistry methods}} |
||
The programs used in computational chemistry are based on many different [[quantum chemistry|quantum-chemical]] methods that solve the molecular [[Schrödinger equation]] associated with the [[molecular Hamiltonian]]. Methods that do not include any empirical or semi-empirical parameters in their equations – being derived directly from theoretical principles, with no inclusion of experimental data – are called ''[[ab initio quantum chemistry methods|ab initio methods]]''.<ref name=":9">{{Cite journal |date=2024-01-01 |title=The Effective Fragment Potential: An Ab Initio Force Field |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782001410 |language=en-US |pages=153–161 |doi=10.1016/B978-0-12-821978-2.00141-0}}</ref> This does not imply that the solution is an exact one; they are all approximate quantum mechanical calculations. It means that a particular approximation is rigorously defined on first principles (quantum theory) and then solved within an error margin that is qualitatively known beforehand. If numerical iterative methods must be used, the aim is to iterate until full machine accuracy is obtained (the best that is possible with a finite [[word length]] on the computer, and within the mathematical and/or physical approximations made). |
The programs used in computational chemistry are based on many different [[quantum chemistry|quantum-chemical]] methods that solve the molecular [[Schrödinger equation]] associated with the [[molecular Hamiltonian]]. Methods that do not include any empirical or semi-empirical parameters in their equations – being derived directly from theoretical principles, with no inclusion of experimental data – are called ''[[ab initio quantum chemistry methods|ab initio methods]]''.<ref name=":9">{{Cite journal |date=2024-01-01 |title=The Effective Fragment Potential: An Ab Initio Force Field |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782001410 |journal=Comprehensive Computational Chemistry |language=en-US |pages=153–161 |doi=10.1016/B978-0-12-821978-2.00141-0}}</ref> This does not imply that the solution is an exact one; they are all approximate quantum mechanical calculations. It means that a particular approximation is rigorously defined on first principles (quantum theory) and then solved within an error margin that is qualitatively known beforehand. If numerical iterative methods must be used, the aim is to iterate until full machine accuracy is obtained (the best that is possible with a finite [[word length]] on the computer, and within the mathematical and/or physical approximations made). |
||
[[File:Electron correlation.svg|thumb|right|300px|Diagram illustrating various ''ab initio'' electronic structure methods in terms of energy. Spacings are not to scale.]] |
[[File:Electron correlation.svg|thumb|right|300px|Diagram illustrating various ''ab initio'' electronic structure methods in terms of energy. Spacings are not to scale.]] |
||
| Line 306: | Line 306: | ||
{{Main article|Density functional theory}} |
{{Main article|Density functional theory}} |
||
Density functional theory (DFT) methods are often considered to be ''[[ab initio quantum chemistry methods|ab initio methods]]'' for determining the molecular electronic structure, even though many of the most common [[Functional (mathematics)|functionals]] use parameters derived from empirical data, or from more complex calculations. In DFT, the total energy is expressed in terms of the total one-[[electronic density|electron density]] rather than the wave function.<ref>{{Cite journal |date=2024-01-01 |title=Conceptual Density Functional Theory |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782000258 |language=en-US |pages=306–321 |doi=10.1016/B978-0-12-821978-2.00025-8}}</ref> In this type of calculation, there is an approximate [[Hamiltonian (quantum mechanics)|Hamiltonian]] and an approximate expression for the total electron density. DFT methods can be very accurate for little computational cost. Some methods combine the density functional exchange functional with the Hartree–Fock exchange term and are termed [[hybrid functional]] methods. |
Density functional theory (DFT) methods are often considered to be ''[[ab initio quantum chemistry methods|ab initio methods]]'' for determining the molecular electronic structure, even though many of the most common [[Functional (mathematics)|functionals]] use parameters derived from empirical data, or from more complex calculations. In DFT, the total energy is expressed in terms of the total one-[[electronic density|electron density]] rather than the wave function.<ref>{{Cite journal |date=2024-01-01 |title=Conceptual Density Functional Theory |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782000258 |journal=Comprehensive Computational Chemistry |language=en-US |pages=306–321 |doi=10.1016/B978-0-12-821978-2.00025-8}}</ref> In this type of calculation, there is an approximate [[Hamiltonian (quantum mechanics)|Hamiltonian]] and an approximate expression for the total electron density. DFT methods can be very accurate for little computational cost. Some methods combine the density functional exchange functional with the Hartree–Fock exchange term and are termed [[hybrid functional]] methods. |
||
=== Semi-empirical methods === |
=== Semi-empirical methods === |
||
| Line 359: | Line 359: | ||
{{Main article|Molecular dynamics}} |
{{Main article|Molecular dynamics}} |
||
Molecular dynamics (MD) use either [[quantum mechanics]], [[molecular mechanics]] or a [[QM/MM|mixture of both]] to calculate forces which are then used to solve [[Newton's laws of motion]] to examine the time-dependent behavior of systems.<ref>{{Cite journal |date=2024-01-01 |title=Ab Initio Molecular Dynamics: A Guide to Applications |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782000969 |language=en-US |pages=493–517 |doi=10.1016/B978-0-12-821978-2.00096-9}}</ref> The result of a molecular dynamics simulation is a trajectory that describes how the position and velocity of particles varies with time. The phase point of a system described by the positions and momenta of all its particles on a previous time point will determine the next phase point in time by integrating over Newton's laws of motion. |
Molecular dynamics (MD) use either [[quantum mechanics]], [[molecular mechanics]] or a [[QM/MM|mixture of both]] to calculate forces which are then used to solve [[Newton's laws of motion]] to examine the time-dependent behavior of systems.<ref>{{Cite journal |date=2024-01-01 |title=Ab Initio Molecular Dynamics: A Guide to Applications |url=https://www.sciencedirect.com/science/article/abs/pii/B9780128219782000969 |journal=Comprehensive Computational Chemistry |language=en-US |pages=493–517 |doi=10.1016/B978-0-12-821978-2.00096-9}}</ref> The result of a molecular dynamics simulation is a trajectory that describes how the position and velocity of particles varies with time. The phase point of a system described by the positions and momenta of all its particles on a previous time point will determine the next phase point in time by integrating over Newton's laws of motion. |
||
=== Monte Carlo === |
=== Monte Carlo === |
||
| Line 367: | Line 367: | ||
{{Main article|QM/MM}} |
{{Main article|QM/MM}} |
||
QM/MM is a hybrid method that attempts to combine the accuracy of quantum mechanics with the speed of molecular mechanics.<ref>{{Cite journal |date=2024-01-01 |title=Molecular Dynamics and QM/MM to Understand Genome Organization and Reproduction in Emerging RNA Viruses |url=https://www.sciencedirect.com/science/article/abs/pii/B978012821978200101X |language=en-US |pages=895–909 |doi=10.1016/B978-0-12-821978-2.00101-X}}</ref> It is useful for simulating very large molecules such as [[enzyme]]s. |
QM/MM is a hybrid method that attempts to combine the accuracy of quantum mechanics with the speed of molecular mechanics.<ref>{{Cite journal |date=2024-01-01 |title=Molecular Dynamics and QM/MM to Understand Genome Organization and Reproduction in Emerging RNA Viruses |url=https://www.sciencedirect.com/science/article/abs/pii/B978012821978200101X |journal=Comprehensive Computational Chemistry |language=en-US |pages=895–909 |doi=10.1016/B978-0-12-821978-2.00101-X}}</ref> It is useful for simulating very large molecules such as [[enzyme]]s. |
||
== Accuracy == |
== Accuracy == |
||
| Line 414: | Line 414: | ||
== References == |
== References == |
||
{{Reflist}} |
{{Reflist}} |
||
* |
|||
== General bibliography == |
|||
* C. J. Cramer ''Essentials of Computational Chemistry'', John Wiley & Sons (2002). |
|||
* T. Clark ''A Handbook of Computational Chemistry'', Wiley, New York (1985). |
|||
* {{cite book|year=2005|last1=Dronskowski|first1=Richard|s2cid=99908474|title=Computational Chemistry of Solid State Materials: A Guide for Materials Scientists, Chemists, Physicists and others|doi=10.1002/9783527612277|isbn=978-3-527-31410-2}} |
|||
* A.K. Hartmann, [https://web.archive.org/web/20090211113048/http://worldscibooks.com/physics/6988.html Practical Guide to Computer Simulations], [[World Scientific]] (2009) |
|||
* F. Jensen ''Introduction to Computational Chemistry'', John Wiley & Sons (1999). |
|||
* K.I. Ramachandran, G Deepa and Krishnan Namboori. P.K. ''Computational Chemistry and Molecular Modeling Principles and applications'' Springer-Verlag GmbH {{ISBN|978-3-540-77302-3}}. |
|||
* {{cite book|doi=10.1002/0471474908|title=Computational Chemistry Using the PC|url=https://archive.org/details/computationalche0000roge|url-access=registration|year=2003|last1=Rogers|first1=Donald W.|isbn=0-471-42800-0}} |
|||
* P. v. R. Schleyer (Editor-in-Chief). ''[http://eu.wiley.com/WileyCDA/WileyTitle/productCd-047196588X.html Encyclopedia of Computational Chemistry]''. Wiley, '''1998'''. {{ISBN|0-471-96588-X}}. |
|||
* D. Sherrill. [http://vergil.chemistry.gatech.edu/notes/ Notes on Quantum Mechanics and Computational Chemistry]. |
|||
* J. Simons ''An introduction to Theoretical Chemistry'', Cambridge (2003) {{ISBN|978-0-521-53047-7}}. |
|||
* A. Szabo, N.S. Ostlund, ''Modern Quantum Chemistry'', McGraw-Hill (1982). |
|||
* D. Young ''Computational Chemistry: A Practical Guide for Applying Techniques to Real World Problems'', John Wiley & Sons (2001). |
|||
* D. Young's [http://www.ccl.net/cca/documents/dyoung/topics-orig/compchem.html Introduction to Computational Chemistry]. |
|||
* {{cite book|doi=10.1007/978-90-481-3862-3|publisher= Springer |location=Heidelberg|title= Computational Chemistry |year= 2011 |last1= Lewars |first1= Errol G. |bibcode= 2011coch.book.....L |isbn= 978-90-481-3860-9}} |
|||
== Specialized journals on computational chemistry == |
== Specialized journals on computational chemistry == |
||
Revision as of 09:30, 3 December 2023
Computational chemistry is a branch of chemistry that uses computer simulation to assist in solving chemical problems. It uses methods of theoretical chemistry, incorporated into computer programs, to calculate the structures and properties of molecules, groups of molecules, and solids. It is essential because, apart from relatively recent results concerning the hydrogen molecular ion (dihydrogen cation, see references therein for more details), the quantum many-body problem cannot be solved analytically, much less in closed form. While computational results normally complement the information obtained by chemical experiments, it can in some cases predict hitherto unobserved chemical phenomena. It is widely used in the design of new drugs and materials.[1]
Examples of such properties are structure (i.e., the expected positions of the constituent atoms), absolute and relative (interaction) energies, electronic charge density distributions, dipoles and higher multipole moments, vibrational frequencies, reactivity, or other spectroscopic quantities, and cross sections for collision with other particles.
The methods used cover both static and dynamic situations. In all cases, the computer time and other resources (such as memory and disk space) increase quickly with the size of the system being studied. That system can be a molecule, a group of molecules, or a solid. Computational chemistry methods range from very approximate to highly accurate; the latter is usually feasible for small systems only. Ab initio methods are based entirely on quantum mechanics and basic physical constants. Other methods are called empirical or semi-empirical because they use additional empirical parameters.
Both ab initio and semi-empirical approaches involve approximations. These range from simplified forms of the first-principles equations that are easier or faster to solve, to approximations limiting the size of the system (for example, periodic boundary conditions), to fundamental approximations to the underlying equations that are required to achieve any solution to them at all. For example, most ab initio calculations make the Born–Oppenheimer approximation, which greatly simplifies the underlying Schrödinger equation by assuming that the nuclei remain in place during the calculation. In principle, ab initio methods eventually converge to the exact solution of the underlying equations as the number of approximations is reduced. In practice, however, it is impossible to eliminate all approximations, and residual error inevitably remains. The goal of computational chemistry is to minimize this residual error while keeping the calculations tractable.
In some cases, the details of electronic structure are less important than the long-time phase space behavior of molecules. This is the case in conformational studies of proteins and protein-ligand binding thermodynamics. Classical approximations to the potential energy surface are used, typically with molecular mechanics force fields, as they are computationally less intensive than electronic calculations, to enable longer simulations of molecular dynamics. Furthermore, cheminformatics uses even more empirical (and computationally cheaper) methods like machine learning based on physicochemical properties. One typical problem in cheminformatics is to predict the binding affinity of drug molecules to a given target. Other problems include predicting binding specificity, off-target effects, toxicity, and pharmacokinetic properties.
History
Building on the founding discoveries and theories in the history of quantum mechanics, the first theoretical calculations in chemistry were those of Walter Heitler and Fritz London in 1927, using valence bond theory. The books that were influential in the early development of computational quantum chemistry include Linus Pauling and E. Bright Wilson's 1935 Introduction to Quantum Mechanics – with Applications to Chemistry, Eyring, Walter and Kimball's 1944 Quantum Chemistry, Heitler's 1945 Elementary Wave Mechanics – with Applications to Quantum Chemistry, and later Coulson's 1952 textbook Valence, each of which served as primary references for chemists in the decades to follow.
With the development of efficient computer technology in the 1940s, the solutions of elaborate wave equations for complex atomic systems began to be a realizable objective. In the early 1950s, the first semi-empirical atomic orbital calculations were performed. Theoretical chemists became extensive users of the early digital computers. One major advance came with the 1951 paper in Reviews of Modern Physics by Clemens C. J. Roothaan in 1951, largely on the "LCAO MO" approach (Linear Combination of Atomic Orbitals Molecular Orbitals), for many years the second-most cited paper in that journal. A very detailed account of such use in the United Kingdom is given by Smith and Sutcliffe.[2] The first ab initio Hartree–Fock method calculations on diatomic molecules were performed in 1956 at MIT, using a basis set of Slater orbitals. For diatomic molecules, a systematic study using a minimum basis set and the first calculation with a larger basis set were published by Ransil and Nesbet respectively in 1960.[3] The first polyatomic calculations using Gaussian orbitals were performed in the late 1950s. The first configuration interaction calculations were performed in Cambridge on the EDSAC computer in the 1950s using Gaussian orbitals by Boys and coworkers.[4] By 1971, when a bibliography of ab initio calculations was published,[5] the largest molecules included were naphthalene and azulene.[6][7] Abstracts of many earlier developments in ab initio theory have been published by Schaefer.[8]
In 1964, Hückel method calculations (using a simple linear combination of atomic orbitals (LCAO) method to determine electron energies of molecular orbitals of π electrons in conjugated hydrocarbon systems) of molecules, ranging in complexity from butadiene and benzene to ovalene, were generated on computers at Berkeley and Oxford.[9] These empirical methods were replaced in the 1960s by semi-empirical methods such as CNDO.[10]
In the early 1970s, efficient ab initio computer programs such as ATMOL, Gaussian, IBMOL, and POLYAYTOM, began to be used to speed ab initio calculations of molecular orbitals. Of these four programs, only Gaussian, now vastly expanded, is still in use, but many other programs are now in use. At the same time, the methods of molecular mechanics, such as MM2 force field, were developed, primarily by Norman Allinger.[11]
One of the first mentions of the term computational chemistry can be found in the 1970 book Computers and Their Role in the Physical Sciences by Sidney Fernbach and Abraham Haskell Taub, where they state "It seems, therefore, that 'computational chemistry' can finally be more and more of a reality."[12] During the 1970s, widely different methods began to be seen as part of a new emerging discipline of computational chemistry.[13] The Journal of Computational Chemistry was first published in 1980.
Computational chemistry has featured in several Nobel Prize awards, most notably in 1998 and 2013. Walter Kohn, "for his development of the density-functional theory", and John Pople, "for his development of computational methods in quantum chemistry", received the 1998 Nobel Prize in Chemistry.[14] Martin Karplus, Michael Levitt and Arieh Warshel received the 2013 Nobel Prize in Chemistry for "the development of multiscale models for complex chemical systems".[15]
Fields of application
The term theoretical chemistry may be defined as a mathematical description of chemistry, whereas computational chemistry is usually used when a mathematical method is sufficiently well developed that it can be automated for implementation on a computer. In theoretical chemistry, chemists, physicists, and mathematicians develop algorithms and computer programs to predict atomic and molecular properties and reaction paths for chemical reactions[16]. Computational chemists, in contrast, may simply apply existing computer programs and methodologies to specific chemical questions.
Computational chemistry has two different aspects:
- Computational studies, used to find a starting point for a laboratory synthesis or to assist in understanding experimental data, such as the position and source of spectroscopic peaks[17].
- Computational studies, used to predict the possibility of so far entirely unknown molecules or to explore reaction mechanisms not readily studied via experiments[17].
Thus, computational chemistry can assist the experimental chemist or it can challenge the experimental chemist to find entirely new chemical objects.
Several major areas may be distinguished within computational chemistry:
- The prediction of the molecular structure of molecules by the use of the simulation of forces, or more accurate quantum chemical methods, to find stationary points on the energy surface as the position of the nuclei is varied.[18]
- Storing and searching for data on chemical entities (see chemical databases)[19].
- Identifying correlations between chemical structures and properties (see quantitative structure–property relationship (QSPR) and quantitative structure–activity relationship (QSAR))[20] .
- Computational approaches to help in the efficient synthesis of compounds.
- Computational approaches to design molecules that interact in specific ways with other molecules (e.g. drug design and catalysis).
Catalysis
Computational chemistry is a tool for analyzing catalytic systems without doing experiments. Modern electronic structure theory and density functional theory has allowed researchers to discover and understand catalysts.[21] Computational studies apply theoretical chemistry to catalysis research. Density functional theory methods calculate the energies and orbitals of molecules to give models of those structures.[22] Using these methods, researchers can predict values like activation energy, site reactivity[23] and other thermodynamic properties.[22]
Data that is difficult to obtain experimentally can be found using computational methods to model the mechanisms of catalytic cycles.[23] Skilled computational chemists provide predictions that are close to experimental data with proper considerations of methods and basis sets.[22] With good computational data, researchers can predict how catalysts can be improved to lower the cost and increase the efficiency of these reactions.
Drug Development
Computational chemistry is used in drug development to model potentially useful drug molecules and help companies save time and cost in drug development. The drug discovery process involves analyzing data, finding ways to improve current molecules, finding synthetic routes, and testing those molecules.[24] Computational chemistry helps with this process by giving predictions of which experiments would be best to do without conducting other experiments. Computational methods can also find values that are difficult to find experimentally like pKa's of compounds.[25] Methods like density functional theory can be used to model drug molecules and find their properties, like their HOMO and LUMO energies and molecular orbitals.[26] Computational chemists also help companies with developing informatics, infrastructure and designs of drugs.
Aside from drug synthesis, drug carriers are also researched by computational chemists for nanomaterials. It allows researchers to simulate environments to test the effectiveness and stability of drug carriers.[27] Understanding how water interacts with these nanomaterials ensures stability of the material in human bodies. These computational simulations help researchers optimize the material find the best way to structure these nanomaterials before making them.
Computational Chemistry Databases
Databases are useful for both computational and non computational chemists in research and verifying the validity of computational methods. Empirical data is used to analyze the error of computational methods against experimental data.[28] Empirical data helps researchers with their methods and basis sets to have greater confidence in the researchers results. Computational chemistry databases are also used in testing software or hardware for computational chemistry.
Databases can also use purely calculated data.[28] Purely calculated data uses calculated values over experimental values for databases. Purely calculated data avoids dealing with these adjusting for different experimental conditions like zero-point energy. These calculations can also avoid experimental errors for difficult to test molecules. Though purely calculated data is often not perfect, identifying issues is often easier for calculated data than experimental.
Databases also give public access to information for researchers to use. They contain data that other researchers have found and uploaded to these databases so that anyone can search for them. Researchers use these databases to find information on molecules of interest and learn what can be done with those molecules.[28] Some publicly available chemistry databases include:
- BindingDB: Contains experimental information about protein-small molecule interactions.[29]
- RCSB: Stores publically available 3D models of macromolecules (proteins, nucleic acids) and small molecules (drugs, inhibitors)[30]
- ChEMBL: Contains data from research on drug development such as assay results.[28]
- DrugBank: Data about mechanisms of drugs can be found here.[28]
Computational Costs in Chemistry Algorithms
Also see: Computational Complexity
For types of computational complexity classes: List of complexity classes
The computational cost and algorithmic complexity in chemistry are used to help understand and predict chemical phenomena. This section focuses on the scaling of computational complexity with molecule size and details the algorithms commonly used in both domains.
In quantum chemistry, particularly, the complexity can grow exponentially with the number of electrons involved in the system.[31] This exponential growth is a significant barrier to simulating large or complex systems accurately.
Advanced algorithms in both fields strive to balance accuracy with computational efficiency. For instance, in MD, methods like Verlet integration or Beeman's algorithm are employed for their computational efficiency.[32] In quantum chemistry, hybrid methods combining different computational approaches (like QM/MM) are increasingly used to tackle large biomolecular systems.
Algorithmic Complexity Examples
1. Molecular Dynamics (MD)
see: Molecular dynamics
Algorithm: Solves Newton's equations of motion for atoms and molecules.[33]
Complexity: The standard pairwise interaction calculation in MD leads to an complexity for particles. This is because each particle interacts with every other particle, resulting in interactions.[34] Advanced algorithms, such as the Ewald summation or Fast Multipole Method, reduce this to or even by grouping distant particles and treating them as a single entity or using clever mathematical approximations.[35][36]

2. Quantum Mechanics/Molecular Mechanics (QM/MM)
see: QM/MM
Algorithm: Combines quantum mechanical calculations for a small region with molecular mechanics for the larger environment.[37]
Complexity: The complexity of QM/MM methods depends on both the size of the quantum region and the method used for quantum calculations.[38] For example, if a Hartree-Fock method is used for the quantum part, the complexity can be approximated as , where is the number of basis functions in the quantum region.[38] This complexity arises from the need to solve a set of coupled equations iteratively until self-consistency is achieved.

3. Hartree-Fock Method
Algorithm: Finds a single Fock state that minimizes the energy.
Complexity: NP-hard or NP-complete as demonstrated by embedding instances of the Ising model into Hartree-Fock calculations.[39] The Hartree-Fock method involves solving the Roothaan-Hall equations, which scales as to depending on implementation, with being the number of basis functions.[39] The computational cost mainly comes from evaluating and transforming the two-electron integrals. This proof of NP-hardness or NP-completeness comes from embedding problems like the Ising model into the Hartree-Fock formalism.

4. Density Functional Theory (DFT)
Algorithm: Investigate the electronic structure (or nuclear structure) (principally the ground state) of many-body systems, in particular atoms, molecules, and the condensed phases.
Complexity: Traditional implementations of DFT typically scale as , mainly due to the need to diagonalize the Kohn-Sham matrix.[40] The diagonalization step, which finds the eigenvalues and eigenvectors of the matrix, contributes most to this scaling.[41] Recent advances in DFT aim to reduce this complexity through various approximations and algorithmic improvements.
5. Standard CCSD and CCSD(T) Method
Algorithm: CCSD and CCSD(T) methods are advanced electronic structure techniques involving single, double, and in the case of CCSD(T), perturbative triple excitations for calculating electronic correlation effects.
Complexity:
CCSD: Scales as where is the number of basis functions. This intense computational demand arises from the inclusion of single and double excitations in the electron correlation calculation.[42]
CCSD(T): With the addition of perturbative triples, the complexity increases to . This elevated complexity restricts practical usage to smaller systems, typically up to 20-25 atoms in conventional implementations.[42]
6. Linear-Scaling CCSD(T) Method
Algorithm: An adaptation of the standard CCSD(T) method using local natural orbitals (NOs) to significantly reduce the computational burden and enable application to larger systems.
Complexity: Achieves linear scaling with the system size, a major improvement over the traditional fifth-power scaling of CCSD.[42] This advancement allows for practical applications to molecules of up to 100 atoms with reasonable basis sets, marking a significant step forward in computational chemistry's capability to handle larger systems with high accuracy.[42]
Proving the complexity classes for algorithms involves a combination of mathematical proof and computational experiments. For example, in the case of the Hartree-Fock method, the proof of NP-hardness is a theoretical result derived from complexity theory, specifically through reductions from known NP-hard problems.[43]
For other methods like MD or DFT, the computational complexity is often empirically observed and supported by algorithm analysis. In these cases, the proof of correctness is less about formal mathematical proofs and more about consistently observing the computational behaviour across various systems and implementations.[43]
Quantum Computational Chemistry
For the foundation of Quantum Chemistry see: Quantum Chemistry
For the electronic structure problem see: Electronic Structure
For a specific recap of quantum computing see: Quantum Computing
Quantum computational chemistry is an emerging field that integrates quantum mechanics with computational methods to simulate chemical systems. Despite quantum mechanics' foundational role in understanding chemical behaviors, traditional computational approaches face significant challenges, largely due to the complexity and computational intensity of quantum mechanical equations. This complexity arises from the exponential growth of a quantum system's wave function with each added particle, making exact simulations on classical computers inefficient.[44]
Efficient quantum algorithms for chemistry problems are expected to have run-times and resource requirements that scale polynomially with system size and desired accuracy. Experimental efforts have validated proof-of-principle chemistry calculations, though currently limited to small systems.
Historical Context for Classical Computational Challenges in Quantum Mechanics
- 1929: Dirac noted the inherent complexity of quantum mechanical equations, underscoring the difficulties in solving these equations using classical computation.[45]
- 1982: Feynman proposed using quantum hardware for simulations, addressing the inefficiency of classical computers in simulating quantum systems.[46]
Methods in Quantum Complexity
Qubitization
One of the problems with hamiltonian simulation is the computational complexity inherent to its formation. Qubitization is a mathematical and algorithmic concept in quantum computing to the simulation of quantum systems via Hamiltonian dynamics. The core idea of qubitization is to encode the problem of Hamiltonian simulation in a way that is more efficiently processable by quantum algorithms.[47]
Qubitization involves a transformation of the Hamiltonian operator, a central object in quantum mechanics representing the total energy of a system. In classical computational terms, a Hamiltonian can be thought of as a matrix describing the energy interactions within a quantum system. The goal of qubitization is to embed this Hamiltonian into a larger, unitary operator, which is a type of operator in quantum mechanics that preserves the norm of vectors upon which it acts.[47] This embedding is crucial for enabling the Hamiltonian's dynamics to be simulated on a quantum computer.
Mathematically, the process of qubitization constructs a unitary operator such that a specific projection of proportional to the Hamiltonian H of interest. This relationship can often be represented as , where is a specific quantum state and is its conjugate transpose. The efficiency of this method comes from the fact that the unitary operator can be implemented on a quantum computer with fewer resources (like qubits and quantum gates) than would be required for directly simulating [47]
A key feature of qubitization is in simulating Hamiltonian dynamics with high precision while reducing the quantum resource overhead. This efficiency is especially beneficial in quantum algorithms where the simulation of complex quantum systems is necessary, such as in quantum chemistry and materials science simulations. Qubitization also develops quantum algorithms for solving certain types of problems more efficiently than classical algorithms. For instance, it has implications for the Quantum Phase Estimation algorithm, which is fundamental in various quantum computing applications, including factoring and solving linear systems of equations.
Applications of Qubization in chemistry
Gaussian Orbital Basis Sets
In Gaussian orbital basis sets, phase estimation algorithms have been optimized empirically from to where is the number of basis sets. Advanced Hamiltonian simulation algorithms have further reduced the scaling, with the introduction of techniques like Taylor series methods and qubitization, providing more efficient algorithms with reduced computational requirements[48].
Plane Wave Basis Sets
Plane wave basis sets, suitable for periodic systems, have also seen advancements in algorithm efficiency, with improvements in product formula-based approaches and Taylor series methods[47].
Quantum Phase Estimation in Chemistry
For the foundational recap of the quantum fourier transform Quantum Fourier Transform
Overview
Phase estimation, as proposed by Kitaev in 1996[49], identifies the lowest energy eigenstate ( ) and excited states ( ) of a physical Hamiltonian, as detailed by Abrams and Lloyd in 1999[50]. In quantum computational chemistry, this technique is employed to encode fermionic Hamiltonians into a qubit framework.
Brief Methodology
1. Initialization: The qubit register is initialized in a state , which has a nonzero overlap with the Full Configuration Interaction (FCI) target eigenstate of the system.[51] This state is expressed as a sum over the energy eigenstates of the Hamiltonian , , where represents complex coefficients [51] .
2. Application of Hadamard Gates: Each ancilla qubit undergoes a Hadamard gate application, placing the ancilla register in a superposed state.[51] Subsequently, controlled gates, as shown above, modify this state.

3. Inverse Quantum Fourier Transform: This transform is applied to the ancilla qubits, revealing the phase information that encodes the energy eigenvalues.[51]
4. Measurement: The ancilla qubits are measured in the Z basis, collapsing the main register into the corresponding energy eigenstate based on the probability .[51]
Requirements
The algorithm requires ancilla qubits, with their number determined by the desired precision and success probability of the energy estimate. Obtaining a binary energy estimate precise to n bits with a success probability necessitates ancilla qubits. This phase estimation has been validated experimentally across various quantum architectures.[51]
Applications of QPEs in chemistry
Time Evolution and Error Analysis
The total coherent time evolution required for the algorithm is approximately .[53] The total evolution time is related to the binary precision , with an expected repeat of the procedure for accurate ground state estimation. Errors in the algorithm include errors in energy eigenvalue estimation (), unitary evolutions (), and circuit synthesis errors (), which can be quantified using techniques like the Solovay-Kitaev theorem.[54]
The phase estimation algorithm can be enhanced or altered in several ways, such as using a single ancilla qubit for sequential measurements, increasing efficiency, parallelization, or enhancing noise resilience in analytical chemistry.[55] The algorithm can also be scaled using classically obtained knowledge about energy gaps between states.
Limitations
Effective state preparation is needed, as a randomly chosen state would exponentially decrease the probability of collapsing to the desired ground state. Various methods for state preparation have been proposed, including classical approaches and quantum techniques like adiabatic state preparation.[56]
Variational Quantum Eigensolver
Overview:
The Variational Quantum Eigensolver is an innovative algorithm in quantum computing, crucial for near-term quantum hardware.[57] Initially proposed by Peruzzo et al. in 2014 and further developed by McClean et al. in 2016, VQE is integral in finding the lowest eigenvalue of Hamiltonians, particularly those in chemical systems [58]. It employs the variational method (quantum mechanics), which guarantees that the expectation value of the Hamiltonian for any parameterized trial wave function is at least the lowest energy eigenvalue of that Hamiltonian.[59] This principle is fundamental in VQE's strategy to optimize parameters and find the ground state energy. VQE is a hybrid algorithm that utilizes both quantum and classical computers. The quantum computer prepares and measures the quantum state, while the classical computer processes these measurements and updates the system. This synergy allows VQE to overcome some limitations of purely quantum methods.
Applications of VQEs in chemistry
1-RDM and 2-RDM Calculation:
For terminology see: Density Matrix
The reduced density matrices (1-RDM and 2-RDM) can be used to extrapolate the electronic structure of a system.[60]
Ground State Energy Extrapolation:
In the Hamiltonian variational ansatz, the initial state is prepared to represent the ground state of the molecular Hamiltonian without electron correlations. The evolution of this state under the Hamiltonian, split into commuting segments , is given by:
where are variational parameters optimized to minimize the energy, providing insights into the electronic structure of the molecule.
Measurement Scaling:
McClean et al. (2016) and Romero et al. (2019) proposed a formula to estimate the number of measurements ( ) required for energy precision. The formula is given by , where are coefficients of each Pauli string in the Hamiltonian. This leads to a scaling of in a Gaussian orbital basis and in a plane wave dual basis.[61][62] Note that is the number of basis functions in the chosen basis set.
Fermionic Level Grouping:
A method by Bonet-Monroig, Babbush, and O'Brien (2019) focuses on grouping terms at a fermionic level rather than a qubit level, leading to a measurement requirement of only circuits with an additional gate depth of .[63]
Limitations of VQE
While VQE's application in solving the electronic Schrödinger equation for small molecules has shown success, its scalability is hindered by two main challenges: the complexity of the quantum circuits required and the intricacies involved in the classical optimization process[64]. These challenges are significantly influenced by the choice of the variational ansatz, which is used to construct the trial wave function. Consequently, the development of an efficient ansatz is a key focus in current research. Modern quantum computers face limitations in running deep quantum circuits, especially when using the existing ansatzes for problems that exceed several qubits.
Jordan-Wigner Encoding
Also see: Jordan-Wigner Transformations
Jordan-Wigner encoding is a fundamental method in quantum computing, extensively used for simulating fermionic systems like molecular orbitals and electron interactions in quantum chemistry.[65]
Overview:
In quantum chemistry, electrons are modeled as fermions with antisymmetric wave functions. The Jordan-Wigner encoding maps these fermionic orbitals to qubits, preserving their antisymmetric nature. Mathematically, this is achieved by associating each fermionic creation and annihilation operator with corresponding qubit operators through the Jordan-Wigner transformation:
Where , , and are Pauli matrices acting on the qubit.
Applications of Jordan-Wigner Encoding in Chemistry
Electron Hopping
Electron hopping between orbitals, central to chemical bonding and reactions, is represented by terms like . Under Jordan-Wigner encoding, these transform as follows:[65]
Computational Complexity in Molecular Systems
The complexity of simulating a molecular system using Jordan-Wigner encoding is influenced by the structure of the molecule and the nature of electron interactions. For a molecular system with orbitals, the number of required qubits scales linearly with , but the complexity of gate operations depends on the specific interactions being modeled.
Limitations of Jordan–Wigner Encoding
The Jordan-Wigner transformation encodes fermionic operators into qubit operators, but it introduces non-local string operators that can make simulations inefficient.[67] The FSWAP gate is used to mitigate this inefficiency by rearranging the ordering of fermions (or their qubit representations), thus simplifying the implementation of fermionic operations.
Fermionic SWAP (FSWAP) Network
FSWAP networks rearrange qubits to efficiently simulate electron dynamics in molecules.[68] These networks are essential for reducing the gate complexity in simulations, especially for non-neighboring electron interactions.
When two fermionic modes (represented as qubits after the Jordan-Wigner transformation) are swapped, the FSWAP gate not only exchanges their states but also correctly updates the phase of the wavefunction to maintain fermionic antisymmetry.[69] This is in contrast to the standard SWAP gate, which does not account for the phase change required in the antisymmetric wavefunctions of fermions.
The use of FSWAP gates can significantly reduce the complexity of quantum circuits for simulating fermionic systems.[70] By intelligently rearranging the fermions, the number of gates required to simulate certain fermionic operations can be reduced, leading to more efficient simulations. This is particularly useful in simulations where fermions need to be moved across large distances within the system, as it can avoid the need for long chains of operations that would otherwise be required.
Interpreting Solutions to Molecular Wavefunctions
The atoms in molecules (QTAIM) model of Richard Bader was developed to effectively link the quantum mechanical model of a molecule, as an electronic wavefunction, to chemically useful concepts such as atoms in molecules, functional groups, bonding, the theory of Lewis pairs, and the valence bond model. Bader has demonstrated that these empirically useful chemistry concepts can be related to the topology of the observable charge density distribution, whether measured or calculated from a quantum mechanical wavefunction. QTAIM analysis of molecular wavefunctions is implemented, for example, in the AIMAll software package.
Methods
One molecular formula can represent more than one molecular isomer: a set of isomers. Each isomer is a local minimum on the energy surface (called the potential energy surface) created from the total energy (i.e., the electronic energy, plus the repulsion energy between the nuclei) as a function of the coordinates of all the nuclei.[71] A stationary point is a geometry such that the derivative of the energy with respect to all displacements of the nuclei is zero. A local (energy) minimum is a stationary point where all such displacements lead to an increase in energy. The local minimum that is lowest is called the global minimum and corresponds to the most stable isomer. If there is one particular coordinate change that leads to a decrease in the total energy in both directions, the stationary point is a transition structure and the coordinate is the reaction coordinate. This process of determining stationary points is called geometry optimization.
The determination of molecular structure by geometry optimization became routine only after efficient methods for calculating the first derivatives of the energy with respect to all atomic coordinates became available.[71] Evaluation of the related second derivatives allows the prediction of vibrational frequencies if harmonic motion is estimated. More importantly, it allows for the characterization of stationary points. The frequencies are related to the eigenvalues of the Hessian matrix, which contains second derivatives. If the eigenvalues are all positive, then the frequencies are all real and the stationary point is a local minimum. If one eigenvalue is negative (i.e., an imaginary frequency), then the stationary point is a transition structure. If more than one eigenvalue is negative, then the stationary point is a more complex one and is usually of little interest. When one of these is found, it is necessary to move the search away from it if the experimenter is looking solely for local minima and transition structures.
The total energy is determined by approximate solutions of the time-dependent Schrödinger equation, usually with no relativistic terms included, and by making use of the Born–Oppenheimer approximation, which allows for the separation of electronic and nuclear motions, thereby simplifying the Schrödinger equation.[71] This leads to the evaluation of the total energy as a sum of the electronic energy at fixed nuclei positions and the repulsion energy of the nuclei. A notable exception is certain approaches called direct quantum chemistry, which treat electrons and nuclei on a common footing. Density functional methods and semi-empirical methods are variants of the major theme. For very large systems, the relative total energies can be compared using molecular mechanics. The ways of determining the total energy to predict molecular structures are:
Ab initio methods
The programs used in computational chemistry are based on many different quantum-chemical methods that solve the molecular Schrödinger equation associated with the molecular Hamiltonian. Methods that do not include any empirical or semi-empirical parameters in their equations – being derived directly from theoretical principles, with no inclusion of experimental data – are called ab initio methods.[72] This does not imply that the solution is an exact one; they are all approximate quantum mechanical calculations. It means that a particular approximation is rigorously defined on first principles (quantum theory) and then solved within an error margin that is qualitatively known beforehand. If numerical iterative methods must be used, the aim is to iterate until full machine accuracy is obtained (the best that is possible with a finite word length on the computer, and within the mathematical and/or physical approximations made).
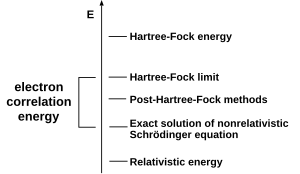
The simplest type of ab initio electronic structure calculation is the Hartree–Fock method (HF), an extension of molecular orbital theory, in which the correlated electron-electron repulsion is not specifically taken into account; only its average effect is included in the calculation.[72] As the basis set size is increased, the energy and wave function tend towards a limit called the Hartree–Fock limit. Many types of calculations (termed post-Hartree–Fock methods) begin with a Hartree–Fock calculation and subsequently correct for electron-electron repulsion, referred to also as electronic correlation. As these methods are pushed to the limit, they approach the exact solution of the non-relativistic Schrödinger equation. To obtain exact agreement with the experiment, it is necessary to include relativistic and spin orbit terms, both of which are far more important for heavy atoms. In all of these approaches, along with a choice of method, it is necessary to choose a basis set. This is a set of functions, usually centered on the different atoms in the molecule, which are used to expand the molecular orbitals with the linear combination of atomic orbitals (LCAO) molecular orbital method ansatz. Ab initio methods need to define a level of theory (the method) and a basis set.
The Hartree–Fock wave function is a single configuration or determinant. In some cases, particularly for bond-breaking processes, this is inadequate, and several configurations must be used. Here, the coefficients of the configurations, and of the basis functions, are optimized together.
The total molecular energy can be evaluated as a function of the molecular geometry; in other words, the potential energy surface. Such a surface can be used for reaction dynamics. The stationary points of the surface lead to predictions of different isomers and the transition structures for conversion between isomers, but these can be determined without full knowledge of the complete surface.
A particularly important objective, called computational thermochemistry, is to calculate thermochemical quantities such as the enthalpy of formation to chemical accuracy. Chemical accuracy is the accuracy required to make realistic chemical predictions and is generally considered to be 1 kcal/mol or 4 kJ/mol. To reach that accuracy in an economic way it is necessary to use a series of post-Hartree–Fock methods and combine the results. These methods are called quantum chemistry composite methods.
Density functional methods
Density functional theory (DFT) methods are often considered to be ab initio methods for determining the molecular electronic structure, even though many of the most common functionals use parameters derived from empirical data, or from more complex calculations. In DFT, the total energy is expressed in terms of the total one-electron density rather than the wave function.[73] In this type of calculation, there is an approximate Hamiltonian and an approximate expression for the total electron density. DFT methods can be very accurate for little computational cost. Some methods combine the density functional exchange functional with the Hartree–Fock exchange term and are termed hybrid functional methods.
Semi-empirical methods
Semi-empirical quantum chemistry methods are based on the Hartree–Fock method formalism, but make many approximations and obtain some parameters from empirical data.[71] They were very important in computational chemistry from the 60s to the 90s, especially for treating large molecules where the full Hartree–Fock method without the approximations were too costly. The use of empirical parameters appears to allow some inclusion of correlation effects into the methods.
Primitive semi-empirical methods were designed even before, where the two-electron part of the Hamiltonian is not explicitly included. For π-electron systems, this was the Hückel method proposed by Erich Hückel, and for all valence electron systems, the extended Hückel method proposed by Roald Hoffmann. Sometimes, Hückel methods are referred to as "completely emprirical" because they do not derive from a Hamiltonian.[74] Yet, the term "empirical methods", or "empirical force fields" is usually used to describe Molecular Mechanics.[75]
Molecular mechanics
In many cases, large molecular systems can be modeled successfully while avoiding quantum mechanical calculations entirely. Molecular mechanics simulations, for example, use one classical expression for the energy of a compound, for instance, the harmonic oscillator.[71] All constants appearing in the equations must be obtained beforehand from experimental data or ab initio calculations.
The database of compounds used for parameterization, i.e., the resulting set of parameters and functions is called the force field, is crucial to the success of molecular mechanics calculations.[71] A force field parameterized against a specific class of molecules, for instance, proteins, would be expected to only have any relevance when describing other molecules of the same class.
These methods can be applied to proteins and other large biological molecules, and allow studies of the approach and interaction (docking) of potential drug molecules.[76][77]
Methods for solids
Computational chemical methods can be applied to solid-state physics problems. The electronic structure of a crystal is in general described by a band structure, which defines the energies of electron orbitals for each point in the Brillouin zone.[71] Ab initio and semi-empirical calculations yield orbital energies; therefore, they can be applied to band structure calculations. Since it is time-consuming to calculate the energy for a molecule, it is even more time-consuming to calculate them for the entire list of points in the Brillouin zone.
Chemical dynamics
Once the electronic and nuclear variables are separated (within the Born–Oppenheimer representation), in the time-dependent approach, the wave packet corresponding to the nuclear degrees of freedom is propagated via the time evolution operator (physics) associated to the time-dependent Schrödinger equation (for the full molecular Hamiltonian). In the complementary energy-dependent approach, the time-independent Schrödinger equation is solved using the scattering theory formalism. The potential representing the interatomic interaction is given by the potential energy surfaces. In general, the potential energy surfaces are coupled via the vibronic coupling terms.
The most popular methods for propagating the wave packet associated to the molecular geometry are:
- the split operator technique,
- the Chebyshev (real) polynomial,
- the multi-configuration time-dependent Hartree method (MCTDH),
- the semiclassical method.
To better understand the split operator technique, an explanation is provided below.
Split Operator Technique
How a computational method solves quantum equations impacts the accuracy and efficiency of the method. The split operator technique is one of these methods for solving differential equations. In computational chemistry, split operator technique reduces computational costs of simulating chemical systems.[78] Computational costs are about how much time it takes for computers to caclulate these chemical systems, as it can take days for more complex systems. Quantum systems are difficult and time consuming to solve for humans. Split operator methods help computers calculate these systems quickly by solving the sub problems in a quantum differential equation. The method does this by separating the differential equation into 2 different equations, like when there are more than two operators. Once solved, the split equations are combined into one equation again to give an easily calculable solution. For example:
This method is used in many fields that require solving differential equations, such as biology.[78] However, the technique comes with a splitting error. For example, for the following solution for a differential equation:
The equation can be split, but the solutions will not be exact, only similar.[78] This is an example of first order splitting.
There are ways to reduce this error, which include taking an average of two split equations.[78]
Another way to increase accuracy is to use higher order splitting.[78] Usually, second order splitting is the most that is done because higher order splitting requires much more time to calculate and is not worth the cost. Higher order methods become too difficult to impliment, and are not useful for solving differential equations despite the higher accuracy.
Computational chemists spend much time trying to find ways to make systems calculated with split operator technique more accurate while minimizing the computational cost. Finding that middle ground of accurate and plausible to calculate is a massive challenge for many chemists trying to simulate molecules or chemical environments.
Molecular dynamics
Molecular dynamics (MD) use either quantum mechanics, molecular mechanics or a mixture of both to calculate forces which are then used to solve Newton's laws of motion to examine the time-dependent behavior of systems.[79] The result of a molecular dynamics simulation is a trajectory that describes how the position and velocity of particles varies with time. The phase point of a system described by the positions and momenta of all its particles on a previous time point will determine the next phase point in time by integrating over Newton's laws of motion.
Monte Carlo
Monte Carlo (MC) generates configurations of a system by making random changes to the positions of its particles, together with their orientations and conformations where appropriate.[80] It is a random sampling method, which makes use of the so-called importance sampling. Importance sampling methods are able to generate low energy states, as this enables properties to be calculated accurately. The potential energy of each configuration of the system can be calculated, together with the values of other properties, from the positions of the atoms.[81][82]
Quantum mechanics/Molecular mechanics (QM/MM)
QM/MM is a hybrid method that attempts to combine the accuracy of quantum mechanics with the speed of molecular mechanics.[83] It is useful for simulating very large molecules such as enzymes.
Accuracy
Computational chemistry is not an exact description of real-life chemistry, as our mathematical models of the physical laws of nature can only provide us with an approximation. However, the majority of chemical phenomena can be described to a certain degree in a qualitative or approximate quantitative computational scheme.
Molecules consist of nuclei and electrons, so the methods of quantum mechanics apply. Computational chemists often attempt to solve the non-relativistic Schrödinger equation, with relativistic corrections added, although some progress has been made in solving the fully relativistic Dirac equation.[84] In principle, it is possible to solve the Schrödinger equation in either its time-dependent or time-independent form, as appropriate for the problem in hand; in practice, this is not possible except for very small systems. Therefore, a great number of approximate methods strive to achieve the best trade-off between accuracy and computational cost.
Accuracy can always be improved with greater computational cost. Significant errors can present themselves in ab initio models comprising many electrons, due to the computational cost of full relativistic-inclusive methods.[42] This complicates the study of molecules interacting with high atomic mass unit atoms, such as transitional metals and their catalytic properties. Present algorithms in computational chemistry can routinely calculate the properties of small molecules that contain up to about 40 electrons with errors for energies less than a few kJ/mol. For geometries, bond lengths can be predicted within a few picometers and bond angles within 0.5 degrees.[85] The treatment of larger molecules that contain a few dozen atoms is computationally tractable by more approximate methods such as density functional theory (DFT).
There is some dispute within the field whether or not the latter methods are sufficient to describe complex chemical reactions, such as those in biochemistry. Large molecules can be studied by semi-empirical approximate methods. Even larger molecules are treated by classical mechanics methods that use what are called molecular mechanics (MM).[86] In QM-MM methods, small parts of large complexes are treated quantum mechanically (QM), and the remainder is treated approximately (MM).[87]
Software packages
Many self-sufficient computational chemistry software packages exist. Some include many methods covering a wide range, while others concentrate on a very specific range or even on one method. Details of most of them can be found in:
- Biomolecular modelling programs: proteins, nucleic acid.
- Molecular mechanics programs.
- Quantum chemistry and solid state-physics software supporting several methods.
- Molecular design software
- Semi-empirical programs.
- Valence bond programs.
See also
- List of computational chemists
- Bioinformatics
- Computational biology
- Computational Chemistry List
- Efficient code generation by computer algebra
- Comparison of force field implementations
- Important publications in computational chemistry
- In silico
- International Academy of Quantum Molecular Science
- Mathematical chemistry
- Molecular graphics
- Molecular modelling
- Molecular modeling on GPUs
- Monte Carlo molecular modeling
- Car–Parrinello molecular dynamics
- Protein dynamics
- Scientific computing
- Statistical mechanics
- Solvent models
References
- ^ Willems, Henriëtte; De Cesco, Stephane; Svensson, Fredrik (2020-09-24). "Computational Chemistry on a Budget: Supporting Drug Discovery with Limited Resources: Miniperspective". Journal of Medicinal Chemistry. 63 (18): 10158–10169. doi:10.1021/acs.jmedchem.9b02126. ISSN 0022-2623. PMID 32298123. S2CID 215802432.
- ^ Smith, S. J.; Sutcliffe, B. T. (1997). "The development of Computational Chemistry in the United Kingdom". Reviews in Computational Chemistry. 10: 271–316.
- ^ Schaefer, Henry F. III (1972). The electronic structure of atoms and molecules. Reading, Massachusetts: Addison-Wesley Publishing Co. p. 146.
- ^ Boys, S. F.; Cook, G. B.; Reeves, C. M.; Shavitt, I. (1956). "Automatic fundamental calculations of molecular structure". Nature. 178 (2): 1207. Bibcode:1956Natur.178.1207B. doi:10.1038/1781207a0. S2CID 4218995.
- ^ Richards, W. G.; Walker, T. E. H.; Hinkley R. K. (1971). A bibliography of ab initio molecular wave functions. Oxford: Clarendon Press.
- ^ Preuss, H. (1968). "DasSCF-MO-P(LCGO)-Verfahren und seine Varianten". International Journal of Quantum Chemistry. 2 (5): 651. Bibcode:1968IJQC....2..651P. doi:10.1002/qua.560020506.
- ^ Buenker, R. J.; Peyerimhoff, S. D. (1969). "Ab initio SCF calculations for azulene and naphthalene". Chemical Physics Letters. 3 (1): 37. Bibcode:1969CPL.....3...37B. doi:10.1016/0009-2614(69)80014-X.
- ^ Schaefer, Henry F. III (1984). Quantum Chemistry. Oxford: Clarendon Press.
- ^ Streitwieser, A.; Brauman, J. I.; Coulson, C. A. (1965). Supplementary Tables of Molecular Orbital Calculations. Oxford: Pergamon Press.
- ^ Pople, John A.; Beveridge, David L. (1970). Approximate Molecular Orbital Theory. New York: McGraw Hill.
- ^ Allinger, Norman (1977). "Conformational analysis. 130. MM2. A hydrocarbon force field utilizing V1 and V2 torsional terms". Journal of the American Chemical Society. 99 (25): 8127–8134. doi:10.1021/ja00467a001.
- ^ Fernbach, Sidney; Taub, Abraham Haskell (1970). Computers and Their Role in the Physical Sciences. Routledge. ISBN 978-0-677-14030-8.
- ^ "vol 1, preface". Reviews in Computational Chemistry. Vol. 1. 1990. doi:10.1002/9780470125786. ISBN 978-0-470-12578-6.[permanent dead link]
- ^ "The Nobel Prize in Chemistry 1998".
- ^ "The Nobel Prize in Chemistry 2013" (Press release). Royal Swedish Academy of Sciences. October 9, 2013. Retrieved October 9, 2013.
- ^ Cramer, Christopher J. (2014). Essentials of computational chemistry: theories and models. Chichester: Wiley. ISBN 978-0-470-09182-1.
- ^ a b Patel, Prajay; Melin, Timothé R. L.; North, Sasha C.; Wilson, Angela K. (2021-01-01), Dixon, David A. (ed.), "Chapter Four - Ab initio composite methodologies: Their significance for the chemistry community", Annual Reports in Computational Chemistry, vol. 17, Elsevier, pp. 113–161, doi:10.1016/bs.arcc.2021.09.002, retrieved 2023-12-03
- ^ Musil, Felix; Grisafi, Andrea; Bartók, Albert P.; Ortner, Christoph; Csányi, Gábor; Ceriotti, Michele (2021-08-25). "Physics-Inspired Structural Representations for Molecules and Materials". Chemical Reviews. 121 (16): 9759–9815. doi:10.1021/acs.chemrev.1c00021. ISSN 0009-2665. PMID 34310133.
- ^ Muresan, Sorel; Sitzmann, Markus; Southan, Christopher (2012), Larson, Richard S. (ed.), "Mapping Between Databases of Compounds and Protein Targets", Bioinformatics and Drug Discovery, Methods in Molecular Biology, Totowa, NJ: Humana Press, pp. 145–164, doi:10.1007/978-1-61779-965-5_8, ISBN 978-1-61779-965-5, PMC 7449375, PMID 22821596, retrieved 2023-12-03
{{citation}}: CS1 maint: PMC format (link) - ^ Roy, Kunal; Kar, Supratik; Das, Rudra Narayan (2015). Understanding the basics of QSAR for applications in pharmaceutical sciences and risk assessment. Amsterdam Boston: Elsevier/Academic Press. ISBN 978-0-12-801505-6.
- ^ Elnabawy, Ahmed O.; Rangarajan, Srinivas; Mavrikakis, Manos (2015-08-01). "Computational chemistry for NH3 synthesis, hydrotreating, and NOx reduction: Three topics of special interest to Haldor Topsøe". Journal of Catalysis. Special Issue: The Impact of Haldor Topsøe on Catalysis. 328: 26–35. doi:10.1016/j.jcat.2014.12.018. ISSN 0021-9517.
- ^ a b c Patel, Prajay; Wilson, Angela K. (2020-12-01). "Computational chemistry considerations in catalysis: Regioselectivity and metal-ligand dissociation". Catalysis Today. Proceedings of 3rd International Conference on Catalysis and Chemical Engineering. 358: 422–429. doi:10.1016/j.cattod.2020.07.057. ISSN 0920-5861. S2CID 225472601.
- ^ a b van Santen, R. A. (1996-05-06). "Computational-chemical advances in heterogeneous catalysis". Journal of Molecular Catalysis A: Chemical. Proceedings of the 8th International Symposium on the Relations between Homogeneous and Heterogeneous Catalysis. 107 (1): 5–12. doi:10.1016/1381-1169(95)00161-1. ISSN 1381-1169. S2CID 59580128.
- ^ Tsui, Vickie; Ortwine, Daniel F.; Blaney, Jeffrey M. (2017-03-01). "Enabling drug discovery project decisions with integrated computational chemistry and informatics". Journal of Computer-Aided Molecular Design. 31 (3): 287–291. Bibcode:2017JCAMD..31..287T. doi:10.1007/s10822-016-9988-y. ISSN 1573-4951. PMID 27796615. S2CID 23373414.
- ^ van Vlijmen, Herman; Desjarlais, Renee L.; Mirzadegan, Tara (March 2017). "Computational chemistry at Janssen". Journal of Computer-Aided Molecular Design. 31 (3): 267–273. Bibcode:2017JCAMD..31..267V. doi:10.1007/s10822-016-9998-9. ISSN 1573-4951. PMID 27995515. S2CID 207166545.
- ^ Ahmad, Imad; Kuznetsov, Aleksey E.; Pirzada, Abdul Saboor; Alsharif, Khalaf F.; Daglia, Maria; Khan, Haroon (2023). "Computational pharmacology and computational chemistry of 4-hydroxyisoleucine: Physicochemical, pharmacokinetic, and DFT-based approaches". Frontiers in Chemistry. 11. Bibcode:2023FrCh...1145974A. doi:10.3389/fchem.2023.1145974. ISSN 2296-2646. PMC 10133580. PMID 37123881.
- ^ El-Mageed, H. R. Abd; Mustafa, F. M.; Abdel-Latif, Mahmoud K. (2022-01-02). "Boron nitride nanoclusters, nanoparticles and nanotubes as a drug carrier for isoniazid anti-tuberculosis drug, computational chemistry approaches". Journal of Biomolecular Structure and Dynamics. 40 (1): 226–235. doi:10.1080/07391102.2020.1814871. ISSN 0739-1102. PMID 32870128. S2CID 221403943.
- ^ a b c d e Muresan, Sorel; Sitzmann, Markus; Southan, Christopher (2012), Larson, Richard S. (ed.), "Mapping Between Databases of Compounds and Protein Targets", Bioinformatics and Drug Discovery, Methods in Molecular Biology, vol. 910, Totowa, NJ: Humana Press, pp. 145–164, doi:10.1007/978-1-61779-965-5_8, ISBN 978-1-61779-964-8, PMC 7449375, PMID 22821596
- ^ Gilson, Michael K.; Liu, Tiqing; Baitaluk, Michael; Nicola, George; Hwang, Linda; Chong, Jenny (2016-01-04). "BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology". Nucleic Acids Research. 44 (D1): D1045–1053. doi:10.1093/nar/gkv1072. ISSN 1362-4962. PMC 4702793. PMID 26481362.
- ^ Zardecki, Christine; Dutta, Shuchismita; Goodsell, David S.; Voigt, Maria; Burley, Stephen K. (2016-03-08). "RCSB Protein Data Bank: A Resource for Chemical, Biochemical, and Structural Explorations of Large and Small Biomolecules". Journal of Chemical Education. 93 (3): 569–575. Bibcode:2016JChEd..93..569Z. doi:10.1021/acs.jchemed.5b00404. ISSN 0021-9584.
- ^ Modern electronic structure theory. 1. Advanced series in physical chemistry. Singapore: World Scientific. 1995. ISBN 978-981-02-2987-0.
- ^ Adcock, Stewart A.; McCammon, J. Andrew (2006-05-01). "Molecular Dynamics: Survey of Methods for Simulating the Activity of Proteins". Chemical Reviews. 106 (5): 1589–1615. doi:10.1021/cr040426m. ISSN 0009-2665. PMC 2547409. PMID 16683746.
- ^ Durrant, Jacob D.; McCammon, J. Andrew (2011-10-28). "Molecular dynamics simulations and drug discovery". BMC Biology. 9 (1): 71. doi:10.1186/1741-7007-9-71. ISSN 1741-7007. PMC 3203851. PMID 22035460.
- ^ Stephan, Simon; Horsch, Martin T.; Vrabec, Jadran; Hasse, Hans (2019-07-03). "MolMod – an open access database of force fields for molecular simulations of fluids". Molecular Simulation. 45 (10): 806–814. doi:10.1080/08927022.2019.1601191. ISSN 0892-7022. S2CID 119199372.
- ^ Kurzak, J.; Pettitt, B. M. (September 2006). "Fast multipole methods for particle dynamics". Molecular Simulation. 32 (10–11): 775–790. doi:10.1080/08927020600991161. ISSN 0892-7022. PMC 2634295. PMID 19194526.
- ^ Giese, Timothy J.; Panteva, Maria T.; Chen, Haoyuan; York, Darrin M. (2015-02-10). "Multipolar Ewald Methods, 1: Theory, Accuracy, and Performance". Journal of Chemical Theory and Computation. 11 (2): 436–450. doi:10.1021/ct5007983. ISSN 1549-9618. PMC 4325605. PMID 25691829.
- ^ Groenhof, Gerrit (2013), Monticelli, Luca; Salonen, Emppu (eds.), "Introduction to QM/MM Simulations", Biomolecular Simulations: Methods and Protocols, Methods in Molecular Biology, vol. 924, Totowa, NJ: Humana Press, pp. 43–66, doi:10.1007/978-1-62703-017-5_3, hdl:11858/00-001M-0000-0010-15DF-C, ISBN 978-1-62703-017-5, PMID 23034745
- ^ a b Tzeliou, Christina Eleftheria; Mermigki, Markella Aliki; Tzeli, Demeter (January 2022). "Review on the QM/MM Methodologies and Their Application to Metalloproteins". Molecules. 27 (9): 2660. doi:10.3390/molecules27092660. ISSN 1420-3049. PMC 9105939. PMID 35566011.
- ^ a b Lucas, Andrew (2014). "Ising formulations of many NP problems". Frontiers in Physics. 2: 5. arXiv:1302.5843. Bibcode:2014FrP.....2....5L. doi:10.3389/fphy.2014.00005. ISSN 2296-424X.
- ^ Michaud-Rioux, Vincent; Zhang, Lei; Guo, Hong (2016-02-15). "RESCU: A real space electronic structure method". Journal of Computational Physics. 307: 593–613. arXiv:1509.05746. Bibcode:2016JCoPh.307..593M. doi:10.1016/j.jcp.2015.12.014. ISSN 0021-9991. S2CID 28836129.
- ^ Motamarri, Phani; Das, Sambit; Rudraraju, Shiva; Ghosh, Krishnendu; Davydov, Denis; Gavini, Vikram (2020-01-01). "DFT-FE – A massively parallel adaptive finite-element code for large-scale density functional theory calculations". Computer Physics Communications. 246: 106853. arXiv:1903.10959. Bibcode:2020CoPhC.24606853M. doi:10.1016/j.cpc.2019.07.016. ISSN 0010-4655. S2CID 85517990.
- ^ a b c d e Sengupta, Arkajyoti; Ramabhadran, Raghunath O.; Raghavachari, Krishnan (2016-01-15). "Breaking a bottleneck: Accurate extrapolation to "gold standard" CCSD(T) energies for large open shell organic radicals at reduced computational cost". Journal of Computational Chemistry. 37 (2): 286–295. doi:10.1002/jcc.24050. ISSN 0192-8651. PMID 26280676. S2CID 23011794.
- ^ a b Whitfield, James Daniel; Love, Peter John; Aspuru-Guzik, Alán (2013). "Computational complexity in electronic structure". Phys. Chem. Chem. Phys. 15 (2): 397–411. arXiv:1208.3334. Bibcode:2013PCCP...15..397W. doi:10.1039/C2CP42695A. ISSN 1463-9076. PMID 23172634. S2CID 12351374.
- ^ Gieres, François (2000). "Mathematical surprises and Dirac's formalism in quantum mechanics". Reports on Progress in Physics. 63 (12): 1893–1931. arXiv:quant-ph/9907069. Bibcode:2000RPPh...63.1893G. doi:10.1088/0034-4885/63/12/201. S2CID 250880658.
- ^ Dirac, P. A. M. (1929-04-06). "Quantum mechanics of many-electron systems". Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character. 123 (792): 714–733. Bibcode:1929RSPSA.123..714D. doi:10.1098/rspa.1929.0094. ISSN 0950-1207. S2CID 121992478.
- ^ Feynman, Richard P. (2019-06-17). Hey, Tony; Allen, Robin W. (eds.). Feynman Lectures On Computation. Boca Raton: CRC Press. doi:10.1201/9780429500442. ISBN 978-0-429-50044-2. S2CID 53898623.
- ^ a b c d Low, Guang Hao; Chuang, Isaac L. (2019-07-12). "Hamiltonian Simulation by Qubitization". Quantum. 3: 163. Bibcode:2019Quant...3..163L. doi:10.22331/q-2019-07-12-163. S2CID 119109921.
- ^ Kwon, Hyuk-Yong; Curtin, Gregory M.; Morrow, Zachary; Kelley, C. T.; Jakubikova, Elena (2022). "Adaptive Basis Sets for Practical Quantum Computing". arXiv:2211.06471.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Kitaev, Alexei (1996-01-17). Quantum measurements and the Abelian Stabilizer Problem (Report). Electronic Colloquium on Computational Complexity (ECCC).
- ^ Abrams, Daniel S.; Lloyd, Seth (1999-12-13). "Quantum Algorithm Providing Exponential Speed Increase for Finding Eigenvalues and Eigenvectors". Physical Review Letters. 83 (24): 5162–5165. arXiv:quant-ph/9807070. Bibcode:1999PhRvL..83.5162A. doi:10.1103/PhysRevLett.83.5162. S2CID 118937256.
- ^ a b c d e f Nielsen, Michael A.; Chuang, Isaac L. (2010). Quantum computation and quantum information (10th anniversary ed.). Cambridge: Cambridge university press. ISBN 978-1-107-00217-3.
- ^ McArdle, Sam; Endo, Suguru; Aspuru-Guzik, Alán; Benjamin, Simon C.; Yuan, Xiao (2020-03-30). "Quantum computational chemistry". Reviews of Modern Physics. 92 (1): 015003. Bibcode:2020RvMP...92a5003M. doi:10.1103/RevModPhys.92.015003. S2CID 119476644.
- ^ Du, Jiangfeng; Xu, Nanyang; Peng, Xinhua; Wang, Pengfei; Wu, Sanfeng; Lu, Dawei (2010-01-22). "NMR Implementation of a Molecular Hydrogen Quantum Simulation with Adiabatic State Preparation". Physical Review Letters. 104 (3): 030502. Bibcode:2010PhRvL.104c0502D. doi:10.1103/PhysRevLett.104.030502. ISSN 0031-9007. PMID 20366636.
- ^ Lanyon, B. P.; Whitfield, J. D.; Gillett, G. G.; Goggin, M. E.; Almeida, M. P.; Kassal, I.; Biamonte, J. D.; Mohseni, M.; Powell, B. J.; Barbieri, M.; Aspuru-Guzik, A.; White, A. G. (2010). "Towards quantum chemistry on a quantum computer". Nature Chemistry. 2 (2): 106–111. arXiv:0905.0887. Bibcode:2010NatCh...2..106L. doi:10.1038/nchem.483. ISSN 1755-4349. PMID 21124400. S2CID 640752.
- ^ Wang, Youle; Zhang, Lei; Yu, Zhan; Wang, Xin (2022). "Quantum Phase Processing and its Applications in Estimating Phase and Entropies". arXiv:2209.14278 [quant-ph].
- ^ Sugisaki, Kenji; Toyota, Kazuo; Sato, Kazunobu; Shiomi, Daisuke; Takui, Takeji (2022-07-25). "Adiabatic state preparation of correlated wave functions with nonlinear scheduling functions and broken-symmetry wave functions". Communications Chemistry. 5 (1): 84. doi:10.1038/s42004-022-00701-8. ISSN 2399-3669. PMC 9814591. PMID 36698020.
- ^ Peruzzo, Alberto; McClean, Jarrod; Shadbolt, Peter; Yung, Man-Hong; Zhou, Xiao-Qi; Love, Peter J.; Aspuru-Guzik, Alán; O'Brien, Jeremy L. (2014-07-23). "A variational eigenvalue solver on a photonic quantum processor". Nature Communications. 5 (1): 4213. arXiv:1304.3061. Bibcode:2014NatCo...5.4213P. doi:10.1038/ncomms5213. ISSN 2041-1723. PMC 4124861. PMID 25055053.
- ^ Peruzzo, Alberto; McClean, Jarrod; Shadbolt, Peter; Yung, Man-Hong; Zhou, Xiao-Qi; Love, Peter J.; Aspuru-Guzik, Alán; O’Brien, Jeremy L. (2014-07-23). "A variational eigenvalue solver on a photonic quantum processor". Nature Communications. 5 (1): 4213. Bibcode:2014NatCo...5.4213P. doi:10.1038/ncomms5213. ISSN 2041-1723. PMC 4124861. PMID 25055053.
- ^ Chan, Albie; Shi, Zheng; Dellantonio, Luca; Dur, Wolfgang; Muschik, Christine A (2023). "Hybrid variational quantum eigensolvers: merging computational models". arXiv:2305.19200 [quant-ph].
- ^ Liu, Jie; Li, Zhenyu; Yang, Jinlong (2021-06-28). "An efficient adaptive variational quantum solver of the Schrödinger equation based on reduced density matrices". The Journal of Chemical Physics. 154 (24). arXiv:2012.07047. Bibcode:2021JChPh.154x4112L. doi:10.1063/5.0054822. ISSN 0021-9606. PMID 34241330. S2CID 229156865.
- ^ Romero, Jonathan; Babbush, Ryan; McClean, Jarrod R; Hempel, Cornelius; Love, Peter J; Aspuru-Guzik, Alán (2018-10-19). "Strategies for quantum computing molecular energies using the unitary coupled cluster ansatz". Quantum Science and Technology. 4 (1): 014008. arXiv:1701.02691. doi:10.1088/2058-9565/aad3e4. ISSN 2058-9565. S2CID 4175437.
- ^ McClean, Jarrod R; Romero, Jonathan; Babbush, Ryan; Aspuru-Guzik, Alán (2016-02-04). "The theory of variational hybrid quantum-classical algorithms". New Journal of Physics. 18 (2): 023023. arXiv:1509.04279. Bibcode:2016NJPh...18b3023M. doi:10.1088/1367-2630/18/2/023023. ISSN 1367-2630. S2CID 92988541.
- ^ Bonet-Monroig, Xavier; Babbush, Ryan; O'Brien, Thomas E. (2020-09-22). "Nearly Optimal Measurement Scheduling for Partial Tomography of Quantum States". Physical Review X. 10 (3): 031064. arXiv:1908.05628. Bibcode:2020PhRvX..10c1064B. doi:10.1103/PhysRevX.10.031064. S2CID 199668962.
- ^ Grimsley, Harper R.; Barron, George S.; Barnes, Edwin; Economou, Sophia E.; Mayhall, Nicholas J. (2023-03-01). "Adaptive, problem-tailored variational quantum eigensolver mitigates rough parameter landscapes and barren plateaus". npj Quantum Information. 9 (1): 19. arXiv:2204.07179. Bibcode:2023npjQI...9...19G. doi:10.1038/s41534-023-00681-0. ISSN 2056-6387. S2CID 257236255.
- ^ a b Jiang, Zhang; Sung, Kevin J.; Kechedzhi, Kostyantyn; Smelyanskiy, Vadim N.; Boixo, Sergio (2018-04-26). "Quantum Algorithms to Simulate Many-Body Physics of Correlated Fermions". Physical Review Applied. 9 (4): 044036. Bibcode:2018PhRvP...9d4036J. doi:10.1103/PhysRevApplied.9.044036. ISSN 2331-7019. S2CID 54064506.
- ^ Li, Qing-Song; Liu, Huan-Yu; Wang, Qingchun; Wu, Yu-Chun; Guo, Guo-Ping (2022). "A unified framework of transformations based on the Jordan–Wigner transformation". The Journal of Chemical Physics. 157 (13). arXiv:2108.01725. Bibcode:2022JChPh.157m4104L. doi:10.1063/5.0107546. PMID 36209000. S2CID 236912625. Retrieved 2023-11-13.
- ^ "Custom Fermionic Codes for Quantum Simulation | Perimeter Institute". www2.perimeterinstitute.ca. Retrieved 2023-11-13.
- ^ Kivlichan, Ian D.; McClean, Jarrod; Wiebe, Nathan; Gidney, Craig; Aspuru-Guzik, Alán; Chan, Garnet Kin-Lic; Babbush, Ryan (2018-03-13). "Quantum Simulation of Electronic Structure with Linear Depth and Connectivity". Physical Review Letters. 120 (11): 110501. arXiv:1711.04789. Bibcode:2018PhRvL.120k0501K. doi:10.1103/PhysRevLett.120.110501. PMID 29601758. S2CID 4219888.
- ^ Hashim, Akel; Rines, Rich; Omole, Victory; Naik, Ravi K; John MarkKreikebaum, John Mark; Santiago, David I; Chong, Frederic T.; Siddiqi, Irfan; Gokhale, Pranav (2021). "Optimized fermionic SWAP networks with equivalent circuit averaging for QAOA". arXiv:2111.04572 [quant-ph].
- ^ Rubin, Nicholas C.; Gunst, Klaas; White, Alec; Freitag, Leon; Throssell, Kyle; Chan, Garnet Kin-Lic; Babbush, Ryan; Shiozaki, Toru (2021-10-27). "The Fermionic Quantum Emulator". Quantum. 5: 568. arXiv:2104.13944. Bibcode:2021Quant...5..568R. doi:10.22331/q-2021-10-27-568. S2CID 233443911.
- ^ a b c d e f g Ramachandran, K. I.; Deepa, G.; Namboori, K. (2008). Computational chemistry and molecular modeling: principles and applications. Berlin: Springer. ISBN 978-3-540-77304-7.
- ^ a b "The Effective Fragment Potential: An Ab Initio Force Field". Comprehensive Computational Chemistry: 153–161. 2024-01-01. doi:10.1016/B978-0-12-821978-2.00141-0.
- ^ "Conceptual Density Functional Theory". Comprehensive Computational Chemistry: 306–321. 2024-01-01. doi:10.1016/B978-0-12-821978-2.00025-8.
- ^ Counts, Richard W. (1987-07-01). "Strategies I". Journal of Computer-Aided Molecular Design. 1 (2): 177–178. Bibcode:1987JCAMD...1..177C. doi:10.1007/bf01676961. ISSN 0920-654X. PMID 3504968. S2CID 40429116.
- ^ Dinur, Uri; Hagler, Arnold T. (1991). Lipkowitz, Kenny B.; Boyd, Donald B. (eds.). Reviews in Computational Chemistry. John Wiley & Sons, Inc. pp. 99–164. doi:10.1002/9780470125793.ch4. ISBN 978-0-470-12579-3.
- ^ Rubenstein, Lester A.; Zauhar, Randy J.; Lanzara, Richard G. (2006). "Molecular dynamics of a biophysical model for β2-adrenergic and G protein-coupled receptor activation" (PDF). Journal of Molecular Graphics and Modelling. 25 (4): 396–409. doi:10.1016/j.jmgm.2006.02.008. PMID 16574446. Archived (PDF) from the original on 2008-02-27.
- ^ Rubenstein, Lester A.; Lanzara, Richard G. (1998). "Activation of G protein-coupled receptors entails cysteine modulation of agonist binding" (PDF). Journal of Molecular Structure: THEOCHEM. 430: 57–71. doi:10.1016/S0166-1280(98)90217-2. Archived (PDF) from the original on 2004-05-30.
- ^ a b c d e Lukassen, Axel Ariaan; Kiehl, Martin (2018-12-15). "Operator splitting for chemical reaction systems with fast chemistry". Journal of Computational and Applied Mathematics. 344: 495–511. doi:10.1016/j.cam.2018.06.001. ISSN 0377-0427. S2CID 49612142.
- ^ "Ab Initio Molecular Dynamics: A Guide to Applications". Comprehensive Computational Chemistry: 493–517. 2024-01-01. doi:10.1016/B978-0-12-821978-2.00096-9.
- ^ Satoh, A. (2003-01-01), Satoh, A. (ed.), "Chapter 3 - Monte Carlo Methods", Studies in Interface Science, Introduction to Molecular-Microsimulation of Colloidal Dispersions, vol. 17, Elsevier, pp. 19–63, retrieved 2023-12-03
- ^ Allen, M. P. (1987). Computer simulation of liquids. D. J. Tildesley. Oxford [England]: Clarendon Press. ISBN 0-19-855375-7. OCLC 15132676.
- ^ McArdle, Sam; Endo, Suguru; Aspuru-Guzik, Alán; Benjamin, Simon C.; Yuan, Xiao (2020-03-30). "Quantum computational chemistry". Reviews of Modern Physics. 92 (1): 015003. Bibcode:2020RvMP...92a5003M. doi:10.1103/RevModPhys.92.015003. ISSN 0034-6861.
- ^ "Molecular Dynamics and QM/MM to Understand Genome Organization and Reproduction in Emerging RNA Viruses". Comprehensive Computational Chemistry: 895–909. 2024-01-01. doi:10.1016/B978-0-12-821978-2.00101-X.
- ^ Visscher, Lucas (June 2002). "The Dirac equation in quantum chemistry: Strategies to overcome the current computational problems". Journal of Computational Chemistry. 23 (8): 759–766. doi:10.1002/jcc.10036. ISSN 0192-8651. PMID 12012352. S2CID 19427995.
- ^ Sax, Alexander F. (2008-04-01). "Computational Chemistry techniques: covering orders of magnitude in space, time, and accuracy". Monatshefte für Chemie - Chemical Monthly. 139 (4): 299–308. doi:10.1007/s00706-007-0827-7. ISSN 1434-4475. S2CID 85451980.
- ^ Vanommeslaeghe, Kenno; Guvench, Olgun; Jr, Alexander D. MacKerell (2014). "Molecular Mechanics". Current Pharmaceutical Design. 20 (20): 3281–3292. doi:10.2174/13816128113199990600. PMC 4026342. PMID 23947650.
- ^ Friesner, R. (2003-03-01). "How iron-containing proteins control dioxygen chemistry: a detailed atomic level description via accurate quantum chemical and mixed quantum mechanics/molecular mechanics calculations". Coordination Chemistry Reviews. 238–239: 267–290. doi:10.1016/S0010-8545(02)00284-9. ISSN 0010-8545.
Specialized journals on computational chemistry
- Annual Reports in Computational Chemistry
- Computational and Theoretical Chemistry
- Computational and Theoretical Polymer Science
- Computers & Chemical Engineering
- Journal of Chemical Information and Modeling
- Journal of Chemical Information and Modeling
- Journal of Chemical Software
- Journal of Chemical Theory and Computation
- Journal of Cheminformatics
- Journal of Computational Chemistry
- Journal of Computer Aided Chemistry
- Journal of Computer Chemistry Japan
- Journal of Computer-aided Molecular Design
- Journal of Theoretical and Computational Chemistry
- Molecular Informatics
- Theoretical Chemistry Accounts
External links
- NIST Computational Chemistry Comparison and Benchmark DataBase – Contains a database of thousands of computational and experimental results for hundreds of systems
- American Chemical Society Division of Computers in Chemistry – American Chemical Society Computers in Chemistry Division, resources for grants, awards, contacts and meetings.
- CSTB report Mathematical Research in Materials Science: Opportunities and Perspectives – CSTB Report
- 3.320 Atomistic Computer Modeling of Materials (SMA 5107) Free MIT Course
- Chem 4021/8021 Computational Chemistry Free University of Minnesota Course
- Technology Roadmap for Computational Chemistry
- Applications of molecular and materials modelling.
- Impact of Advances in Computing and Communications Technologies on Chemical Science and Technology CSTB Report
- MD and Computational Chemistry applications on GPUs
- Susi Lehtola, Antti J. Karttunen:"Free and open source software for computational chemistry education", First published: 23 March 2022, https://doi.org/10.1002/wcms.1610 (Open Access) Archived 9 August 2022 at the Wayback Machine
- CCL.NET: Computational Chemistry List, Ltd.

























































