Choanoflagellate: Difference between revisions
CodeBadger (talk | contribs) m Added month of publication to cite ('Cyst Formation in a Freshwater Strain of the Choanoflagellate Desmarella moniliformis Kent'). |
Citation bot (talk | contribs) Added bibcode. | Use this bot. Report bugs. | Suggested by Abductive | Category:Wikipedia articles needing clarification from April 2024 | #UCB_Category 703/818 |
||
| (146 intermediate revisions by 77 users not shown) | |||
| Line 1: | Line 1: | ||
{{short description|Group of eukaryotes considered the closest living relatives of animals}} |
|||
{{Taxobox |
|||
{{redirect|Choanoflagellates|the clade that groups these organisms with animals|Choanozoa}} |
|||
| color = {{taxobox color|[[animalia]]}} |
|||
{{Automatic taxobox |
|||
| fossil_range = {{long fossil range|900|0}} <small>No fossils known, molecular clock evidence for origin 1050-800Ma<ref name="Wegener-Parfrey et al. 2011">{{cite journal|last=Wegener-Parfrey|first=Laura|author2=Lahr DJ|author3=Knoll AH|author4=Katz LA.|title=Estimating the timing of early eukaryotic diversification with multigene molecular clocks|journal=PNAS|date=August 16, 2011|volume=108|issue=33|pages=13624–13629|doi=10.1073/pnas.1110633108|url=http://www.pnas.org/content/108/33/13624.abstract|pmid=21810989|pmc=3158185}}</ref></small> |
|||
| name = Choanoflagellates |
| name = Choanoflagellates |
||
| fossil_range = {{long fossil range|100.5|0|earliest=900}} <small>Only possible fossils are known from [[Cretaceous]] ([[Cenomanian]]/[[Turonian]]),<ref>{{Cite journal |last1=Fonseca |first1=Carolina |last2=Mendonça Filho |first2=João Graciano |last3=Reolid |first3=Matías |last4=Duarte |first4=Luís V. |last5=de Oliveira |first5=António Donizeti |last6=Souza |first6=Jaqueline Torres |last7=Lézin |first7=Carine |date=2023-01-23 |title=First putative occurrence in the fossil record of choanoflagellates, the sister group of Metazoa |journal=Scientific Reports |language=en |volume=13 |issue=1 |pages=1242 |doi=10.1038/s41598-022-26972-8 |pmid=36690681 |pmc=9870899 |bibcode=2023NatSR..13.1242F |issn=2045-2322}}</ref> molecular clock evidence for origin 1050-800 Ma<ref name="Wegener-Parfrey et al. 2011">{{cite journal | vauthors = Parfrey LW, Lahr DJ, Knoll AH, Katz LA | title = Estimating the timing of early eukaryotic diversification with multigene molecular clocks | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 108 | issue = 33 | pages = 13624–9 | date = August 2011 | pmid = 21810989 | pmc = 3158185 | doi = 10.1073/pnas.1110633108 | bibcode = 2011PNAS..10813624P | doi-access = free }}</ref></small> |
|||
| image = Cronoflagelado2.svg |
|||
| |
| image = 1singlelate.jpg |
||
| image_caption = ''Codosiga'' sp. |
|||
| image_width = 150px |
|||
| taxon = Choanoflagellata |
|||
| domain = [[Eukarya]] |
|||
| display_parents = 7 |
|||
| unranked_phylum = [[Opisthokonta]] |
|||
| authority = [[William Saville-Kent|Kent]], 1880–1882<ref>[[Saville-Kent, W.]] (1880). ''A manual of Infusoria''. London, vol. 1, p. 324, [https://archive.org/stream/manualofinfusori18081kent#page/324/mode/2up].</ref><ref name="Adl 2019">{{cite journal|vauthors=Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, ((Lahr DJG)), Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, ((Mitchell EAD)), Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q|year=2019|title=Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes|journal=Journal of Eukaryotic Microbiology|volume=66|issue=1 |pages=4–119|doi=10.1111/jeu.12691|pmid=30257078 |pmc=6492006 }}</ref> |
|||
| unranked_subphylum = [[Choanozoa]] or [[Holozoa]] |
|||
| type_species = ''[[Monosiga brevicollis]]''<ref>{{cite journal | vauthors = King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D | display-authors = 6 | title = The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans | journal = Nature | volume = 451 | issue = 7180 | pages = 783–8 | date = February 2008 | pmid = 18273011 | pmc = 2562698 | doi = 10.1038/nature06617 | bibcode = 2008Natur.451..783K }}</ref> |
|||
| unranked_superclassis = [[Filozoa]] |
|||
| subdivision_ranks = Orders & families |
|||
| classis = '''Choanoflagellatea''' |
|||
| subdivision = |
|||
| classis_authority = Cavalier-Smith, 1998<ref>Cavalier-Smith, T. 1998. Neomonada and the origin of animals and fungi. In: Coombs GH, Vickerman K, Sleigh MA, Warren A (ed.) ''Evolutionary relationships among protozoa''. Kluwer, London, pp. 375-407.</ref> |
|||
* [[Craspedida]] |
|||
| subdivision_ranks = Families |
|||
** [[Codonosigidae]] |
|||
* [[Acanthoecida]] |
|||
[[Salpingoecidae]]<br> |
|||
** [[Stephanoecidae]] |
|||
[[Acanthoecidae]] |
|||
** [[Acanthoecidae]] |
|||
| synonyms = |
|||
* Craspedmonadina <small>Stein, 1878</small> |
| synonyms = * Craspedmonadina <small>Stein, 1878</small> |
||
* Choanoflagellata <small>[[William Saville-Kent|Kent]], 1880</small><ref>[[Saville-Kent, W.]] (1880). ''A manual of Infusoria''. London, vol. 1, p. 324, [https://archive.org/stream/manualofinfusori18081kent#page/324/mode/2up].</ref> |
|||
* Craspedomonadaceae <small>Senn, 1900</small> |
* Craspedomonadaceae <small>Senn, 1900</small> |
||
* Craspedophyceae <small>Chadefaud, 1960</small> |
* Craspedophyceae <small>Chadefaud, 1960</small> |
||
| Line 26: | Line 25: | ||
* Choanoflagellida <small>Levine et al., 1980, Lee et al., 1985</small> |
* Choanoflagellida <small>Levine et al., 1980, Lee et al., 1985</small> |
||
* Choanoflagellea <small>Cavalier-Smith, 1997</small> |
* Choanoflagellea <small>Cavalier-Smith, 1997</small> |
||
* Choanomonada <small>Adl et al. |
* Choanomonada <small>Adl et al. 2005</small><ref>{{cite journal | vauthors = Nitsche F, Carr M, Arndt H, Leadbeater BS | title = Higher level taxonomy and molecular phylogenetics of the Choanoflagellatea | journal = The Journal of Eukaryotic Microbiology | volume = 58 | issue = 5 | pages = 452–62 | year = 2011 | pmid = 21895836 | doi = 10.1111/j.1550-7408.2011.00572.x | s2cid = 2076733 }}</ref> |
||
* Choanoflagellatea {{au|Cavalier-Smith, 1998}}<ref>{{cite book | vauthors = Cavalier-Smith T | date = 1998 | chapter = Neomonada and the origin of animals and fungi. | veditors = Coombs GH, Vickerman K, Sleigh MA, Warren A | title = Evolutionary relationships among protozoa | publisher = Kluwer | location = London | pages = 375–407 }}</ref><ref name="Leadbeater">{{cite book| last1 = Leadbeater| first1 = Barry S. C. | name-list-style = vanc | year = 2015 | title = The choanoflagellates: evolution, biology, and ecology | url =http://www.cambridge.org/us/academic/subjects/life-sciences/ecology-and-conservation/choanoflagellates-evolution-biology-and-ecology?format=AR | publisher=University of Birmingham | isbn=978-0-521-88444-0}}</ref> |
|||
}} |
}} |
||
The '''choanoflagellates''' are a group of free-living unicellular and colonial [[flagellate]] [[eukaryotes]] considered to be the closest living relatives of the [[animal]]s. Choanoflagellates |
The '''choanoflagellates''' are a group of free-living unicellular and colonial [[flagellate]] [[eukaryotes]] considered to be the closest living relatives of the [[animal]]s. Choanoflagellates are collared flagellates, having a funnel shaped collar of interconnected [[microvilli]] at the base of a [[flagellum]]. Choanoflagellates are capable of both [[Asexual reproduction|asexual]] and sexual reproduction.<ref name="sciencedaily.com">{{Cite web | url=https://www.sciencedaily.com/releases/2017/08/170831123023.htm | title=Bacterial protein acts as aphrodisiac for choanoflagellates}}</ref> They have a distinctive cell [[morphology (biology)|morphology]] characterized by an ovoid or spherical cell body 3–10 [[Micrometre|μm]] in diameter with a single apical flagellum surrounded by a collar of 30–40 microvilli (see figure). Movement of the flagellum creates water currents that can propel [[free-swimming]] choanoflagellates through the water column and trap [[bacteria]] and [[detritus]] against the collar of microvilli, where these foodstuffs are engulfed. This feeding provides a critical link within the global [[carbon cycle]], linking [[trophic level]]s. In addition to their critical ecological roles, choanoflagellates are of particular interest to evolutionary biologists studying the origins of multicellularity in animals. As the closest living relatives of animals, choanoflagellates serve as a useful model for reconstructions of the last unicellular ancestor of animals. |
||
==Etymology== |
==Etymology== |
||
{{wikt}} |
|||
From Greek ''Khoanē'' meaning "'''funnel'''" (due to the shape of the collar) |
|||
''Choanoflagellate'' is a [[hybrid word]] from Greek {{wikt-lang|grc|χοάνη|italic=no}} ''{{Lang|grc-latn|khoánē}}'' meaning "[[funnel]]" (due to the shape of the collar) and the Latin word {{wikt-lang|la|flagellum}} (whence English ''[[flagellum]]'').{{Citation needed|date=February 2021}} |
|||
and the English word "'''[[flagellum]]'''". |
|||
==Appearance |
==Appearance== |
||
[[File:Cronoflagelado2.svg|thumb|150px|left|Cell scheme]] |
|||
[[Image:Salpingoeca sp..jpg|thumb|150px|left|''Salpingoeca'' sp., in transmission electron microscopy ([[Transmission electron microscopy|TEM]]).]] |
|||
Each choanoflagellate has a single [[flagellum]], surrounded by a ring of [[actin]]-filled protrusions called [[microvilli]], forming a cylindrical or conical collar (''choanos'' in Greek). Movement of the [[flagellum]] draws water through the collar, and [[bacteria]] and detritus are captured by the microvilli and ingested.<ref name=King2008>{{cite journal |
|||
| author = [[Nicole King|King, N.]] |author2=Westbrook, M.J. |author3=Young, S.L. |author4=Kuo, A. |author5=Abedin, M. |author6=Chapman, J. |author7=Fairclough, S. |author8=Hellsten, U. |author9=Isogai, Y. |author10=Letunic, I. |
|||
| date = 14 February 2008 |
|||
| title = The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans |
|||
| journal = [[Nature (journal)|Nature]] |
|||
| volume = 451 |
|||
| issue = 7180 |
|||
| pages = 783–8 |
|||
| doi = 10.1038/nature06617 |
|||
| pmid = 18273011 |
|||
| pmc = 2562698 |
|||
|display-authors=etal}}</ref> |
|||
Water currents generated by the [[flagellum]] also push free-swimming cells along, as in [[animal]] [[spermatozoon|sperm]]. In contrast, most other flagellates are ''pulled'' by their flagella. |
|||
Each choanoflagellate has a single [[flagellum]], surrounded by a ring of [[actin]]-filled protrusions called [[Microvillus|microvilli]], forming a cylindrical or conical collar (''{{Lang|grc-latn|choanos}}'' in Greek). Movement of the flagellum draws water through the collar, and bacteria and detritus are captured by the microvilli and ingested.<ref name=King2008>{{cite journal | vauthors = King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D | display-authors = 6 | title = The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans | journal = Nature | volume = 451 | issue = 7180 | pages = 783–8 | date = February 2008 | pmid = 18273011 | pmc = 2562698 | doi = 10.1038/nature06617 | author-link = Nicole King | bibcode = 2008Natur.451..783K }}</ref> Water currents generated by the flagellum also push free-swimming cells along, as in [[animal]] [[spermatozoon|sperm]]. In contrast, most other flagellates are ''pulled'' by their flagella.{{Citation needed|date=February 2021}} |
|||
In addition to the single apical [[flagellum]] surrounded by [[actin]]-filled [[microvilli]] that characterizes choanoflagellates, the internal organization of [[organelles]] in the [[cytoplasm]] is constant.<ref name=Leadbeater2000(1)>{{cite journal |
|||
| author = Leadbeater, B.S.C. |
|||
|author2=Thomsen, H. |
|||
| year = 2000 |
|||
| title = Order Choanoflagellida |
|||
| journal = An Illustrated Guide to the Protozoa, Second Edition. Lawrence : Society of Protozoologists |
|||
| volume = 451 |
|||
| pages = 14–38 |
|||
}}</ref> A flagellar [[basal body]] sits at the base of the [[wikt:apical|apical]] flagellum, and a second, non-flagellar [[basal body]] rests at a right angle to the flagellar base. The [[Cell nucleus|nucleus]] occupies an apical-to-central position in the cell, and [[food vacuole]]s are positioned in the [[Basal (anatomy)#Elongated organisms with distinctive ends|basal]] region of the [[cytoplasm]].<ref name=Leadbeater2000(1)/><ref name=Karpov1998>{{cite journal |
|||
| author = Karpov S. |
|||
|author2=Leadbeater, B.S.C. |
|||
| date = May 1998 |
|||
| title = Cytoskeleton structure and composition in choanoflagellates |
|||
| journal = [[Journal of Eukaryotic Microbiology]] |
|||
| volume = 45 |
|||
| pages = 361–367 |
|||
| doi = 10.1111/j.1550-7408.1998.tb04550.x |
|||
| issue = 3 |
|||
}}</ref> Additionally, the cell body of many choanoflagellates is surrounded by a distinguishing [[extracellular matrix]] or [[Periplasmic space|periplast]]. These cell coverings vary greatly in structure and composition and are used by taxonomists for classification purposes. Many choanoflagellates build complex basket-shaped "houses", called [[Lorica (biology)|lorica]], from several silica strips cemented together.<ref name=Leadbeater2000(1)/> The functional significance of the periplast is unknown, but in sessile organisms, it is thought to aid attachment to the substrate. In planktonic organisms, there is speculation that the periplast increases drag, thereby counteracting the force generated by the flagellum and increasing feeding efficiency.<ref name=Leadbeater2001>{{cite journal |
|||
| author = Leadbeater, B.S.C. |
|||
| year = 2001 |
|||
|author2=Kelly, M. |
|||
| title = Evolution of animals choanoflagellates and sponges |
|||
| journal = Water and Atmosphere Online |
|||
| volume = 9 |
|||
| issue = 2 |
|||
| pages = 9–11 |
|||
}}</ref> |
|||
Choanoflagellates are either free-swimming in the water column or [[Sessility (zoology)|sessile]], adhering to the substrate directly or through either the periplast or a thin pedicel.<ref name=Leadbeater1983>{{cite journal |
|||
| author = Leadbeater, B.S.C. |
|||
| date = February 1983 |
|||
| title = Life-History and Ultrastructure of a New Marine Species of ''Proterospongia'' (Choanoflagellida) |
|||
| journal = [[Journal of the Marine Biological Association of the United Kingdom|J. Mar. Biol. Ass. U.K.]] |
|||
| volume =63 |
|||
| issue = 63 |
|||
| pages = 135–160 |
|||
| doi = 10.1017/S0025315400049857 |
|||
}}</ref> Although choanoflagellates are thought to be strictly free-living and [[heterotrophic]], a number of choanoflagellate relatives, such as members of [[Mesomycetozoea|Ichthyosporea or Mesomycetozoa]], follow a [[parasitic]] or [[pathogenic]] lifestyle.<ref name=Mendoza2002>{{cite journal |
|||
| author = Mendoza L. |
|||
| year = 2002 |author2=Taylor, J. |author3=Ajello, L. |
|||
| title = The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary |
|||
| journal = [[Annu. Rev. Microbiol.]] |
|||
| pmid = 12142489 |
|||
| volume = 56 |
|||
| issue = |
|||
| pages = 315–44 |
|||
| doi = 10.1146/annurev.micro.56.012302.160950 |
|||
}}</ref> The life histories of choanoflagellates are poorly understood. Many species are thought to be solitary; however coloniality seems to have arisen independently several times within the group and colonial species retain a solitary stage.<ref name=Leadbeater1983/> |
|||
In addition to the single apical flagellum surrounded by actin-filled microvilli that characterizes choanoflagellates, the internal organization of [[organelles]] in the [[cytoplasm]] is constant.<ref name=Leadbeater2000(1)>{{cite journal | vauthors = Leadbeater BS, Thomsen H | year = 2000 | title = Order Choanoflagellida | journal = An Illustrated Guide to the Protozoa, Second Edition. Lawrence: Society of Protozoologists | volume = 451 | pages = 14–38}}</ref> A flagellar [[basal body]] sits at the base of the [[wikt:apical|apical]] flagellum, and a second, non-flagellar basal body rests at a right angle to the flagellar base. The [[Cell nucleus|nucleus]] occupies an apical-to-central position in the cell, and [[food vacuole]]s are positioned in the basal region of the cytoplasm.<ref name=Leadbeater2000(1)/><ref name=Karpov1998>{{cite journal | vauthors = Karpov S, Leadbeater BS | date = May 1998 | title = Cytoskeleton structure and composition in choanoflagellates | journal = Journal of Eukaryotic Microbiology | volume = 45 | pages = 361–367 | doi = 10.1111/j.1550-7408.1998.tb04550.x | issue = 3| s2cid = 86287656 }}</ref> Additionally, the cell body of many choanoflagellates is surrounded by a distinguishing [[extracellular matrix]] or [[periplast]]. These cell coverings vary greatly in structure and composition and are used by taxonomists for classification purposes. Many choanoflagellates build complex basket-shaped "houses", called [[Lorica (biology)|lorica]], from several silica strips cemented together.<ref name=Leadbeater2000(1)/> The functional significance of the periplast is unknown, but in sessile organisms, it is thought to aid attachment to the substrate. In planktonic organisms, there is speculation that the periplast increases drag, thereby counteracting the force generated by the flagellum and increasing feeding efficiency.<ref name=Leadbeater2001>{{cite journal | vauthors = Leadbeater BS, Kelly M | year = 2001 | title = Evolution of animals choanoflagellates and sponges | journal = Water and Atmosphere Online | volume = 9 | issue = 2 | pages = 9–11}}</ref> |
|||
Choanoflagellates grow vegetatively, with many species undergoing longitudinal fission;<ref name=Karpov1998/> however, the reproductive life cycle of choanoflagellates remains to be elucidated. Currently, it is unclear whether there is a sexual phase to the choanoflagellate life cycle, and the [[ploidy]] level is unknown;<ref>Claus Nielsen. Animal Evolution: Interrelationships of the Living Phyla. 3rd ed. Claus Nielsen. Oxford, UK: Oxford University Press, 2012, p. 14.</ref> however, the discovery of both retrotransposons and key genes involved in meiosis |
|||
<ref name=Carr2010>{{cite journal |
|||
| author = Carr M. |
|||
| year = 2002 |author2=Leadbeater B. |author3=Baldauf, S. |
|||
| title = Conserved Meiotic Genes Point to Sex in the Choanoflagellates |
|||
| journal = [[J. Eukaryot. Microbiol.]] |
|||
| volume = 57 |
|||
| issue = 1 |
|||
| pages = 56–62 |
|||
| doi = 10.1111/j.1550-7408.2009.00450.x |
|||
}}</ref> suggests that they are cryptically sexual. Interestingly, some choanoflagellates can undergo encystment, which involves the retraction of the flagellum and collar and encasement in an electron dense fibrillar wall. On transfer to fresh media, excystment occurs; though it remains to be directly observed.<ref name=Leadbeater2000(2)>{{cite journal |
|||
| author = Leadbeater, B.S.C. |
|||
|author2=Karpov, S. |
|||
| date = September-October 2000 |
|||
| title = Cyst Formation in a Freshwater Strain of the Choanoflagellate ''Desmarella moniliformis'' Kent |
|||
| journal = [[J. Eukaryot. Microbiol.]] |
|||
| volume = 47 |
|||
| pages = 433–439 |
|||
| doi = 10.1111/j.1550-7408.2000.tb00071.x |
|||
| pmid = 11001139 |
|||
| issue = 5 |
|||
}}</ref> Further examination of the choanoflagellate [[Biological life cycle|life cycle]] will be informative about mechanisms of colony formation and attributes present before the [[evolution]] of [[animal]] multicellularity. |
|||
Choanoflagellates are either [[Motility|free-swimming]] in the water column or [[Sessility (zoology)|sessile]], adhering to the substrate directly or through either the periplast or a thin pedicel.<ref name="Leadbeater1983">{{cite journal | vauthors = Leadbeater BS | date = February 1983 | title = Life-History and Ultrastructure of a New Marine Species of ''Proterospongia'' (Choanoflagellida) | journal = [[Journal of the Marine Biological Association of the United Kingdom|J. Mar. Biol. Assoc. U. K.]] | volume = 63 | issue = 1 | pages = 135–160 | doi = 10.1017/S0025315400049857| bibcode = 1983JMBUK..63..135L | s2cid = 84666673 }}</ref> Although choanoflagellates are thought to be strictly free-living and [[heterotrophic]], a number of choanoflagellate relatives, such as members of [[Mesomycetozoea|Ichthyosporea or Mesomycetozoa]], follow a [[parasitic]] or [[pathogenic]] lifestyle.<ref name="Mendoza2002">{{cite journal | vauthors = Mendoza L, Taylor JW, Ajello L | s2cid = 14764188 | title = The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary | journal = Annual Review of Microbiology | volume = 56 | pages = 315–44 | year = 2002 | pmid = 12142489 | doi = 10.1146/annurev.micro.56.012302.160950 }}</ref> The life histories of choanoflagellates are poorly understood. Many species are thought to be solitary; however, coloniality seems to have arisen independently several times within the group, and colonial species still retain a solitary stage.<ref name="Leadbeater1983" /> |
|||
==Classification== |
|||
==Ecology== |
|||
===Phylogenetic relationships=== |
|||
[[File:Choanoflagellates (Méchnikov).png|thumb|250px|right| Drawing of a choanoflagellate colony by [[Metchnikoff]], 1886]] |
|||
The choanoflagellates were included in [[Chrysophyceae]] until Hibberd, 1975.<ref>Reviers, B. de. (2006). ''[http://books.google.com.br/books?id=6fw4AgAAQBAJ&lpg=PA13&hl=pt-BR&pg=PA156#v=onepage&q&f=false Biologia e Filogenia das Algas]''. Editora Artmed, Porto Alegre, p. 156.</ref> Recent molecular [[phylogenetic]] reconstruction of the internal relationships of choanoflagellates allows the polarization of character evolution within the clade. Large fragments of the nuclear [[18S ribosomal RNA|SSU]] and [[28S ribosomal RNA|LSU]] [[ribosomal RNA]], [[tubulin|alpha tubulin]], and [[heat-shock protein]] 90 coding genes were used to resolve the internal relationships and character polarity within choanoflagellates.<ref name=Carr2008/> Each of the four genes showed similar results independently and analysis of the combined data set ([[concatenated]]) along with sequences from other closely related species ([[animal]]s and [[fungi]]) demonstrate that choanoflagellates are strongly supported as [[monophyletic]] and confirm their position as the closest known unicellular living relative of animals. |
|||
Over 125 extant species of choanoflagellates<ref name=King2008/> are known, distributed globally in [[marine (ocean)|marine]], [[brackish]] and [[freshwater]] environments from the Arctic to the tropics, occupying both [[pelagic]] and [[benthic]] zones. Although most sampling of choanoflagellates has occurred between {{Convert|0 and 25|m|ft|abbr=on|sp=us}}, they have been recovered from as deep as {{Convert|300|m|ft|abbr=on|sp=us}} in open water<ref name=Thomsen1982>{{cite book | vauthors = Thomsen H | year = 1982 | title = Planktonic choanoflagellates from Disko Bugt, West Greenland, with a survey of the marine nanoplankton of the area | series= Meddelelser om Gronland, Bioscience | volume = 8 | pages = 3–63 | isbn = 978-87-635-1149-0}}</ref> and {{Convert|100|m|ft|abbr=on|sp=us}} under Antarctic ice sheets.<ref name="Buck1988">{{cite journal | vauthors = Buck KR, Garrison DL | date = June 1988 | title = Distribution and abundance of choanoflagellates (Acanthoecidae) across the ice-edge zone in the Weddell Sea, Antarctica | journal = [[Marine Biology (journal)|Mar. Biol.]] | volume = 98 | issue = 2 | pages = 263–269 | doi = 10.1007/BF00391204 | bibcode = 1988MarBi..98..263B | s2cid = 84931348 }}</ref> Many species are hypothesized to be [[cosmopolitan (biology)|cosmopolitan]] on a global scale [e.g., ''[[Diaphanoeca grandis]]'' has been reported from [[North America]], [[Europe]] and [[Australia]] (OBIS)], while other species are reported to have restricted regional distributions.<ref name=Thomsen1991>{{cite journal | vauthors = Thomsen H, Buck K, Chavez F | year = 1991 | title = Choanoflagellates of the central California waters: Taxonomy, morphology and species assemblages | journal = [[Ophelia (journal)|Ophelia]] | volume = 33 | pages = 131–164 | doi =10.1080/00785326.1991.10429736 | issue = 2}}</ref> Co-distributed choanoflagellate species can occupy quite different microenvironments, but in general, the factors that influence the distribution and dispersion of choanoflagellates remain to be elucidated.{{Citation needed|date=February 2021}} |
|||
Previously, Choanoflagellida was divided into these three families based on the composition and structure of their periplast: Codonosigidae, Salpingoecidae and Acanthoecidae. Members of the family Codonosigidae appear to lack a periplast when examined by light microscopy, but may have a fine outer coat visible only by [[electron microscopy]]. The family Salpingoecidae consists of species whose cells are encased in a firm theca that is visible by both light and electron microscopy. The [[theca]] is a secreted covering predominately composed of [[cellulose]] or other [[polysaccharides]] (Adl, et al., 2005). These divisions are now known to be [[paraphyletic]], with convergent evolution of these forms widespread. The third family of choanoflagellates, the Acanthoecidae, has been supported as a monophyletic group. This clade possess a [[synapomorphy]] of the cells being found within a basket-like lorica, providing the alternative name of "Loricate Choanoflagellates". The Acanthoecid lorica is composed of a series of [[siliceous]] costal strips arranged into a species-specific lorica pattern."<ref name=Leadbeater2000(1)/><ref name=Leadbeater2001/> |
|||
A number of [[species]], such as those in the [[genus]] ''[[Proterospongia]]'', form simple [[Colony (biology)|colonies]],<ref name=King2008/> [[planktonic]] clumps that resemble a miniature cluster of grapes in which each cell in the colony is flagellated or clusters of cells on a single stalk.<ref name=Leadbeater2000(1)/><ref name=Carr2008>{{cite journal | vauthors = Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL | title = Molecular phylogeny of choanoflagellates, the sister group to Metazoa | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 105 | issue = 43 | pages = 16641–6 | date = October 2008 | pmid = 18922774 | pmc = 2575473 | doi = 10.1073/pnas.0801667105 | bibcode = 2008PNAS..10516641C | doi-access = free }}</ref> In October 2019, scientists found a new band behaviour of choanoflagellates: they apparently can coordinate to respond to light.<ref>{{Cite web|url=https://www.hhmi.org/news/newly-discovered-microorganisms-band-together-flip-out|title=Newly Discovered Microorganisms Band Together, 'Flip Out'|website=HHMI.org|language=en|access-date=2019-10-29}}</ref> |
|||
The choanoflagellate tree based on molecular phylogenetics divides into three well supported [[clades]].<ref name=Carr2008/> Clade 1 and Clade 2 each consist of a combination of species traditionally attributed to the Codonosigidae and Salpingoecidae, while Clade 3 comprises species from the group taxonomically classified as Acanthoecidae.<ref name=Carr2008/> The mapping of character traits on to this phylogeny indicates that the [[last common ancestor]] of choanoflagellates was a marine organism with a differentiated [[Biological life cycle|life cycle]] with [[sessility (zoology)|sedentary]] and [[Motility|motile]] stages.<ref name=Carr2008/> |
|||
The choanoflagellates feed on [[bacteria]] and link otherwise inaccessible forms of [[carbon]] to organisms higher in the trophic chain.<ref name=Butterfield1997>{{cite journal | vauthors = Butterfield NJ | date = April 1, 1997 | title = Plankton ecology and the Proterozoic-Phanerozoic transition | journal = [[Paleobiology (journal)|Paleobiology]] | volume = 23 | issue = 2 | pages = 247–262 | url = http://paleobiol.geoscienceworld.org/cgi/content/abstract/23/2/247 | doi = 10.1017/S009483730001681X | bibcode = 1997Pbio...23..247B | s2cid = 140642074 }}</ref> Even today, they are important in the [[carbon cycle]] and [[microbial]] [[food web]].<ref name=King2008/> There is some evidence that choanoflagellates feast on viruses as well.<ref>{{cite news|work=The New York Times|title=Nothing Eats Viruses, Right? Meet Some Hungry Protists: New genetic evidence builds the case that single-celled marine microbes might chow down on viruses|first=Katherine J.|last=Wu |url=https://www.nytimes.com/2020/09/24/science/virus-eaters-protists.html|date=September 24, 2020|page=D3 (September 29, 2020 print ed.)|access-date=December 1, 2020}}</ref> |
|||
===Relationship of choanoflagellates to metazoans=== |
|||
[[Félix Dujardin|Dujardin]], a French biologist interested in protozoan evolution, recorded the morphological similarities of choanoflagellates and sponge choanocytes and proposed the possibility of a close relationship as early as 1841.<ref name=Leadbeater2001/> Over the past decade, this hypothesized relationship between choanoflagellates and animals has been upheld by independent analyses of multiple unlinked sequences: 18S rDNA, nuclear protein-coding genes, and mitochondrial genomes (Steenkamp, et al., 2006; Burger, et al., 2003;<ref name=Mendoza2002/> Wainright, et al., 1993). Importantly, comparisons of mitochondrial genome sequences from a choanoflagellate and three sponges confirm the placement of choanoflagellates as an outgroup to [[Metazoa]] and negate the possibility that choanoflagellates evolved from metazoans (Lavrov, et al., 2005). Finally, recent studies of genes expressed in choanoflagellates have revealed that choanoflagellates synthesize homologues of metazoan cell signaling and adhesion genes.<ref name=King2001>{{cite journal |
|||
| author = [[Nicole King|King, N.]] |
|||
|author2=Carroll, S.B. |authorlink2=Sean B. Carroll |
|||
| date = 18 December 2001 |
|||
| title = A receptor tyrosine kinase from choanoflagellates: Molecular insights into early animal evolution |
|||
| journal = [[Proceedings of the National Academy of Sciences|PNAS]] |
|||
| pmid = 11752452 |
|||
| volume =98| issue = 26 |
|||
| pmc = 64978 |
|||
| pages = 15032–15037 |
|||
| doi = 10.1073/pnas.261477698 |
|||
}}</ref> (King, 2003) Genome sequencing shows that, among living organisms, the choanoflagellates are most closely related to animals.<ref name=King2008/> |
|||
Because choanoflagellates and metazoans are closely related, comparisons between the two groups promise to provide insights into the biology of their [[last common ancestor]] and the earliest events in [[metazoan]] evolution. The [[choanocyte]]s (also known as "collared cells") of [[sea sponge|sponges]] (considered among the most basal metazoa) have the same basic structure as choanoflagellates. Collared cells are found in other [[animal]] groups, such as [[Nemertea|ribbon worms]],<ref>{{cite journal |
|||
| last1 = Cantell |
|||
| first1 = Carl-Erik |
|||
| last2 = Franzén |
|||
| first2 = Åke |
|||
| last3 = Sensenbaugh |
|||
| first3 = Terry |
|||
| title = Ultrastructure of multiciliated collar cells in the pilidium larva of ''Lineus bilineatus'' (Nemertini) |
|||
| journal = [[Zoomorphology]] |
|||
| volume = 101 |
|||
| issue = 1 |
|||
| year = 1982 |
|||
| doi = 10.1007/BF00312027 |
|||
| pages = 1–15 |
|||
}}</ref> suggesting this was the [[morphology (biology)|morphology]] of their [[last common ancestor]]. The [[last common ancestor]] of [[animal]]s and choanoflagellates was unicellular, perhaps forming simple colonies; in contrast, the [[last common ancestor]] of all [[eumetazoan|eumetazoan animals]] was a multicellular organism, with differentiated tissues, a definite "body plan", and embryonic development (including gastrulation).<ref name=King2008/> The timing of the splitting of these lineages is difficult to constrain, but was probably in the late Precambrian, >{{ma|600}}.<ref name=King2008/> |
|||
== |
==Life cycle== |
||
[[File:Choanoflagellate and human spermatozoon.jpg|thumb|upright=1.9| The [[calcium homeostasis]] of a modern [[sperm cell]] (B) looks very similar to that of an ancient choanoflagellate (A). [[Farnesol]] is very ancient in evolution, and its use goes back at least as far as the choanoflagellates which preceded the animals.<ref>{{Cite journal|doi = 10.3389/fnins.2019.00141|doi-access = free|title = Mode of Action of Farnesol, the "Noble Unknown" in Particular in Ca2+ Homeostasis, and its Juvenile Hormone-Esters in Evolutionary Retrospect|year = 2019|last1 = De Loof|first1 = Arnold|last2 = Schoofs|first2 = Liliane|journal = Frontiers in Neuroscience|volume = 13|page = 141|pmid = 30858798|pmc = 6397838}}</ref>{{Clarify|reason=What does farnesol have to do with choanoflagellates?|date=April 2024}}]] |
|||
A number of [[species]], such as those in the [[genus]] ''[[Proterospongia]]'', form simple [[Colony (biology)|colonies]],<ref name=King2008/> [[planktonic]] clumps that resemble a miniature cluster of grapes in which each cell in the colony is flagellated or clusters of cells on a single stalk.<ref name=Leadbeater2000(1)/><ref name=Carr2008>{{cite journal |
|||
| author = Carr, M. |
|||
Choanoflagellates grow vegetatively, with multiple species undergoing longitudinal fission;<ref name="Karpov1998" /> however, the reproductive life cycle of choanoflagellates remains to be elucidated. A paper released in August 2017 showed that environmental changes, including the presence of certain bacteria, trigger the swarming and subsequent sexual reproduction of choanoflagellates.<ref name="sciencedaily.com"/> The [[ploidy]] level is unknown;<ref>Claus Nielsen. Animal Evolution: Interrelationships of the Living Phyla. 3rd ed. Claus Nielsen. Oxford, UK: Oxford University Press, 2012, p. 14.</ref> however, the discovery of both retrotransposons and key genes involved in meiosis<ref name="Carr2010">{{cite journal | vauthors = Carr M, Leadbeater BS, Baldauf SL | title = Conserved meiotic genes point to sex in the choanoflagellates | journal = The Journal of Eukaryotic Microbiology | volume = 57 | issue = 1 | pages = 56–62 | year = 2002 | pmid = 20015185 | doi = 10.1111/j.1550-7408.2009.00450.x | s2cid = 205759832 }}</ref> previously suggested that they used [[sexual reproduction]] as part of their life cycle. Some choanoflagellates can undergo encystment, which involves the retraction of the flagellum and collar and encasement in an electron dense fibrillar wall. On transfer to fresh media, excystment occurs; though it remains to be directly observed.<ref name="Leadbeater2000(2)">{{cite journal | vauthors = Leadbeater BS, Karpov SA | title = Cyst formation in a freshwater strain of the choanoflagellate Desmarella moniliformis Kent | journal = The Journal of Eukaryotic Microbiology | volume = 47 | issue = 5 | pages = 433–9 | date = September–October 2000 | pmid = 11001139 | doi = 10.1111/j.1550-7408.2000.tb00071.x | s2cid = 23357186 }}</ref> |
|||
| date = 28 October 2008 |
|||
| title = Molecular phylogeny of choanoflagellates, the sister group to Metazoa |
|||
Evidence for sexual reproduction has been reported in the choanoflagellate species ''[[Salpingoeca rosetta]]''.<ref name="pmid28867285">{{cite journal | vauthors = Woznica A, Gerdt JP, Hulett RE, Clardy J, King N | title = Mating in the Closest Living Relatives of Animals Is Induced by a Bacterial Chondroitinase | journal = Cell | volume = 170 | issue = 6 | pages = 1175–1183.e11 | date = September 2017 | pmid = 28867285 | pmc = 5599222 | doi = 10.1016/j.cell.2017.08.005 }}</ref><ref name="pmid24139741">{{cite journal | vauthors = Levin TC, King N | title = Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta | journal = Current Biology | volume = 23 | issue = 21 | pages = 2176–80 | date = November 2013 | pmid = 24139741 | pmc = 3909816 | doi = 10.1016/j.cub.2013.08.061 | bibcode = 2013CBio...23.2176L }}</ref> Evidence has also been reported for the presence of conserved [[meiosis|meiotic genes]] in the choanoflagellates ''Monosiga brevicollis'' and ''Monosiga ovata''.<ref name="pmid20015185"/> |
|||
| journal = [[Proceedings of the National Academy of Sciences|PNAS]] |
|||
| volume = 105 |
|||
{{clear}} |
|||
| pmid = 18922774 |
|||
| issue = 43 |
|||
| pmc = 2575473 |
|||
| pages = 16641–16646 |
|||
| doi = 10.1073/pnas.0801667105 |
|||
| last2 = Leadbeater |
|||
| first2 = B. S. C. |
|||
| last3 = Hassan |
|||
| first3 = R. |
|||
| last4 = Nelson |
|||
| first4 = M. |
|||
| last5 = Baldauf |
|||
| first5 = S. L. |
|||
}}</ref> |
|||
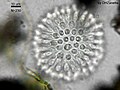
[[Image:Sphaeroeca-colony.jpg|thumb|400px|rifht|''Sphaeroeca'', a colony of choanoflagellates (approx. 230 individuals), in [[light microscopy]].]] |
|||
==Silicon biomineralization== |
==Silicon biomineralization== |
||
The Acanthoecid choanoflagellates produce an extracellular basket structure known as a lorica. The lorica is composed of individual costal strips, made of a silica-protein biocomposite. Each costal strip is formed within the choanoflagellate cell and is then secreted to the cell surface. In nudiform choanoflagellates, lorica assembly takes place using a number of tentacles once sufficient costal strips have been produced to comprise a full lorica. In tectiform choanoflagellates, costal strips are accumulated in a set arrangement below the collar. During cell division, the new cell takes these costal strips as part of [[cytokinesis]] and assembles its own lorica using only these previously produced strips.<ref name="Leadbeater et al. 2009">{{cite journal| |
The Acanthoecid choanoflagellates produce an extracellular basket structure known as a lorica. The lorica is composed of individual costal strips, made of a silica-protein biocomposite. Each costal strip is formed within the choanoflagellate cell and is then secreted to the cell surface. In nudiform choanoflagellates, lorica assembly takes place using a number of tentacles once sufficient costal strips have been produced to comprise a full lorica. In tectiform choanoflagellates, costal strips are accumulated in a set arrangement below the collar. During cell division, the new cell takes these costal strips as part of [[cytokinesis]] and assembles its own lorica using only these previously produced strips.<ref name="Leadbeater et al. 2009">{{cite journal | vauthors = Leadbeater BS, Yu Q, Kent J, Stekel DJ | title = Three-dimensional images of choanoflagellate loricae | journal = Proceedings. Biological Sciences | volume = 276 | issue = 1654 | pages = 3–11 | date = January 2009 | pmid = 18755674 | pmc = 2581655 | doi = 10.1098/rspb.2008.0844 }}</ref> |
||
Choanoflagellate biosilicification requires the concentration of [[silicic acid]] within the cell. This is carried out by |
Choanoflagellate biosilicification requires the concentration of [[silicic acid]] within the cell. This is carried out by [[silicon transporter]] (SiT) proteins. Analysis of choanoflagellate SiTs shows that they are similar to the SiT-type silicon transporters of [[diatoms]] and other silica-forming [[stramenopiles]]. The SiT gene family shows little or no homology to any other genes, even to genes in non-siliceous choanoflagellates or stramenopiles. This suggests that the SiT gene family evolved via a lateral gene transfer event between Acanthoecids and Stramenopiles. This is a remarkable case of [[horizontal gene transfer]] between two distantly related eukaryotic groups, and has provided clues to the biochemistry and silicon-protein interactions of the unique SiT gene family.<ref name=Marron2013>{{cite journal | vauthors = Marron AO, Alston MJ, Heavens D, Akam M, Caccamo M, Holland PW, Walker G | title = A family of diatom-like silicon transporters in the siliceous loricate choanoflagellates | journal = Proceedings. Biological Sciences | volume = 280 | issue = 1756 | pages = 20122543 | date = April 2013 | pmid = 23407828 | pmc = 3574361 | doi = 10.1098/rspb.2012.2543 | author6-link = Peter Holland (zoologist) }}</ref> |
||
== |
==Classification== |
||
{{More citations needed section|date=February 2021}} |
|||
There are over 125 extant species of choanoflagellates<ref name=King2008/> distributed globally in [[marine (ocean)|marine]], [[brackish]] and [[freshwater]] environments from the Arctic to the tropics, occupying both [[pelagic]] and [[benthic]] zones. Although most sampling of choanoflagellates has occurred between 0 m and 25 m, they have been recovered from as deep as 300 m in open water<ref name=Thomsen1982>{{cite book |
|||
| author = Thomsen, H. |
|||
| year = 1982 |
|||
| title = Planktonic choanoflagellates from Disko Bugt, West Greenland, with a survey of the marine nanoplankton of the area |
|||
| series= Meddelelser om Gronland, Bioscience |
|||
| volume = 8 |
|||
| pages = 3–63 |
|||
| isbn = 978-87-635-1149-0 |
|||
}}</ref> and 100 m under Antarctic ice sheets.<ref name=Buck1988>{{cite journal |
|||
| author = Buck, K. |
|||
|author2=Garrison, D |
|||
| year = 1988 |
|||
| title = Distribution and abundance of choanoflagellates (Acanthoecidae) across the ice-edge zone in the Weddell Sea, Antarctica |
|||
| journal = [[Marine Biology (journal)|Mar. Biol.]] |
|||
| volume = 98 |
|||
| pages = 263–269 |
|||
| doi = 10.1007/BF00391204 |
|||
| url = http://www.springerlink.com/index/N141343025V6736T.pdf |
|||
| issue = 2 |
|||
}}</ref> Many species are hypothesized to be [[cosmopolitan (biology)|cosmopolitan]] on a global scale [e.g., ''[[Diaphanoeca grandis]]'' has been reported from [[North America]], [[Europe]] and [[Australia]] (OBIS)], while other species are reported to have restricted regional distributions.<ref name=Thomsen1991>{{cite journal |
|||
| author = Thomsen, H. |author2=Buck, K. |author3=Chavez, F. |
|||
| year = 1991 |
|||
| title = Choanoflagellates of the central California waters: Taxonomy, morphology and species assemblages |
|||
| journal = [[Ophelia (journal)|Ophelia]] |
|||
| volume = 33 |
|||
| pages = 131–164 |
|||
| doi =10.1080/00785326.1991.10429736 |
|||
| issue = 2 |
|||
}}</ref> Co-distributed choanoflagellate species can occupy quite different microenvironments, but in general, the factors that influence the distribution and dispersion of choanoflagellates remain to be elucidated. |
|||
===Relationship to metazoans=== |
|||
The choanoflagellates feed on [[bacteria]] and link otherwise inaccessible forms of [[carbon]] to organisms higher in the [[trophic level|trophic]] chain.<ref name=Butterfield1997>{{cite journal |
|||
[[Félix Dujardin|Dujardin]], a French biologist interested in protozoan evolution, recorded the morphological similarities of choanoflagellates and sponge [[choanocytes]] and proposed the possibility of a close relationship as early as 1841.<ref name=Leadbeater2001/> Over the past decade, this hypothesized relationship between choanoflagellates and animals has been upheld by independent analyses of multiple unlinked sequences: 18S rDNA, nuclear protein-coding genes, and mitochondrial genomes (Steenkamp, et al., 2006; Burger, et al., 2003;<ref name=Mendoza2002/> Wainright, et al., 1993). Importantly, comparisons of mitochondrial genome sequences from a choanoflagellate and three sponges confirm the placement of choanoflagellates as an outgroup to [[Metazoa]] and negate the possibility that choanoflagellates evolved from metazoans (Lavrov, et al., 2005). Finally, a 2001 study of genes expressed in choanoflagellates have revealed that choanoflagellates synthesize homologues of metazoan cell signaling and adhesion genes.<ref name=King2001>{{cite journal | vauthors = King N, Carroll SB | title = A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution | journal = Proceedings of the National Academy of Sciences of the United States of America | volume = 98 | issue = 26 | pages = 15032–7 | date = December 2001 | pmid = 11752452 | pmc = 64978 | doi = 10.1073/pnas.261477698 | author-link = Nicole King | author-link2 = Sean B. Carroll | bibcode = 2001PNAS...9815032K | doi-access = free }}</ref> Genome sequencing shows that, among living organisms, the choanoflagellates are most closely related to animals.<ref name=King2008/> |
|||
| author = Butterfield, N.J. |
|||
Because choanoflagellates and metazoans are closely related, comparisons between the two groups promise to provide insights into the biology of their last common ancestor and the earliest events in [[metazoan]] evolution. The [[choanocyte]]s (also known as "collared cells") of [[sea sponge|sponges]] (considered among the most basal metazoa) have the same basic structure as choanoflagellates. Collared cells are found in other animal groups, such as [[Nemertea|ribbon worms]],<ref>{{cite journal | last1 = Cantell | first1 = Carl-Erik | last2 = Franzén | first2 = Åke | last3 = Sensenbaugh | first3 = Terry | name-list-style = vanc | title = Ultrastructure of multiciliated collar cells in the pilidium larva of ''Lineus bilineatus'' (Nemertini) | journal = [[Zoomorphology]] | volume = 101 | issue = 1 | date = October 1982 | doi = 10.1007/BF00312027 | pages = 1–15| s2cid = 42242685 }}</ref> suggesting this was the [[morphology (biology)|morphology]] of their last common ancestor. The last common ancestor of animals and choanoflagellates was unicellular, perhaps forming simple colonies; in contrast, the last common ancestor of all [[eumetazoan|eumetazoan animals]] was a multicellular organism, with differentiated tissues, a definite "body plan", and embryonic development (including gastrulation).<ref name=King2008/> The timing of the splitting of these lineages is difficult to constrain, but was probably in the late Precambrian, >{{ma|600}}.<ref name=King2008/> |
|||
| date = 1 April 1997 |
|||
| title = Plankton ecology and the Proterozoic-Phanerozoic transition |
|||
| journal = [[Paleobiology (journal)|Paleobiology]] |
|||
| volume = 23 |
|||
| issue = 2 |
|||
| pages = 247–262 |
|||
| url = http://paleobiol.geoscienceworld.org/cgi/content/abstract/23/2/247 |
|||
}}</ref> Even today they are important in the [[carbon cycle]] and [[microbial]] [[food web]].<ref name=King2008/> |
|||
External relationships of Choanoflagellatea.<ref>{{Cite journal|last1=Torruella|first1=Guifré|last2=Mendoza|first2=Alex de|last3=Grau-Bové|first3=Xavier|last4=Antó|first4=Meritxell|last5=Chaplin|first5=Mark A.|last6=Campo|first6=Javier del|last7=Eme|first7=Laura|last8=Pérez-Cordón|first8=Gregorio|last9=Whipps|first9=Christopher M.|title=Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi|journal=Current Biology|volume=25|issue=18|pages=2404–2410|doi=10.1016/j.cub.2015.07.053|pmid=26365255|year=2015|doi-access=free|bibcode=2015CBio...25.2404T }}</ref> |
|||
==Genomes and transcriptomes== |
|||
{{Clade |
|||
|style=font-size:80%; line-height:80% |
|||
|label1=[[Opisthokonta]] |
|||
|1={{clade |
|||
|label1=[[Holomycota]] |
|||
|1={{clade |
|||
|1=[[Cristidiscoidea]] |
|||
|2=[[Fungi]] |
|||
}} |
|||
|label2='''[[Holozoa]]''' |
|||
|2={{clade |
|||
|1=[[Ichthyosporea]] |
|||
|2={{clade |
|||
|1=[[Corallochytrea]] |
|||
|label2=[[Filozoa]] |
|||
|2={{clade |
|||
|1=[[Filasterea]] |
|||
|label2=[[Choanozoa]] |
|||
|2={{clade |
|||
|1=[[Animal]]ia |
|||
|2='''Choanoflagellatea''' |
|||
}} |
|||
}} |
|||
}} |
|||
}} |
|||
}} |
|||
}} |
|||
===Phylogenetic relationships=== |
|||
Two choanoflagellate species have had their genomes fully sequenced, with another two species having had [[transcriptome]] data published. |
|||
The choanoflagellates were included in [[Chrysophyceae]] until Hibberd, 1975.<ref>Reviers, B. de. (2006). ''[https://books.google.com/books?id=6fw4AgAAQBAJ&pg=PA156 Biologia e Filogenia das Algas]''. Editora Artmed, Porto Alegre, p. 156.</ref> Recent molecular [[phylogenetic]] reconstruction of the internal relationships of choanoflagellates allows the polarization of character evolution within the clade. Large fragments of the nuclear [[18S ribosomal RNA|SSU]] and [[28S ribosomal RNA|LSU]] [[ribosomal RNA]], [[tubulin|alpha tubulin]], and [[heat-shock protein]] 90 coding genes were used to resolve the internal relationships and character polarity within choanoflagellates.<ref name=Carr2008/> Each of the four genes showed similar results independently and analysis of the combined data set ([[concatenated]]) along with sequences from other closely related species ([[animal]]s and [[fungi]]) demonstrate that choanoflagellates are strongly supported as [[monophyletic]] and confirm their position as the closest known unicellular living relative of animals. |
|||
Previously, Choanoflagellida was divided into these three families based on the composition and structure of their periplast: Codonosigidae, Salpingoecidae and Acanthoecidae. Members of the family Codonosigidae appear to lack a periplast when examined by light microscopy, but may have a fine outer coat visible only by [[electron microscopy]]. The family Salpingoecidae consists of species whose cells are encased in a firm theca that is visible by both light and electron microscopy. The [[theca]] is a secreted covering predominately composed of [[cellulose]] or other [[polysaccharides]].<ref>(Adl, et al., 2005)</ref> These divisions are now known to be [[paraphyletic]], with convergent evolution of these forms widespread. The third family of choanoflagellates, the Acanthoecidae, has been supported as a monophyletic group. This clade possess a [[synapomorphy]] of the cells being found within a basket-like lorica, providing the alternative name of "Loricate Choanoflagellates". The Acanthoecid lorica is composed of a series of [[siliceous]] costal strips arranged into a species-specific lorica pattern."<ref name=Leadbeater2000(1)/><ref name=Leadbeater2001/> |
|||
===''Monosiga brevicollis'' genome=== |
|||
The choanoflagellate tree based on molecular phylogenetics divides into three well supported [[clades]].<ref name=Carr2008/> Clade 1 and Clade 2 each consist of a combination of species traditionally attributed to the Codonosigidae and Salpingoecidae, while Clade 3 comprises species from the group taxonomically classified as Acanthoecidae.<ref name=Carr2008/> The mapping of character traits on to this phylogeny indicates that the [[last common ancestor]] of choanoflagellates was a marine organism with a differentiated [[Biological life cycle|life cycle]] with [[sessility (zoology)|sedentary]] and [[Motility|motile]] stages.<ref name=Carr2008/> |
|||
The genome of ''[[Monosiga brevicollis]]'', with 41.6 million base pairs,<ref name=King2008/> is similar in size to filamentous fungi and other free-living unicellular eukaryotes, but far smaller than that of typical animals.<ref name=King2008/> In 2010, a phylogenomic study revealed that several algal genes are present in the genome of ''Monosiga brevicollis''. This could be due to the fact that, in early evolutionary history, choanoflagellates consumed algae as food through [[phagocytosis]].<ref>{{cite journal|last=Sun|first=Guiling|author2=Arjun Ishwar |author3=Zefeng Yang |author4=Jinling Huang |title=Algal genes in the closest relatives of animals|journal=Molecular Biology and Evolution|date=July 13, 2010|volume=27|issue=12|doi=10.1093/molbev/msq175|pmid=20627874|url=http://mbe.oxfordjournals.org/content/27/12/2879.abstract|pages=2879–89}}</ref> |
|||
===Taxonomy=== |
|||
Choanoflagellates;<ref name="Leadbeater" /> |
|||
* '''Order [[Craspedida]]''' <small>Cavalier-Smith 1997 em. Nitsche et al. 2011</small> |
|||
** '''Family [[Salpingoecidae]]''' <small>Kent 1880-1882</small> |
|||
*** ?''[[Dicraspedella]]'' <small>Ellis 1930</small> |
|||
*** ?''[[Diploeca]]'' <small>Ellis 1930</small> |
|||
*** ?''[[Diplosigopsis]]'' <small>Francé 1897</small> |
|||
*** ?''[[Pachysoeca]]'' <small>Ellis 1930</small> |
|||
*** ?''[[Piropsis]]'' <small>Meunier 1910</small> |
|||
*** ?''[[Salpingorhiza]]'' <small>Klug 1936</small> |
|||
*** ?''[[Sphaerodendron]]'' <small>Zhukov, Mylnikov & Moiseev 1976 non Seemann 1865</small> |
|||
*** ?''[[Stelexomonas]]'' <small>Lackey 1942</small> |
|||
*** ''[[Astrosiga]]'' <small>Kent 1880-1882</small> |
|||
*** ''[[Aulomonas]]'' <small>Lackey 1942</small> |
|||
*** ''[[Choanoeca]]'' <small>Ellis 1930</small> |
|||
*** ''[[Cladospongia (Chaonozoa)|Cladospongia]]'' <small>Iyengar & Ramathan 1940</small> |
|||
*** ''[[Codonosigopsis]]'' <small>Senn 1900</small> |
|||
*** ''[[Diplosiga]]'' <small>Frenzel 1891</small> |
|||
*** ''[[Hartaetosiga]]'' <small>Carr, Richter & Nitsche 2017</small> |
|||
*** ''[[Mylnosiga]]'' <small>Carr, Richter & Nitsche 2017</small> |
|||
*** ''[[Lagenoeca]]'' <small>Kent 1881</small> |
|||
*** ''[[Microstomoeca]]'' <small>Carr, Richter & Nitsche 2017</small> |
|||
*** ''[[Paramonosiga]]'' <small>Jeuck, Arndt & Nitsche 2014</small> |
|||
*** ''[[Salpingoeca]]'' <small>James-Clark 1868 non Ellis 1933</small> |
|||
*** ''[[Stagondoeca]]'' <small>Carr, Richter & Nitsche 2017</small> |
|||
** '''Family [[Codonosigaceae]]''' <small>Kent 1880-1882</small> |
|||
*** ''[[Codosiga]]'' <small>James-Clark 1866</small> |
|||
*** ''[[Desmarella]]'' <small>Kent 1880-1882</small> |
|||
*** ''[[Kentrosiga]]'' <small>Schiller 1953</small> |
|||
*** ''[[Monosiga]]'' <small>Kent 1880-1882</small> |
|||
*** ''[[Proterospongia]]'' <small>Kent 1882</small> |
|||
*** ''[[Sphaeroeca]]'' <small>Lauterborn 1894 non Meyrick 1895</small> |
|||
*** ''[[Stylochromonas]]'' <small>Lackey 1940</small> |
|||
* '''Order [[Acanthoecida]]''' <small>Norris 1965 em. Nitsche et al. 2011</small> (Loricate choanoflagellates) |
|||
** ''[[Conioeca]]'' <small>Thomsen & Ostergaard 2019</small> |
|||
** '''Family [[Acanthoecidae]]''' <small>Norris 1965 em. Nitsche et al. 2011</small> (Nudiform choanoflagellates) |
|||
*** ''[[Acanthoeca]]'' <small>Ellis 1930</small> |
|||
*** ''[[Enibas]]'' <small>Schiwitza, Arndt & Nitsche 2019</small> |
|||
*** ''[[Helgoeca]]'' <small>Leadbeater 2008</small> |
|||
*** ''[[Polyoeca]]'' <small>Kent 1880</small> |
|||
*** ''[[Savillea]]'' <small>Loeblich III 1967</small> |
|||
** '''Family [[Stephanoecidae]]''' <small>Leadbeater 2011</small> (Tectiform choanoflagellates) |
|||
*** ?''[[Conion]]'' <small>Thomsen 1982</small> |
|||
*** ?''[[Spiraloecion]]'' <small>Marchant & Perrin 1986</small> |
|||
*** ''[[Acanthocorbis]]'' <small>Hara & Takahashi 1984</small> |
|||
*** ''[[Amoenoscopa]]'' <small>Hara & Takahashi 1987</small> |
|||
*** ''[[Apheloecion]]'' <small>Thomsen 1983</small> |
|||
*** ''[[Bicosta]]'' <small>Leadbeater 1978</small> |
|||
*** ''[[Calliacantha]]'' <small>Leadbeater 1978</small> |
|||
*** ''[[Calotheca]]'' <small>Thomsen & Moestrup 1983 non Desv. 1810 non Spreng. 1817 non Heyden 1887</small> |
|||
*** ''[[Cosmoeca]]'' <small>Thomsen 1984</small> |
|||
*** ''[[Crinolina]]'' <small>Thomsen 1976 non Smetana 1982</small> |
|||
*** ''[[Crucispina]]'' <small>Espeland & Throndsen 1986</small> |
|||
*** ''[[Diaphanoeca]]'' <small>Ellis 1930</small> |
|||
*** ''[[Didymoeca]]'' <small>Doweld 2003</small> |
|||
*** ''[[Kakoeca]]'' <small>Buck & Marchant 1991</small> |
|||
*** ''[[Monocosta]]'' <small>Thomsen 1979 non Monocostus Schumann 1904</small> |
|||
*** ''[[Nannoeca]]'' <small>Thomsen 1988</small> |
|||
*** ''[[Parvicorbicula]]'' <small>Deflandre 1960</small> |
|||
*** ''[[Pleurasiga]]'' <small>Schiller 1925</small> |
|||
*** ''[[Polyfibula]]'' <small>Manton 1981</small> |
|||
*** ''[[Saepicula]]'' <small>Leadbeater 1980</small> |
|||
*** ''[[Saroeca]]'' <small>Thomsen 1979</small> |
|||
*** ''[[Spinoeca]]'' <small>Thomsen, Ostergaard & Hansen 1995 non Poulsen 1973</small> |
|||
*** ''[[Stephanacantha]]'' <small>Thomsen 1983</small> |
|||
*** ''[[Stephanoeca]]'' <small>Ellis 1930</small> |
|||
*** ''[[Syndetophyllum]]'' <small>Thomsen & Moestrup 1983</small> |
|||
*** ''[[Thomsenella]]'' <small>Özdikmen 2009</small> |
|||
==Genomes and transcriptomes== |
|||
===''Monosiga brevicollis'' genome=== |
|||
The genome of ''[[Monosiga brevicollis]]'', with 41.6 million base pairs,<ref name=King2008/> is similar in size to filamentous fungi and other free-living unicellular eukaryotes, but far smaller than that of typical animals.<ref name=King2008/> In 2010, a phylogenomic study revealed that several algal genes are present in the genome of ''Monosiga brevicollis''. This could be due to the fact that, in early evolutionary history, choanoflagellates consumed algae as food through [[phagocytosis]].<ref>{{cite journal | vauthors = Sun G, Yang Z, Ishwar A, Huang J | title = Algal genes in the closest relatives of animals | journal = Molecular Biology and Evolution | volume = 27 | issue = 12 | pages = 2879–89 | date = December 2010 | pmid = 20627874 | doi = 10.1093/molbev/msq175 | doi-access = free }}</ref> Carr et al. (2010)<ref name="pmid20015185">{{cite journal | vauthors = Carr M, Leadbeater BS, Baldauf SL | title = Conserved meiotic genes point to sex in the choanoflagellates | journal = The Journal of Eukaryotic Microbiology | volume = 57 | issue = 1 | pages = 56–62 | year = 2010 | pmid = 20015185 | doi = 10.1111/j.1550-7408.2009.00450.x | s2cid = 205759832 }}</ref> screened the ''M. brevicollis'' genome for known [[eukaryote|eukaryotic]] meiosis genes. Of 19 known eukaryotic meiotic genes tested (including 8 that function in no other process than meiosis), 18 were identified in ''M. brevicollis''. The presence of meiotic genes, including meiosis specific genes, indicates that meiosis, and by implication, [[sexual reproduction|sex]] is present within the choanoflagellates. |
|||
===''Salpingoeca rosetta'' genome=== |
===''Salpingoeca rosetta'' genome=== |
||
The genome of ''[[Salpingoeca rosetta]]'' is 55 megabases in size.<ref name=Fairclough2013>{{cite journal | vauthors = Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, Manning G, Lang BF, Haas B, Nusbaum C, King N | title = Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta | journal = Genome Biology | volume = 14 | issue = 2 | pages = R15 | date = February 2013 | pmid = 23419129 | pmc = 4054682 | doi = 10.1186/gb-2013-14-2-r15 | doi-access = free }}</ref> Homologs of cell adhesion, neuropeptide and glycosphingolipid metabolism genes are present in the genome. |
|||
''S. rosetta'' has a sexual life cycle and transitions between [[ploidy|haploid and diploid stages]].<ref name="pmid24139741" /> In response to nutrient limitation, haploid cultures of ''S. rosetta'' become diploid. This ploidy shift coincides with mating during which small, flagellated cells fuse with larger flagellated cells. There is also evidence of historical mating and [[genetic recombination|recombination]] in ''S. rosetta''. |
|||
''S. rosetta'' is induced to undergo sexual reproduction by the marine bacterium ''[[Aliivibrio fischeri|Vibrio fischeri]]''.<ref name="pmid28867285" /> A single ''V. fischeri'' protein, EroS fully recapitulates the aphrodisiac-like activity of live ''V. fisheri''. |
|||
The genome is 55 megabases in size.<ref name=Fairclough2013>{{cite journal |last=Fairclough |first=S. R. |last2=et al. |year=2013 |title=Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate ''Salpingoeca rosetta'' |journal=[[Genome Biology|Genome Biol]] |volume=14 |issue=2 |pages=R15 |doi=10.1186/gb-2013-14-2-r15 |first2=Zehua |last3=Kramer |first3=Eric |last4=Zeng |first4=Qiandong |last5=Young |first5=Sarah |last6=Robertson |first6=Hugh M |last7=Begovic |first7=Emina |last8=Richter |first8=Daniel J |last9=Russ |first9=Carsten |last10=Westbrook |first10=M Jody |last11=Manning |first11=Gerard |last12=Lang |first12=B Franz |last13=Haas |first13=Brian |last14=Nusbaum |first14=Chad |last15=King |first15=Nicole |display-authors=8 }}</ref> Homologs of cell adhesion, neuropeptide and glycosphingolipid metabolism genes are present in the genome. |
|||
=== Other genomes === |
|||
===''Monosiga ovata'' transcriptome=== |
|||
The single-cell amplified genomes of four uncultured marine choanoflagellates, tentatively called UC1–UC4, were sequenced in 2019. The genomes of UC1 and UC4 are relatively complete.<ref>{{cite journal |last1=López-Escardó |first1=D |last2=Grau-Bové |first2=X |last3=Guillaumet-Adkins |first3=A |last4=Gut |first4=M |last5=Sieracki |first5=ME |last6=Ruiz-Trillo |first6=I |title=Reconstruction of protein domain evolution using single-cell amplified genomes of uncultured choanoflagellates sheds light on the origin of animals. |journal=Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences |date=25 November 2019 |volume=374 |issue=1786 |pages=20190088 |doi=10.1098/rstb.2019.0088 |pmid=31587642 |pmc=6792448 |doi-access=free}}</ref> |
|||
An EST dataset from ''Monosiga ovata'' was published in 2006.<ref name="Snell et al. 2006">{{cite journal|last=Snell|first=Elizabeth A|author2=Nina M Brooke |author3=William R Taylor |author4=Didier Casane |author5=Hervé Philippe |author6=[[Peter Holland (zoologist)|Peter W.H Holland]] |title=An unusual choanoflagellate protein released by Hedgehog autocatalytic processing|journal=Proceedings of the Royal Society B|date=February 22, 2006|volume=273|issue=1585|pages=401–407| doi=10.1098/rspb.2005.3263 |url=http://rspb.royalsocietypublishing.org/content/273/1585/401.abstract|pmid=16615205|pmc=1560198}}</ref> The major finding of this transcriptome was the choanoflagellate Hoglet domain and shed light on the role of domain shuffling in the evolution of the [[Hedgehog signaling pathway]]. |
|||
===Transcriptomes=== |
|||
===''Stephanoeca diplocostata'' transcriptome=== |
|||
{{anchor|Monosiga ovata transcriptome}} |
|||
The first transcriptome of a loricate choanoflagellate <ref name=Marron2013/> led to the discovery of choanoflagellate silicon transporters. Subsequently, similar genes were identified in a second loricate species, ''Diaphanoeca grandis''. Analysis of these genes found that the choanoflagellate SITs show homology to the SIT-type silicon transporters of [[diatoms]] and have evolved through [[horizontal gene transfer]]. |
|||
An EST dataset from ''Monosiga ovata'' was published in 2006.<ref name="Snell et al. 2006">{{cite journal | vauthors = Snell EA, Brooke NM, Taylor WR, Casane D, Philippe H, Holland PW | title = An unusual choanoflagellate protein released by Hedgehog autocatalytic processing | journal = Proceedings. Biological Sciences | volume = 273 | issue = 1585 | pages = 401–7 | date = February 2006 | pmid = 16615205 | pmc = 1560198 | doi = 10.1098/rspb.2005.3263 | author6-link = Peter Holland (zoologist) }}</ref> The major finding of this transcriptome was the choanoflagellate Hoglet domain and shed light on the role of domain shuffling in the evolution of the [[Hedgehog signaling pathway]]. ''M. ovata'' has at least four eukaryotic meiotic genes.<ref name="pmid20015185" /> |
|||
{{anchor|Stephanoeca diplocostata transcriptome}} |
|||
The transcriptiome of ''Stephanoeca diplocostata'' was published in 2013. This first transcriptome of a loricate choanoflagellate<ref name=Marron2013/> led to the discovery of choanoflagellate silicon transporters. Subsequently, similar genes were identified in a second loricate species, ''Diaphanoeca grandis''. Analysis of these genes found that the choanoflagellate SITs show homology to the SIT-type silicon transporters of [[diatoms]] and have evolved through [[horizontal gene transfer]]. |
|||
An additional 19 transcriptomes were published in 2018. A large number of [[gene families]] previously thought to be animal-only were found.<ref>{{cite journal |last1=Richter |first1=Daniel J |last2=Fozouni |first2=Parinaz |last3=Eisen |first3=Michael B |last4=King |first4=Nicole |title=Gene family innovation, conservation and loss on the animal stem lineage |journal=eLife |date=31 May 2018 |volume=7 |pages=e34226 |doi=10.7554/eLife.34226 |pmid=29848444 |pmc=6040629 |doi-access=free}}</ref> |
|||
==Gallery== |
|||
<gallery class="center"> |
|||
File:Monosiga Brevicollis Phase.jpg|''[[Monosiga|Monosiga brevicollis]]'' under [[phase-contrast microscopy|PCM]] |
|||
File:Protero-7.png|''[[Salpingoeca]]'' under PCM |
|||
Image:Salpingoeca sp..jpg|''Salpingoeca'' sp. section under [[Transmission electron microscopy|TEM]] |
|||
File:Desmarella moniliformis.jpg|''[[Desmarella|Desmarella moniliformis]]'' colony under PCM |
|||
File:0803col.jpg|''[[Codosiga]]'' colony under [[light microscopy]] |
|||
File:Sphaeroeca-colony.jpg|''[[Sphaeroeca]]'' colony (approx. 230 individuals) under light microscopy. |
|||
</gallery> |
|||
==References== |
==References== |
||
{{ |
{{Reflist|30em}} |
||
==External links== |
==External links== |
||
{{ |
{{Commons category|Choanoflagellatea}} |
||
* [http://www.choano.org ChoanoWiki] a collaborative resource maintained by the Choanoflagellate research community |
* [https://archive.today/20130414120247/http://www.choano.org/ ChoanoWiki] a collaborative resource maintained by the Choanoflagellate research community |
||
* [http://www.tolweb.org/Choanoflagellates/2375/ Tree of Life Webpage for Choanoflagellates] |
* [http://www.tolweb.org/Choanoflagellates/2375/ Tree of Life Webpage for Choanoflagellates] |
||
* [http://genomeportal.jgi-psf.org/Monbr1/Monbr1.home.html ''Monosiga brevicollis'' genome browser] |
* [https://web.archive.org/web/20140407084821/http://genomeportal.jgi-psf.org/Monbr1/Monbr1.home.html ''Monosiga brevicollis'' genome browser] |
||
* [ |
* [https://www.newscientist.com/article/mg21128283.800-your-brain-chemistry-existed-before-animals-did.html?full=true&print=true Your brain chemistry existed before animals did] |
||
* [http://mcb.berkeley.edu/labs/king/choano/ Choanobase], the Choanoflagellate genetic library, developed and maintained by the [[Nicole King]] laboratory at the [[University of California, Berkeley]] |
* [https://web.archive.org/web/20040804013410/http://mcb.berkeley.edu/labs/king/choano/ Choanobase], the Choanoflagellate genetic library, developed and maintained by the [[Nicole King]] laboratory at the [[University of California, Berkeley]] |
||
* [http://www.cambridge.org/us/academic/subjects/life-sciences/ecology-and-conservation/choanoflagellates-evolution-biology-and-ecology?format=AR] A 2014 book bringing together the evolution, biology and ecology of the choanoflagellates. |
|||
{{Eukaryota classification}} |
{{Eukaryota classification}} |
||
| Line 265: | Line 227: | ||
{{plankton}} |
{{plankton}} |
||
{{Taxonbar|from1=Q129012|from2=Q21446923}} |
|||
[[Category:Protista]] |
|||
[[Category:Flagellates]] |
|||
[[Category:Ambiregnal protists]] |
|||
[[Category:Proterozoic first appearances]] |
[[Category:Proterozoic first appearances]] |
||
[[Category:Choanoflagellatea| ]] |
|||
Revision as of 04:21, 2 May 2024
| Choanoflagellates Temporal range: Only possible fossils are known from Cretaceous (Cenomanian/Turonian),[1] molecular clock evidence for origin 1050-800 Ma[2]
| |
|---|---|

| |
| Codosiga sp. | |
| Scientific classification | |
| Domain: | Eukaryota |
| Clade: | Amorphea |
| Clade: | Obazoa |
| (unranked): | Opisthokonta |
| (unranked): | Holozoa |
| (unranked): | Filozoa |
| Clade: | Choanozoa |
| Class: | Choanoflagellata Kent, 1880–1882[3][4] |
| Type species | |
| Monosiga brevicollis[5] | |
| Orders & families | |
| Synonyms | |
| |
The choanoflagellates are a group of free-living unicellular and colonial flagellate eukaryotes considered to be the closest living relatives of the animals. Choanoflagellates are collared flagellates, having a funnel shaped collar of interconnected microvilli at the base of a flagellum. Choanoflagellates are capable of both asexual and sexual reproduction.[9] They have a distinctive cell morphology characterized by an ovoid or spherical cell body 3–10 μm in diameter with a single apical flagellum surrounded by a collar of 30–40 microvilli (see figure). Movement of the flagellum creates water currents that can propel free-swimming choanoflagellates through the water column and trap bacteria and detritus against the collar of microvilli, where these foodstuffs are engulfed. This feeding provides a critical link within the global carbon cycle, linking trophic levels. In addition to their critical ecological roles, choanoflagellates are of particular interest to evolutionary biologists studying the origins of multicellularity in animals. As the closest living relatives of animals, choanoflagellates serve as a useful model for reconstructions of the last unicellular ancestor of animals.
Etymology
Choanoflagellate is a hybrid word from Greek χοάνη khoánē meaning "funnel" (due to the shape of the collar) and the Latin word flagellum (whence English flagellum).[citation needed]
Appearance

Each choanoflagellate has a single flagellum, surrounded by a ring of actin-filled protrusions called microvilli, forming a cylindrical or conical collar (choanos in Greek). Movement of the flagellum draws water through the collar, and bacteria and detritus are captured by the microvilli and ingested.[10] Water currents generated by the flagellum also push free-swimming cells along, as in animal sperm. In contrast, most other flagellates are pulled by their flagella.[citation needed]
In addition to the single apical flagellum surrounded by actin-filled microvilli that characterizes choanoflagellates, the internal organization of organelles in the cytoplasm is constant.[11] A flagellar basal body sits at the base of the apical flagellum, and a second, non-flagellar basal body rests at a right angle to the flagellar base. The nucleus occupies an apical-to-central position in the cell, and food vacuoles are positioned in the basal region of the cytoplasm.[11][12] Additionally, the cell body of many choanoflagellates is surrounded by a distinguishing extracellular matrix or periplast. These cell coverings vary greatly in structure and composition and are used by taxonomists for classification purposes. Many choanoflagellates build complex basket-shaped "houses", called lorica, from several silica strips cemented together.[11] The functional significance of the periplast is unknown, but in sessile organisms, it is thought to aid attachment to the substrate. In planktonic organisms, there is speculation that the periplast increases drag, thereby counteracting the force generated by the flagellum and increasing feeding efficiency.[13]
Choanoflagellates are either free-swimming in the water column or sessile, adhering to the substrate directly or through either the periplast or a thin pedicel.[14] Although choanoflagellates are thought to be strictly free-living and heterotrophic, a number of choanoflagellate relatives, such as members of Ichthyosporea or Mesomycetozoa, follow a parasitic or pathogenic lifestyle.[15] The life histories of choanoflagellates are poorly understood. Many species are thought to be solitary; however, coloniality seems to have arisen independently several times within the group, and colonial species still retain a solitary stage.[14]
Ecology

Over 125 extant species of choanoflagellates[10] are known, distributed globally in marine, brackish and freshwater environments from the Arctic to the tropics, occupying both pelagic and benthic zones. Although most sampling of choanoflagellates has occurred between 0 and 25 m (0 and 82 ft), they have been recovered from as deep as 300 m (980 ft) in open water[16] and 100 m (330 ft) under Antarctic ice sheets.[17] Many species are hypothesized to be cosmopolitan on a global scale [e.g., Diaphanoeca grandis has been reported from North America, Europe and Australia (OBIS)], while other species are reported to have restricted regional distributions.[18] Co-distributed choanoflagellate species can occupy quite different microenvironments, but in general, the factors that influence the distribution and dispersion of choanoflagellates remain to be elucidated.[citation needed]
A number of species, such as those in the genus Proterospongia, form simple colonies,[10] planktonic clumps that resemble a miniature cluster of grapes in which each cell in the colony is flagellated or clusters of cells on a single stalk.[11][19] In October 2019, scientists found a new band behaviour of choanoflagellates: they apparently can coordinate to respond to light.[20]
The choanoflagellates feed on bacteria and link otherwise inaccessible forms of carbon to organisms higher in the trophic chain.[21] Even today, they are important in the carbon cycle and microbial food web.[10] There is some evidence that choanoflagellates feast on viruses as well.[22]
Life cycle

Choanoflagellates grow vegetatively, with multiple species undergoing longitudinal fission;[12] however, the reproductive life cycle of choanoflagellates remains to be elucidated. A paper released in August 2017 showed that environmental changes, including the presence of certain bacteria, trigger the swarming and subsequent sexual reproduction of choanoflagellates.[9] The ploidy level is unknown;[24] however, the discovery of both retrotransposons and key genes involved in meiosis[25] previously suggested that they used sexual reproduction as part of their life cycle. Some choanoflagellates can undergo encystment, which involves the retraction of the flagellum and collar and encasement in an electron dense fibrillar wall. On transfer to fresh media, excystment occurs; though it remains to be directly observed.[26]
Evidence for sexual reproduction has been reported in the choanoflagellate species Salpingoeca rosetta.[27][28] Evidence has also been reported for the presence of conserved meiotic genes in the choanoflagellates Monosiga brevicollis and Monosiga ovata.[29]
Silicon biomineralization
The Acanthoecid choanoflagellates produce an extracellular basket structure known as a lorica. The lorica is composed of individual costal strips, made of a silica-protein biocomposite. Each costal strip is formed within the choanoflagellate cell and is then secreted to the cell surface. In nudiform choanoflagellates, lorica assembly takes place using a number of tentacles once sufficient costal strips have been produced to comprise a full lorica. In tectiform choanoflagellates, costal strips are accumulated in a set arrangement below the collar. During cell division, the new cell takes these costal strips as part of cytokinesis and assembles its own lorica using only these previously produced strips.[30]
Choanoflagellate biosilicification requires the concentration of silicic acid within the cell. This is carried out by silicon transporter (SiT) proteins. Analysis of choanoflagellate SiTs shows that they are similar to the SiT-type silicon transporters of diatoms and other silica-forming stramenopiles. The SiT gene family shows little or no homology to any other genes, even to genes in non-siliceous choanoflagellates or stramenopiles. This suggests that the SiT gene family evolved via a lateral gene transfer event between Acanthoecids and Stramenopiles. This is a remarkable case of horizontal gene transfer between two distantly related eukaryotic groups, and has provided clues to the biochemistry and silicon-protein interactions of the unique SiT gene family.[31]
Classification
This section needs additional citations for verification. (February 2021) |
Relationship to metazoans
Dujardin, a French biologist interested in protozoan evolution, recorded the morphological similarities of choanoflagellates and sponge choanocytes and proposed the possibility of a close relationship as early as 1841.[13] Over the past decade, this hypothesized relationship between choanoflagellates and animals has been upheld by independent analyses of multiple unlinked sequences: 18S rDNA, nuclear protein-coding genes, and mitochondrial genomes (Steenkamp, et al., 2006; Burger, et al., 2003;[15] Wainright, et al., 1993). Importantly, comparisons of mitochondrial genome sequences from a choanoflagellate and three sponges confirm the placement of choanoflagellates as an outgroup to Metazoa and negate the possibility that choanoflagellates evolved from metazoans (Lavrov, et al., 2005). Finally, a 2001 study of genes expressed in choanoflagellates have revealed that choanoflagellates synthesize homologues of metazoan cell signaling and adhesion genes.[32] Genome sequencing shows that, among living organisms, the choanoflagellates are most closely related to animals.[10] Because choanoflagellates and metazoans are closely related, comparisons between the two groups promise to provide insights into the biology of their last common ancestor and the earliest events in metazoan evolution. The choanocytes (also known as "collared cells") of sponges (considered among the most basal metazoa) have the same basic structure as choanoflagellates. Collared cells are found in other animal groups, such as ribbon worms,[33] suggesting this was the morphology of their last common ancestor. The last common ancestor of animals and choanoflagellates was unicellular, perhaps forming simple colonies; in contrast, the last common ancestor of all eumetazoan animals was a multicellular organism, with differentiated tissues, a definite "body plan", and embryonic development (including gastrulation).[10] The timing of the splitting of these lineages is difficult to constrain, but was probably in the late Precambrian, >600 million years ago.[10]
External relationships of Choanoflagellatea.[34]
| Opisthokonta |
| ||||||
Phylogenetic relationships
The choanoflagellates were included in Chrysophyceae until Hibberd, 1975.[35] Recent molecular phylogenetic reconstruction of the internal relationships of choanoflagellates allows the polarization of character evolution within the clade. Large fragments of the nuclear SSU and LSU ribosomal RNA, alpha tubulin, and heat-shock protein 90 coding genes were used to resolve the internal relationships and character polarity within choanoflagellates.[19] Each of the four genes showed similar results independently and analysis of the combined data set (concatenated) along with sequences from other closely related species (animals and fungi) demonstrate that choanoflagellates are strongly supported as monophyletic and confirm their position as the closest known unicellular living relative of animals.
Previously, Choanoflagellida was divided into these three families based on the composition and structure of their periplast: Codonosigidae, Salpingoecidae and Acanthoecidae. Members of the family Codonosigidae appear to lack a periplast when examined by light microscopy, but may have a fine outer coat visible only by electron microscopy. The family Salpingoecidae consists of species whose cells are encased in a firm theca that is visible by both light and electron microscopy. The theca is a secreted covering predominately composed of cellulose or other polysaccharides.[36] These divisions are now known to be paraphyletic, with convergent evolution of these forms widespread. The third family of choanoflagellates, the Acanthoecidae, has been supported as a monophyletic group. This clade possess a synapomorphy of the cells being found within a basket-like lorica, providing the alternative name of "Loricate Choanoflagellates". The Acanthoecid lorica is composed of a series of siliceous costal strips arranged into a species-specific lorica pattern."[11][13]
The choanoflagellate tree based on molecular phylogenetics divides into three well supported clades.[19] Clade 1 and Clade 2 each consist of a combination of species traditionally attributed to the Codonosigidae and Salpingoecidae, while Clade 3 comprises species from the group taxonomically classified as Acanthoecidae.[19] The mapping of character traits on to this phylogeny indicates that the last common ancestor of choanoflagellates was a marine organism with a differentiated life cycle with sedentary and motile stages.[19]
Taxonomy
Choanoflagellates;[8]
- Order Craspedida Cavalier-Smith 1997 em. Nitsche et al. 2011
- Family Salpingoecidae Kent 1880-1882
- ?Dicraspedella Ellis 1930
- ?Diploeca Ellis 1930
- ?Diplosigopsis Francé 1897
- ?Pachysoeca Ellis 1930
- ?Piropsis Meunier 1910
- ?Salpingorhiza Klug 1936
- ?Sphaerodendron Zhukov, Mylnikov & Moiseev 1976 non Seemann 1865
- ?Stelexomonas Lackey 1942
- Astrosiga Kent 1880-1882
- Aulomonas Lackey 1942
- Choanoeca Ellis 1930
- Cladospongia Iyengar & Ramathan 1940
- Codonosigopsis Senn 1900
- Diplosiga Frenzel 1891
- Hartaetosiga Carr, Richter & Nitsche 2017
- Mylnosiga Carr, Richter & Nitsche 2017
- Lagenoeca Kent 1881
- Microstomoeca Carr, Richter & Nitsche 2017
- Paramonosiga Jeuck, Arndt & Nitsche 2014
- Salpingoeca James-Clark 1868 non Ellis 1933
- Stagondoeca Carr, Richter & Nitsche 2017
- Family Codonosigaceae Kent 1880-1882
- Codosiga James-Clark 1866
- Desmarella Kent 1880-1882
- Kentrosiga Schiller 1953
- Monosiga Kent 1880-1882
- Proterospongia Kent 1882
- Sphaeroeca Lauterborn 1894 non Meyrick 1895
- Stylochromonas Lackey 1940
- Family Salpingoecidae Kent 1880-1882
- Order Acanthoecida Norris 1965 em. Nitsche et al. 2011 (Loricate choanoflagellates)
- Conioeca Thomsen & Ostergaard 2019
- Family Acanthoecidae Norris 1965 em. Nitsche et al. 2011 (Nudiform choanoflagellates)
- Acanthoeca Ellis 1930
- Enibas Schiwitza, Arndt & Nitsche 2019
- Helgoeca Leadbeater 2008
- Polyoeca Kent 1880
- Savillea Loeblich III 1967
- Family Stephanoecidae Leadbeater 2011 (Tectiform choanoflagellates)
- ?Conion Thomsen 1982
- ?Spiraloecion Marchant & Perrin 1986
- Acanthocorbis Hara & Takahashi 1984
- Amoenoscopa Hara & Takahashi 1987
- Apheloecion Thomsen 1983
- Bicosta Leadbeater 1978
- Calliacantha Leadbeater 1978
- Calotheca Thomsen & Moestrup 1983 non Desv. 1810 non Spreng. 1817 non Heyden 1887
- Cosmoeca Thomsen 1984
- Crinolina Thomsen 1976 non Smetana 1982
- Crucispina Espeland & Throndsen 1986
- Diaphanoeca Ellis 1930
- Didymoeca Doweld 2003
- Kakoeca Buck & Marchant 1991
- Monocosta Thomsen 1979 non Monocostus Schumann 1904
- Nannoeca Thomsen 1988
- Parvicorbicula Deflandre 1960
- Pleurasiga Schiller 1925
- Polyfibula Manton 1981
- Saepicula Leadbeater 1980
- Saroeca Thomsen 1979
- Spinoeca Thomsen, Ostergaard & Hansen 1995 non Poulsen 1973
- Stephanacantha Thomsen 1983
- Stephanoeca Ellis 1930
- Syndetophyllum Thomsen & Moestrup 1983
- Thomsenella Özdikmen 2009
Genomes and transcriptomes
Monosiga brevicollis genome
The genome of Monosiga brevicollis, with 41.6 million base pairs,[10] is similar in size to filamentous fungi and other free-living unicellular eukaryotes, but far smaller than that of typical animals.[10] In 2010, a phylogenomic study revealed that several algal genes are present in the genome of Monosiga brevicollis. This could be due to the fact that, in early evolutionary history, choanoflagellates consumed algae as food through phagocytosis.[37] Carr et al. (2010)[29] screened the M. brevicollis genome for known eukaryotic meiosis genes. Of 19 known eukaryotic meiotic genes tested (including 8 that function in no other process than meiosis), 18 were identified in M. brevicollis. The presence of meiotic genes, including meiosis specific genes, indicates that meiosis, and by implication, sex is present within the choanoflagellates.
Salpingoeca rosetta genome
The genome of Salpingoeca rosetta is 55 megabases in size.[38] Homologs of cell adhesion, neuropeptide and glycosphingolipid metabolism genes are present in the genome. S. rosetta has a sexual life cycle and transitions between haploid and diploid stages.[28] In response to nutrient limitation, haploid cultures of S. rosetta become diploid. This ploidy shift coincides with mating during which small, flagellated cells fuse with larger flagellated cells. There is also evidence of historical mating and recombination in S. rosetta.
S. rosetta is induced to undergo sexual reproduction by the marine bacterium Vibrio fischeri.[27] A single V. fischeri protein, EroS fully recapitulates the aphrodisiac-like activity of live V. fisheri.
Other genomes
The single-cell amplified genomes of four uncultured marine choanoflagellates, tentatively called UC1–UC4, were sequenced in 2019. The genomes of UC1 and UC4 are relatively complete.[39]
Transcriptomes
An EST dataset from Monosiga ovata was published in 2006.[40] The major finding of this transcriptome was the choanoflagellate Hoglet domain and shed light on the role of domain shuffling in the evolution of the Hedgehog signaling pathway. M. ovata has at least four eukaryotic meiotic genes.[29]
The transcriptiome of Stephanoeca diplocostata was published in 2013. This first transcriptome of a loricate choanoflagellate[31] led to the discovery of choanoflagellate silicon transporters. Subsequently, similar genes were identified in a second loricate species, Diaphanoeca grandis. Analysis of these genes found that the choanoflagellate SITs show homology to the SIT-type silicon transporters of diatoms and have evolved through horizontal gene transfer.
An additional 19 transcriptomes were published in 2018. A large number of gene families previously thought to be animal-only were found.[41]
Gallery
-
Monosiga brevicollis under PCM
-
Salpingoeca under PCM
-
Salpingoeca sp. section under TEM
-
Desmarella moniliformis colony under PCM
-
Codosiga colony under light microscopy
-
Sphaeroeca colony (approx. 230 individuals) under light microscopy.
References
- ^ Fonseca, Carolina; Mendonça Filho, João Graciano; Reolid, Matías; Duarte, Luís V.; de Oliveira, António Donizeti; Souza, Jaqueline Torres; Lézin, Carine (2023-01-23). "First putative occurrence in the fossil record of choanoflagellates, the sister group of Metazoa". Scientific Reports. 13 (1): 1242. Bibcode:2023NatSR..13.1242F. doi:10.1038/s41598-022-26972-8. ISSN 2045-2322. PMC 9870899. PMID 36690681.
- ^ Parfrey LW, Lahr DJ, Knoll AH, Katz LA (August 2011). "Estimating the timing of early eukaryotic diversification with multigene molecular clocks". Proceedings of the National Academy of Sciences of the United States of America. 108 (33): 13624–9. Bibcode:2011PNAS..10813624P. doi:10.1073/pnas.1110633108. PMC 3158185. PMID 21810989.
- ^ Saville-Kent, W. (1880). A manual of Infusoria. London, vol. 1, p. 324, [1].
- ^ Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q (2019). "Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes". Journal of Eukaryotic Microbiology. 66 (1): 4–119. doi:10.1111/jeu.12691. PMC 6492006. PMID 30257078.
- ^ King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, et al. (February 2008). "The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans". Nature. 451 (7180): 783–8. Bibcode:2008Natur.451..783K. doi:10.1038/nature06617. PMC 2562698. PMID 18273011.
- ^ Nitsche F, Carr M, Arndt H, Leadbeater BS (2011). "Higher level taxonomy and molecular phylogenetics of the Choanoflagellatea". The Journal of Eukaryotic Microbiology. 58 (5): 452–62. doi:10.1111/j.1550-7408.2011.00572.x. PMID 21895836. S2CID 2076733.
- ^ Cavalier-Smith T (1998). "Neomonada and the origin of animals and fungi.". In Coombs GH, Vickerman K, Sleigh MA, Warren A (eds.). Evolutionary relationships among protozoa. London: Kluwer. pp. 375–407.
- ^ a b Leadbeater BS (2015). The choanoflagellates: evolution, biology, and ecology. University of Birmingham. ISBN 978-0-521-88444-0.
- ^ a b "Bacterial protein acts as aphrodisiac for choanoflagellates".
- ^ a b c d e f g h i King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, et al. (February 2008). "The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans". Nature. 451 (7180): 783–8. Bibcode:2008Natur.451..783K. doi:10.1038/nature06617. PMC 2562698. PMID 18273011.
- ^ a b c d e Leadbeater BS, Thomsen H (2000). "Order Choanoflagellida". An Illustrated Guide to the Protozoa, Second Edition. Lawrence: Society of Protozoologists. 451: 14–38.
- ^ a b Karpov S, Leadbeater BS (May 1998). "Cytoskeleton structure and composition in choanoflagellates". Journal of Eukaryotic Microbiology. 45 (3): 361–367. doi:10.1111/j.1550-7408.1998.tb04550.x. S2CID 86287656.
- ^ a b c Leadbeater BS, Kelly M (2001). "Evolution of animals choanoflagellates and sponges". Water and Atmosphere Online. 9 (2): 9–11.
- ^ a b Leadbeater BS (February 1983). "Life-History and Ultrastructure of a New Marine Species of Proterospongia (Choanoflagellida)". J. Mar. Biol. Assoc. U. K. 63 (1): 135–160. Bibcode:1983JMBUK..63..135L. doi:10.1017/S0025315400049857. S2CID 84666673.
- ^ a b Mendoza L, Taylor JW, Ajello L (2002). "The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary". Annual Review of Microbiology. 56: 315–44. doi:10.1146/annurev.micro.56.012302.160950. PMID 12142489. S2CID 14764188.
- ^ Thomsen H (1982). Planktonic choanoflagellates from Disko Bugt, West Greenland, with a survey of the marine nanoplankton of the area. Meddelelser om Gronland, Bioscience. Vol. 8. pp. 3–63. ISBN 978-87-635-1149-0.
- ^ Buck KR, Garrison DL (June 1988). "Distribution and abundance of choanoflagellates (Acanthoecidae) across the ice-edge zone in the Weddell Sea, Antarctica". Mar. Biol. 98 (2): 263–269. Bibcode:1988MarBi..98..263B. doi:10.1007/BF00391204. S2CID 84931348.
- ^ Thomsen H, Buck K, Chavez F (1991). "Choanoflagellates of the central California waters: Taxonomy, morphology and species assemblages". Ophelia. 33 (2): 131–164. doi:10.1080/00785326.1991.10429736.
- ^ a b c d e Carr M, Leadbeater BS, Hassan R, Nelson M, Baldauf SL (October 2008). "Molecular phylogeny of choanoflagellates, the sister group to Metazoa". Proceedings of the National Academy of Sciences of the United States of America. 105 (43): 16641–6. Bibcode:2008PNAS..10516641C. doi:10.1073/pnas.0801667105. PMC 2575473. PMID 18922774.
- ^ "Newly Discovered Microorganisms Band Together, 'Flip Out'". HHMI.org. Retrieved 2019-10-29.
- ^ Butterfield NJ (April 1, 1997). "Plankton ecology and the Proterozoic-Phanerozoic transition". Paleobiology. 23 (2): 247–262. Bibcode:1997Pbio...23..247B. doi:10.1017/S009483730001681X. S2CID 140642074.
- ^ Wu, Katherine J. (September 24, 2020). "Nothing Eats Viruses, Right? Meet Some Hungry Protists: New genetic evidence builds the case that single-celled marine microbes might chow down on viruses". The New York Times. p. D3 (September 29, 2020 print ed.). Retrieved December 1, 2020.
- ^ De Loof, Arnold; Schoofs, Liliane (2019). "Mode of Action of Farnesol, the "Noble Unknown" in Particular in Ca2+ Homeostasis, and its Juvenile Hormone-Esters in Evolutionary Retrospect". Frontiers in Neuroscience. 13: 141. doi:10.3389/fnins.2019.00141. PMC 6397838. PMID 30858798.
- ^ Claus Nielsen. Animal Evolution: Interrelationships of the Living Phyla. 3rd ed. Claus Nielsen. Oxford, UK: Oxford University Press, 2012, p. 14.
- ^ Carr M, Leadbeater BS, Baldauf SL (2002). "Conserved meiotic genes point to sex in the choanoflagellates". The Journal of Eukaryotic Microbiology. 57 (1): 56–62. doi:10.1111/j.1550-7408.2009.00450.x. PMID 20015185. S2CID 205759832.
- ^ Leadbeater BS, Karpov SA (September–October 2000). "Cyst formation in a freshwater strain of the choanoflagellate Desmarella moniliformis Kent". The Journal of Eukaryotic Microbiology. 47 (5): 433–9. doi:10.1111/j.1550-7408.2000.tb00071.x. PMID 11001139. S2CID 23357186.
- ^ a b Woznica A, Gerdt JP, Hulett RE, Clardy J, King N (September 2017). "Mating in the Closest Living Relatives of Animals Is Induced by a Bacterial Chondroitinase". Cell. 170 (6): 1175–1183.e11. doi:10.1016/j.cell.2017.08.005. PMC 5599222. PMID 28867285.
- ^ a b Levin TC, King N (November 2013). "Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta". Current Biology. 23 (21): 2176–80. Bibcode:2013CBio...23.2176L. doi:10.1016/j.cub.2013.08.061. PMC 3909816. PMID 24139741.
- ^ a b c Carr M, Leadbeater BS, Baldauf SL (2010). "Conserved meiotic genes point to sex in the choanoflagellates". The Journal of Eukaryotic Microbiology. 57 (1): 56–62. doi:10.1111/j.1550-7408.2009.00450.x. PMID 20015185. S2CID 205759832.
- ^ Leadbeater BS, Yu Q, Kent J, Stekel DJ (January 2009). "Three-dimensional images of choanoflagellate loricae". Proceedings. Biological Sciences. 276 (1654): 3–11. doi:10.1098/rspb.2008.0844. PMC 2581655. PMID 18755674.
- ^ a b Marron AO, Alston MJ, Heavens D, Akam M, Caccamo M, Holland PW, Walker G (April 2013). "A family of diatom-like silicon transporters in the siliceous loricate choanoflagellates". Proceedings. Biological Sciences. 280 (1756): 20122543. doi:10.1098/rspb.2012.2543. PMC 3574361. PMID 23407828.
- ^ King N, Carroll SB (December 2001). "A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution". Proceedings of the National Academy of Sciences of the United States of America. 98 (26): 15032–7. Bibcode:2001PNAS...9815032K. doi:10.1073/pnas.261477698. PMC 64978. PMID 11752452.
- ^ Cantell CE, Franzén Å, Sensenbaugh T (October 1982). "Ultrastructure of multiciliated collar cells in the pilidium larva of Lineus bilineatus (Nemertini)". Zoomorphology. 101 (1): 1–15. doi:10.1007/BF00312027. S2CID 42242685.
- ^ Torruella, Guifré; Mendoza, Alex de; Grau-Bové, Xavier; Antó, Meritxell; Chaplin, Mark A.; Campo, Javier del; Eme, Laura; Pérez-Cordón, Gregorio; Whipps, Christopher M. (2015). "Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi". Current Biology. 25 (18): 2404–2410. Bibcode:2015CBio...25.2404T. doi:10.1016/j.cub.2015.07.053. PMID 26365255.
- ^ Reviers, B. de. (2006). Biologia e Filogenia das Algas. Editora Artmed, Porto Alegre, p. 156.
- ^ (Adl, et al., 2005)
- ^ Sun G, Yang Z, Ishwar A, Huang J (December 2010). "Algal genes in the closest relatives of animals". Molecular Biology and Evolution. 27 (12): 2879–89. doi:10.1093/molbev/msq175. PMID 20627874.
- ^ Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, Manning G, Lang BF, Haas B, Nusbaum C, King N (February 2013). "Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta". Genome Biology. 14 (2): R15. doi:10.1186/gb-2013-14-2-r15. PMC 4054682. PMID 23419129.
- ^ López-Escardó, D; Grau-Bové, X; Guillaumet-Adkins, A; Gut, M; Sieracki, ME; Ruiz-Trillo, I (25 November 2019). "Reconstruction of protein domain evolution using single-cell amplified genomes of uncultured choanoflagellates sheds light on the origin of animals". Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 374 (1786): 20190088. doi:10.1098/rstb.2019.0088. PMC 6792448. PMID 31587642.
- ^ Snell EA, Brooke NM, Taylor WR, Casane D, Philippe H, Holland PW (February 2006). "An unusual choanoflagellate protein released by Hedgehog autocatalytic processing". Proceedings. Biological Sciences. 273 (1585): 401–7. doi:10.1098/rspb.2005.3263. PMC 1560198. PMID 16615205.
- ^ Richter, Daniel J; Fozouni, Parinaz; Eisen, Michael B; King, Nicole (31 May 2018). "Gene family innovation, conservation and loss on the animal stem lineage". eLife. 7: e34226. doi:10.7554/eLife.34226. PMC 6040629. PMID 29848444.
External links
- ChoanoWiki a collaborative resource maintained by the Choanoflagellate research community
- Tree of Life Webpage for Choanoflagellates
- Monosiga brevicollis genome browser
- Your brain chemistry existed before animals did
- Choanobase, the Choanoflagellate genetic library, developed and maintained by the Nicole King laboratory at the University of California, Berkeley