Genetic history of Europe: Difference between revisions
Muntuwandi (talk | contribs) note |
|||
| (5 intermediate revisions by the same user not shown) | |||
| Line 103: | Line 103: | ||
* [[Haplogroup N (Y-DNA)|Haplogroup N]] in the form of its N1c1 sub-clade reaches frequencies of approximately 60% among Finns and approximately 40% among Lithuanians. This clade is found also to the East, as far as Siberia, Japan and China. |
* [[Haplogroup N (Y-DNA)|Haplogroup N]] in the form of its N1c1 sub-clade reaches frequencies of approximately 60% among Finns and approximately 40% among Lithuanians. This clade is found also to the East, as far as Siberia, Japan and China. |
||
* Haplogroup [[Haplogroup E1b1b (Y-DNA)|E1b1b1]], mainly in the form of its E1b1b1a2 (E-V13) sub-clade reaches frequencies above 40% around the area of [[Kosovo]].<ref>Peričic et al. (2005), "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations", Mol. Biol. Evol. 22 (10): 1964–75, doi:10.1093/molbev/msi185, PMID 15944443</ref> This clade is thought to have arrived in Europe from Western Asia either in the later Mesolithic,<ref>{{Citation| author=Battaglia et al. | year=2008 | title=Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe | journal=European Journal of Human Genetics | doi=10.1038/ejhg.2008.249}}</ref> or the Neolithic.<ref>{{Citation |
* Haplogroup [[Haplogroup E1b1b (Y-DNA)|E1b1b1]], mainly in the form of its E1b1b1a2 (E-V13) sub-clade reaches frequencies above 40% around the area of [[Kosovo]].<ref>Peričic et al. (2005), "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations", Mol. Biol. Evol. 22 (10): 1964–75, doi:10.1093/molbev/msi185, PMID 15944443</ref> This clade is thought to have arrived in Europe from Western Asia either in the later Mesolithic,<ref>{{Citation| author=Battaglia et al. | year=2008 | title=Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe | journal=European Journal of Human Genetics | doi=10.1038/ejhg.2008.249}}</ref> or the Neolithic.<ref name="cruciani2004">{{Citation |
||
| last = Cruciani et al. |
| last = Cruciani et al. |
||
| title = Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa |
| title = Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa |
||
| Line 110: | Line 110: | ||
| pages = 1014–1022 |
| pages = 1014–1022 |
||
| year = 2004 |
| year = 2004 |
||
| url = http://www. |
| url = http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1181964 |
||
| doi = 10.1086/386294 |
| doi = 10.1086/386294 |
||
| accessdate = 2009-07-22 |
| accessdate = 2009-07-22 |
||
| Line 170: | Line 170: | ||
[[File:HgE1b1b1a2.png|thumb|left|330px|The distribution of the V-13 sub-lineage of haplogroup [[Haplogroup E1b1b (Y-DNA)|E1b1b]] in Europe]] |
[[File:HgE1b1b1a2.png|thumb|left|330px|The distribution of the V-13 sub-lineage of haplogroup [[Haplogroup E1b1b (Y-DNA)|E1b1b]] in Europe]] |
||
Since then, a lot more information has been discovered about this haplgroup family. The vast majority of European E-M35 clades belong to subhaplogroup [[Haplogroup_E1b1b_(Y-DNA)#E1b1b1a2_.28E-V13.29|E-M78 V-13]]. In fact, the highest frequencies of V13 are found in Europe - specifically the southern Balkans.<ref>Cruciani 2007 |
Since then, a lot more information has been discovered about this haplgroup family. The vast majority of European E-M35 clades belong to subhaplogroup [[Haplogroup_E1b1b_(Y-DNA)#E1b1b1a2_.28E-V13.29|E-M78 V-13]]. In fact, the highest frequencies of V13 are found in Europe - specifically the southern Balkans.<ref>Cruciani 2007</ref><ref name="cruciani2004"/> Whilst the parental E-M78 clade probably arose in northeastern Africa, how and when V-13 came to enjoy significant frequencies in Europe is not entirely clear at present. Cruciani proposed that the mutation which differentiated V-13 from other E-M78 clades first arose in the Near East c. 11 kYa, and at some point experienced a low-grade expansion into Europe. At a later time, it enjoyed a rapid expansion within Europe, accounting for its current frequencies. He dated this to 'no earlier than 5.3 kYa'.<ref>Cruciani 2007</ref>. {{Harvcoltxt|Battaglia et al.|2008}} suggest that E-M78 arrived into Europe directly from northern Africa during the humid phase of the Holocene (c. 8.5 kYa). Subsequently, V-13 might have arisen in the region of Macedonia-Peloponesse. They argue that its expansion was due to the 'rapid and robust adoption' of the "Neolithic package" by Balkan Mesolithic peoples, characterised by V-13, who then underwent a rapid demographic expansion. According to either study, the spread of Haplogroup E1b1b into Europe cannot be connected with to large scale migration of 'Neolithic farmers' from the Near East. Other studies, such as those {{Harvcoltxt|Underhill and Kivisild|2007}} and King et al (2008) substantiate the idea that E1b1b clades were already present in Europe prior to the incipient Neolithic.<ref> Underhill's study proposes that E1b1b seems to represent a late-Pleistocene migration from Africa to Europe over the [[Sinai Peninsula]] in [[Egypt]], evidence for which does not show up in mitochondrial DNA. ''Y chromosome data show a signal for a separate late-Pleistocene migration from Africa to Europe via Sinai as evidenced through the distribution of haplogroup E3b lineages, which is not manifested in mtDNA haplogroup distributions.''</ref> Moreoever, whilst Battaglia connected the expansion phase of V-13 with an expansion of indegenous Balkan "foragers-come-farmers", Cruciani sees little connection between its expansion and that of farming in Europe. |
||
=== Bronze and Iron Age migrations === |
=== Bronze and Iron Age migrations === |
||
| Line 190: | Line 190: | ||
E1b1b is thought to have expanded from Sub-Saharan Africa into North Africa, the Levant and then into Europe. In 2000 Rosser, connected the presence of E1b1b1 (E-M35) clades in southern Europe (the Balkans, Iberia and southern Italy) to a 'northern African influence'. However, the author did not directly connect this with any demographic event or time period.<ref>{{Harvcoltxt|Rosser et al|2000}} Here, it is referred to as Hg 21</ref> Subsequently, as discussed above, the vast majority of these clades were found to belong to haplogroup E1b1b1a2 (E-V13), whose current frequencies in Europe are due to a demographic expansion originating from ''within'' Europe during or after the Neolithic. |
E1b1b is thought to have expanded from Sub-Saharan Africa into North Africa, the Levant and then into Europe. In 2000 Rosser, connected the presence of E1b1b1 (E-M35) clades in southern Europe (the Balkans, Iberia and southern Italy) to a 'northern African influence'. However, the author did not directly connect this with any demographic event or time period.<ref>{{Harvcoltxt|Rosser et al|2000}} Here, it is referred to as Hg 21</ref> Subsequently, as discussed above, the vast majority of these clades were found to belong to haplogroup E1b1b1a2 (E-V13), whose current frequencies in Europe are due to a demographic expansion originating from ''within'' Europe during or after the Neolithic. |
||
Another subclade of E1b1b1 (E-M35) is [[E1b1b#E1b1b1b .28E-M81.29.3B formerly E3b1b.2C E3b2|E1b1b1b]] (E-M81). This is often considered to be a "Berber marker",<ref |
Another subclade of E1b1b1 (E-M35) is [[E1b1b#E1b1b1b .28E-M81.29.3B formerly E3b1b.2C E3b2|E1b1b1b]] (E-M81). This is often considered to be a "Berber marker",<ref name="cruciani2004"/> or marker of relatively recent northwestern African migrations into Southern Europe. Unlike E1b1b1a2 (E-V13), notable frequencies are limited to specific areas within the [[Iberian Peninsula]], [[Italy]] and [[Sicily]]. In certain Iberian populations, it is more common than E-M78, at an average frequency of 4-5.6%, with frequencies reaching 9% in [[Galicia (Spain)|Galicia]], 10% in Western [[Andalusia]] and Northwest [[Castile (historical region)|Castile]] and 13 % in [[Cantabria]].<ref>{{Harvcoltxt|Flores et al.|2005}}, {{Harvcoltxt|Beleza et al.|2006}}, {{Harvcoltxt|Adams et al.|2008}}</ref><ref name=Capelli09/> The highest frequency of this clade found so far in Europe has been observed at 40% the [[Pas and Miera valleys|Pasiegos]] from [[Cantabria]].<ref name=Cruciani04>{{Harvcoltxt|Cruciani|2004}}</ref> {{Harvcoltxt|Flores et al.|2004}} propose that the absence of [[microsatellite]] variation suggests a very recent arrival from North Africa consistent with historical exchanges across the Mediterranean during the period of Islamic expansion, namely of [[Berber people|Berber]] populations,<ref name=Semino04>{{Harvcoltxt|Semino et al.|2004}}</ref> although in a study of [[Portugal|Portuguese]] Y-chromosome lineages, {{Harvcoltxt|Gonçalves et al.|2005}} revealed that "The mtDNA and Y data indicate that the Berber presence in that region dates prior to the Moorish expansion in 711 AD... Our data indicate that male Berbers, unlike sub-Saharan immigrants, constituted a long-lasting and continuous community in the country". |
||
In [[Italy]], E1b1b1b is typically more common in parts of Sicily, but it is found also in continental [[Italy]]<ref>{{Harvcoltxt|Gaetano et al.|2008}}</ref> (for example near [[Lucera]], a region where Sicilian Moslems are known to have been settled in the time of [[Frederick II]])<ref name=Capelli09>{{Harvcoltxt|Capelli et al.|2009}}</ref> and [[France]],<ref name=Cruciani04/> possibly due to historic migrations during the [[Islam in Europe|Islamic]], [[Roman Empire|Roman]], and [[Carthage|Carthaginian]] empires, as well as the influence of [[Sephardi Jews|Sephardic]] [[Jew]]s.<ref>{{Harvcoltxt|Gonçalves et al.|2005}}</ref> In all, sequences whose origins lay in northern Africa have been "estimated to contribute to the Sicilian gene pool at a rate of 6%". |
In [[Italy]], E1b1b1b is typically more common in parts of Sicily, but it is found also in continental [[Italy]]<ref>{{Harvcoltxt|Gaetano et al.|2008}}</ref> (for example near [[Lucera]], a region where Sicilian Moslems are known to have been settled in the time of [[Frederick II]])<ref name=Capelli09>{{Harvcoltxt|Capelli et al.|2009}}</ref> and [[France]],<ref name=Cruciani04/> possibly due to historic migrations during the [[Islam in Europe|Islamic]], [[Roman Empire|Roman]], and [[Carthage|Carthaginian]] empires, as well as the influence of [[Sephardi Jews|Sephardic]] [[Jew]]s.<ref>{{Harvcoltxt|Gonçalves et al.|2005}}</ref> In all, sequences whose origins lay in northern Africa have been "estimated to contribute to the Sicilian gene pool at a rate of 6%". |
||
| Line 199: | Line 199: | ||
The Y-DNA data has been supported by mtDNA studies. [[Haplogroup U (mtDNA)|Haplogroups U6]] and [[Haplogroup M (mtDNA)|M1]] are present in Iberian populations in ranges of 0 to 7%.<ref>"Haplogroup U6 is present at frequencies ranging from 0 to 7% in the various Iberian populations, with an average of 1.8%. Given that the frequency of U6 in NW Africa is 10%, the mtDNA contribution of NW Africa to Iberia can be estimated at 18%. This is larger than the contribution estimated with Y-chromosomal lineages (7%) (Bosch et al. 2001)."[http://www.blackwell-synergy.com/links/doi/10.1046/j.1469-1809.2003.00039.x/abs Joining the Pillars of Hercules: mtDNA Sequences Show Multidirectional Gene Flow in the Western Mediterranean (2003)]</ref><ref name=Pereira05>{{cite journal |author=Pereira L, Cunha C, Alves C, Amorim A |title=African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times |journal=Hum. Biol. |volume=77 |issue=2 |pages=213–29 |year=2005 |month=April |pmid=16201138 |quote=Although the absolute value of observed U6 frequency in Iberia is low, it reveals a considerable North African female contribution, if we keep in mind that haplogroup U6 is not very common in North Africa itself and virtually absent in the rest of Europe. Indeed, because the range of variation in western North Africa is 4-28%, the estimated minimum input is 8.54%}}</ref><ref>"Our results clearly reinforce, extend, and clarify the preliminary clues of an "important mtDNA contribution from [[northwest Africa]] into the Iberian Peninsula" (Côrte-Real et al., 1996; Rando et al., 1998; Flores et al., 2000a; Rocha et al., 1999)(...) Our own data allow us to make minimal estimates of the maternal African pre-Neolithic, Neolithic, and/or recent [[History of slavery|slave trade]] input into Iberia. For the former, we consider only the [[Average|mean value]] of the U6 frequency in northern African populations, excluding Saharans, Tuareg, and Mauritanians (16%), as the pre-Neolithic frequency in that area, and the present frequency in the whole Iberian Peninsula (2.3%) as the result of the northwest African gene flow at that time. The value obtained (14%) could be as high as 35% using the data of [[Corte-Real]] et al. (1996), or 27% with our north Portugal sample." [http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&uid=12627534&cmd=showdetailview&indexed=google Mitochondrial DNA affinities at the Atlantic fringe of Europe (2003)]</ref><ref name="Malyarchuk2008"/> However, the presence of U6 in Iberia cannot be equated with certainty to historic Islamic presence given that it happens to be a characteristic genetic marker of the Saami populations of Northern Scandinavia.<ref name=Pereira05/>, it is more frequent in the north of the Iberian Peninsula rather than in the south, and is also attested too in the [[British Islands]], again at their northern and western borders. It may be a trace of a prehistoric neolithic/megalithic expansion along the Atlantic coasts from North Africa, perhaps in conjunction with seaborne trade. A trace of these lineages have even made their way to eastern European populations<ref name="Malyarchuk2008">{{cite journal|year=2008|last=Malyarchuk et al.|doi=10.1038/ejhg.2008.70|url=http://www.nature.com/ejhg/journal/v16/n9/abs/ejhg200870a.html|title=Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs}}</ref> |
The Y-DNA data has been supported by mtDNA studies. [[Haplogroup U (mtDNA)|Haplogroups U6]] and [[Haplogroup M (mtDNA)|M1]] are present in Iberian populations in ranges of 0 to 7%.<ref>"Haplogroup U6 is present at frequencies ranging from 0 to 7% in the various Iberian populations, with an average of 1.8%. Given that the frequency of U6 in NW Africa is 10%, the mtDNA contribution of NW Africa to Iberia can be estimated at 18%. This is larger than the contribution estimated with Y-chromosomal lineages (7%) (Bosch et al. 2001)."[http://www.blackwell-synergy.com/links/doi/10.1046/j.1469-1809.2003.00039.x/abs Joining the Pillars of Hercules: mtDNA Sequences Show Multidirectional Gene Flow in the Western Mediterranean (2003)]</ref><ref name=Pereira05>{{cite journal |author=Pereira L, Cunha C, Alves C, Amorim A |title=African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times |journal=Hum. Biol. |volume=77 |issue=2 |pages=213–29 |year=2005 |month=April |pmid=16201138 |quote=Although the absolute value of observed U6 frequency in Iberia is low, it reveals a considerable North African female contribution, if we keep in mind that haplogroup U6 is not very common in North Africa itself and virtually absent in the rest of Europe. Indeed, because the range of variation in western North Africa is 4-28%, the estimated minimum input is 8.54%}}</ref><ref>"Our results clearly reinforce, extend, and clarify the preliminary clues of an "important mtDNA contribution from [[northwest Africa]] into the Iberian Peninsula" (Côrte-Real et al., 1996; Rando et al., 1998; Flores et al., 2000a; Rocha et al., 1999)(...) Our own data allow us to make minimal estimates of the maternal African pre-Neolithic, Neolithic, and/or recent [[History of slavery|slave trade]] input into Iberia. For the former, we consider only the [[Average|mean value]] of the U6 frequency in northern African populations, excluding Saharans, Tuareg, and Mauritanians (16%), as the pre-Neolithic frequency in that area, and the present frequency in the whole Iberian Peninsula (2.3%) as the result of the northwest African gene flow at that time. The value obtained (14%) could be as high as 35% using the data of [[Corte-Real]] et al. (1996), or 27% with our north Portugal sample." [http://www.ncbi.nlm.nih.gov/sites/entrez?db=pubmed&uid=12627534&cmd=showdetailview&indexed=google Mitochondrial DNA affinities at the Atlantic fringe of Europe (2003)]</ref><ref name="Malyarchuk2008"/> However, the presence of U6 in Iberia cannot be equated with certainty to historic Islamic presence given that it happens to be a characteristic genetic marker of the Saami populations of Northern Scandinavia.<ref name=Pereira05/>, it is more frequent in the north of the Iberian Peninsula rather than in the south, and is also attested too in the [[British Islands]], again at their northern and western borders. It may be a trace of a prehistoric neolithic/megalithic expansion along the Atlantic coasts from North Africa, perhaps in conjunction with seaborne trade. A trace of these lineages have even made their way to eastern European populations<ref name="Malyarchuk2008">{{cite journal|year=2008|last=Malyarchuk et al.|doi=10.1038/ejhg.2008.70|url=http://www.nature.com/ejhg/journal/v16/n9/abs/ejhg200870a.html|title=Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs}}</ref> |
||
=== |
=== Sub-Saharan African admixture === |
||
In general, Sub-Saharan admixture in Europe is distributed along a South-to-North cline. Peak frequencies of admixture have been detected in the Mediterranean region and Iberia. Gene flow from Africa to Europe is thought to have occurred in both recent and prehistoric times. This has either involved direct contact with Sub-Saharan Africans, or indirect contact through North African or Middle Eastern populations with Sub-Saharan affiliations. |
|||
| ⚫ | |||
Y-DNA haplogroups [[Haplogroup A (Y-DNA)|A]], [[Haplogroup B (Y-DNA)|B]] and [[Haplogroup E (Y-DNA)|E]] are the predominant haplogroups in Sub-Saharan Africa. Only Haplogroup E is found at moderate levels(7.2% in one sample<ref name="cruciani2004"/>) in Europe whereas all others haplogroups are present at low levels, usually less than 1%. Haplogroup E which originated in Sub-Saharan Africa is thought to have entered Europe via the Middle East in the late Pleistocene or Early Holocene.<ref name="frudakis2007">{{cite book|title=Molecular photofitting|first=Tony|last=Frudakis|chapter=Apportionment of Autosomal Diversity with Continental Markers|pages=page 326|chapterurl=http://books.google.com/books?id=9vXeydpj7VkC&pg=PA326|year=2007|isbn=0120884925}}</ref><ref group=note>[http://www.human-evol.cam.ac.uk/Members/Lahr/pubs/AHG-65-01.pdf Underhill et al, 2001] state" A mesolithic population carrying Group III lineages with the M35/M215 mutation expanded northwards from '''sub-Saharan''' to north Africa and the Levant. The Levantine population of farmers that dispersed into Europe during and after the Neolithic carried these African Group III M35/M215 lineages, together with a cluster of Group VI lineages characterized by M172 and M201 mutations."</ref><ref group=note>[http://books.google.com/books?id=9vXeydpj7VkC&pg=PA326 Tony Frudakis states] "Underhill et al showed that the frequency of the YAP+ haplogroup commonly referred to as haplogroup E or (III) is relatively high (about 25%) in the Middle East and Mediterranean. This haplogroup E is the major haplogroup in Sub-Saharan Africa(75% of all Y chromosomes). Specifically, Europeans contain the E3b subhaplogroup, which was derived from haplogroup E in sub-Saharan Africa and currently is distributed along the North and East of Africa. Thus we refer to the affiliation as African, West African, or North/East African depending on the time frame we wish to refer to and/or our semantic preference. The West African affiliation for continental Europeans and Middle Eastern populations appears to be anthropoligically meaningful information that might help define partitions of substructure in Europe."</ref> The main clade found in Europe is [[E1b1b]]. Haplogroup [[E1b1a]] dispersed recently in Africa, and therefore its sporadic presence in Europe is usually attributed to recent events, such as the slave trade, the Moorish occupation of Iberia and other historical contacts across the Mediterrenean and Atlantic. Haplogroups A, B and [[haplogroup E1|E1*]] are ancient African lineages that are relatively infrequent in Africa. They have sporadically been detected in Europe but their exact mode of entry into Europe is still not understood. A prehistoric entry of these haplogroups into Europe remains a possibility. <ref name="king">{{cite journal|last=King|title=Africans in Yorkshire? - the deepest-rooting clade of the Y phylogeny within an English genealogy|url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2590664#R7|year=2007}}</ref><ref name="goncalves2005"><cite journal|url=http://www.uma.pt/abrehm/v1.1/docs/downloads/pdfs/Goncalves_Y_Portugal_AnnHumGenet2005.pdf|last=Goncalves|title=Y-chromosome Lineages from Portugal, Madeira and A¸cores Record Elements of Sephardim and Berber Ancestry|year=2005}}</ref> |
|||
MtDNA (haplogroups [[Haplogroup L1 (mtDNA)|L1]], [[Haplogroup L2 (mtDNA)|L2]] and [[Haplogroup L3 (mtDNA)|L3]]) are the principal haplogroups found in Sub-Saharan Africa. Haplogroup L lineages are relatively infrequent(less than 1%) throughout Europe with the exception of Iberia, where frequencies as high as 14.8% percent have been reported. These lineages had previously been associated with the African slave trade.<ref name="pereira2000">{{cite journal |author=Pereira L, Prata MJ, Amorim A |title=Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation |journal=Ann. Hum. Genet. |volume=64 |issue=Pt 6 |pages=491–506 |year=2000 |month=November |pmid=11281213 |doi= |url=http://www3.interscience.wiley.com/cgi-bin/fulltext/119002773/PDFSTART}}</ref> However an analysis by Gonzalez et al revealed that most of the L lineages matched Northwest African L lineages rather than contemporary Sub-Saharan L lineages. The authors suggest this pattern indicates that most of the Sub-Saharan L lineages entered Iberia in prehistoric times rather than in more recent periods.<ref name="gonzalez2003"> {{cite journal|title=Mitochondrial DNA Affinities at the Atlantic Fringe of Europe|year=2003|last=Gonzalez et al|url=http://backintyme.com/admixture/gonzalez02.pdf|doi=10.1002/ajpa.10168 }}</ref> |
|||
| ⚫ | <ref>Salas et al. (2004) [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1182259 The African Diaspora: Mitochondrial DNA and the Atlantic Slave Trade]. Am J Hum Genet; 74(3): 454-465: "African types in Eurasia, unlike those in America, can therefore be attributed to gene flow from both eastern Africa -- perhaps partly via the Arab slave trade -- and western and southeastern Africa, more likely as a result of the Atlantic slave trade. [...] A contribution from the medieval Arab/Berber conquest of Iberia and Sicily is also possible. [...] However, there are also discernible western and southeastern African components to the (relatively few) mtDNAs of recent African ancestry within Europe, which are likely to be mainly attributable to the more recent Atlantic trade. Portuguese western, southwestern, and southeastern Africa were the main sources for the Atlantic slave trade to Europe."Therefore, it seems unlikely that the impact of mtDNAs of recent African ancestry on Iberia was the result of a single process; indeed, earlier gene flow may also have contributed some of the lineages.</ref> <ref>[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1852723 Achilli et al. 2007], [http://www.nature.com/ejhg/journal/v16/n9/abs/ejhg200870a.html Malyarchuk et al. 2008], [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=12627534&query_hl=2 Gonzalez et al. (2003)]</ref><ref name="cruciani2004"/><ref>[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15280900&query_hl=13&itool=pubmed_docsum Flores et al. 2004], [http://www3.interscience.wiley.com/cgi-bin/abstract/112139846/ABSTRACT Brion et al. 2005], [http://dx.doi.org/10.1016/j.forsciint.2005.06.002 Brion et al. 2004], [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=11078479&dopt=Abstract Rosser et al. 2000], [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=15069642 Semino et al. 2004], [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=12927125&dopt=Abstract DiGiacomo et al. 2003]</ref> |
||
<!---The edit war regarding the evidence for SSA contribution has been moved to the talk page, hammer out the differences there and avoid [[WP:COI]] and [[WP:NPOV]] here---> |
|||
African autosomal admixture tends to mirror the South-to-North cline profiles of Y-chromosome and MtDNA haplogroups. Halder et al were able to detect West African admixture in Europe wherever haplogroup E was prevalent. Auton et al detected a South-to-North cline of West African haplotypes in Europe with peak frequencies in Iberia. <ref group="note">[http://www3.interscience.wiley.com/journal/117914842/abstract?CRETRY=1&SRETRY=0 Halder et al] state. "We observed patterns of apportionment similar |
|||
to those described previously using sex and autosomal markers, such as European admixture for African Americans (14.3%) and Mexicans (43.2%), European (65.5%) and East Asian affiliation (27%) for South Asians, and low levels of African admixture (2.8–10.8%) mirroring the distribution of Y E3b haplogroups among various Eurasian populations.</ref> |
to those described previously using sex and autosomal markers, such as European admixture for African Americans (14.3%) and Mexicans (43.2%), European (65.5%) and East Asian affiliation (27%) for South Asians, and low levels of African admixture (2.8–10.8%) mirroring the distribution of Y E3b haplogroups among various Eurasian populations.</ref> |
||
<ref group="note">[http://genome.cshlp.org/content/early/2009/02/12/gr.088898.108.abstract Auton et al state]."These two results therefore suggest that while the initial migrations into Europe came via the Middle East, at least some degree of subsequent gene flow has occurred directly from Africa" "Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa."</ref> |
<ref group="note">[http://genome.cshlp.org/content/early/2009/02/12/gr.088898.108.abstract Auton et al state]."These two results therefore suggest that while the initial migrations into Europe came via the Middle East, at least some degree of subsequent gene flow has occurred directly from Africa" "Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa."</ref> |
||
| Line 332: | Line 340: | ||
| date = |
| date = |
||
| year = 2004 |
| year = 2004 |
||
| url = http://www. |
| url = http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1181964 |
||
| doi = 10.1086/386294 |
| doi = 10.1086/386294 |
||
| id = |
| id = |
||
Revision as of 05:41, 30 July 2009
This article needs attention from an expert in Molecular and Cellular Biology. Please add a reason or a talk parameter to this template to explain the issue with the article. (November 2008) |
This article's factual accuracy is disputed. (July 2007) |
The genetic history of Europe can be inferred by observing the patterns of genetic diversity across the continent and comparing them with the patterns on the adjacent land masses. These patterns can be found by using classical genetic markers or by using molecular genetics (autosomal, Y-chromosome and mitochondrial DNA). Most data is from modern populations, but there is a small amount of information from ancient DNA. European populations have a complicated demographic and genetic history, including many layers of successive migrations between different time periods, from the first appearance of Homo sapiens in the Upper Paleolithic to contemporary immigration.

Genetic Studies
One of the first scholars to perform genetic studies was Luigi Luca Cavalli-Sforza. He used classical genetic markers to analyse DNA by proxy. This method studies differences in the frequencies of particular allelic traits, namely polymorphisms from proteins found within human blood (such as the ABO blood groups, Rhesus blood antigens, HLA loci, immunoglobulins, G-6-P-D isoenzymes, amongst others). Subsequently his team calculated genetic distance between populations, based on the principle that two populations that share similar frequencies of a trait are more closely related than populations that have more divergent frequencies of the trait. From this, he constructed phylogenetic trees which showed genetic distances diagrammatically. His team also performed principal component analyses, which is good at analysing multivariate data with minimal loss of information. The information that is lost can be partly restored by generating a second principal component, and so on.[1] In turn, the information from each individual principal component (PC) can be presented graphically in synthetic maps. These maps show peaks and troughs, which represent populations whose gene frequencies take extreme values compared to others in the studied area.[2] Peaks and troughs usually, but not necessarily, connected by smooth gradients, called clines. Genetic clines can be generated in several ways: including adaptation to environment (natural selection), continuous gene flow between two initially different populations, or a demographic expansion into a scarcely populated environment with little initial admixture with pre-existing populations.[3] Cavalli-Sforza connected these gradients with postulated pre-historic population movements based on known archaeological and linguistic theories. However, given that the time depths of such patterns are not known, “associating them with particular demographic events is usually speculative”.[4]
Studies using direct DNA analysis are now abundant, and may utilize mitochondrial DNA (mtDNA), the non-recombining portion of the Y chromosome (NRY) or autosomal DNA. MtDNA and NRY DNA share some similar features which have made them particularly useful in genetic anthropology. These properties include the direct, unaltered inheritance of mtDNA and NRY DNA from mother to offspring, and father to son, respectively, without the 'scrambling' effects of genetic recombination. We also presume that these genetic loci are not affected by natural selection, and that the major process responsible for changes in base pairs has been mutation (which can calculated).[5] The smaller effective population size of the NRY and mtDNA enhances the consequences of drift and founder effect relative to the autosomes, making NRY and mtDNA variation a potentially sensitive index of population composition.[4][6][7] However, these biologically plausible assumptions are nevertheless not concrete. For example, Rosser suggests that climactic conditions may affect the fertility of certain lineages.[4] Even more problematic, however, is the underlying mutation rate used by the geneticists. They often use different mutation rates, and therefore studies are frequently arriving at vastly different conclusions.[4] Moroever, NRY and mtDNA may be so susceptible to drift that some ancient patterns may have become obscured over time. Another implicit assumption is that population genealogies are approximated by allele genealogies. Barbujani points out that this only holds if population groups develop from a genetically monomorphic set of founders. However, Barbujani argues that there is no reason to believe that Europe was colonized by monomorphic populations. This would result in an overestimation of haplogroup age, thus falsely extending the demographic history of Europe into the Late Paleolithic rather than the Neolithic era.[8] (See also Genetic drift, Founder effect, Population bottleneck.)
Whereas Y-DNA and mtDNA haplogroups represent but a small component of a person’s DNA pool, autosomal DNA has the advantage of containing hundreds and thousands of examinable genetic loci, thus giving a more complete picture of genetic composition (eg see Seldin). However, descent relationships can only to be determined on a statistical basis because autosomal DNA undergoes recombination and is liable to the process of natural selection.
Genetic studies operate on numerous assumptions and suffer from usual methodological limitations such as selection bias and confounding. Furthermore, no matter how accurate the methodology, conclusions derived from such studies are ultimately compiled on the basis of how the author envisages their data fits with established archaeological or linguistic theories.
Relation between Europeans and other populations
| Africa | Oceania | East Asia | Europe | |
|---|---|---|---|---|
| Oceania | 24.7 | |||
| East Asia | 20.6 | 10 | ||
| Europe | 16.6 | 13.5 | 9.7 | |
| America | 22.6 | 14.6 | 8.9 | 9.5 |
According to Cavalli-Sforza's work, all non-African populations are more closely related to each other than to Africans; supporting the hypothesis that all non-Africans descend from a single African population. The genetic distance from Africa to Europe (16.6) was found to be shorter than the genetic distance from Africa to East Asia (20.6), and much shorter than that from Africa to Australia (24.7). He explains: "both Africans and Asians contributed to the settlement of Europe, which began about 40,000 years ago. It seems very reasonable to assume that both continents nearest to Europe contributed to its settlement, even if perhaps at different times and maybe repeatedly. It is reassuring that the analysis of other markers also consistently gives the same results in this case. Moreover, a specific evolutionary model tested, i.e., that Europe is formed by contributions from Asia and Africa, fits the distance matrix perfectly (6). In this simplified model, the migrations postulated to have populated Europe are estimated to have occurred at an early date (30,000 years ago), but it is impossible to distinguish, on the basis of these data, this model from that of several migrations at different times. The overall contributions from Asia and Africa were estimated to be around two-thirds and one-third, respectively".[9][10][11]
This particular model used an Out of Africa migration 100,000 years ago which separated Africans from non-Africans followed by a single admixture event 30,000 years ago leading to the formulation of the European population. The admixture event consisted of a source population that was 35% African and 65% East Asian. However the study notes that a more realistic scenario would include several admixture events occurring over a sustained period. In particular they cite the spread of farming from a source population in West Asia 5000-9000 years ago may have played a role in the genetic relatedness of Africans and Europeans since West Asia is sandwiched in between Africa and Central Asia. The model assumed an out of Africa migration 100kya and a single admixture event 30kya. However, most contemporary studies have more recent dates that place the out of Africa migration 50-70kya. The study also involved a direct comparison between Sub-Saharan Africans (Central Africans and Senegalese) and Europeans. North Africans population were omitted from the study as they are known to have both Eurasian and Sub-Saharan admixture. These considerations might help explain the apparent short genetic distance between Europeans and Africans[9][10][11]
A later study by Bauchet, which utilised ~ 10 thousand autosomal DNA SNPs arrived at similar results. Principal component analysis clearly identified four widely dispersed groupings corresponding to Europe, South Asia, Central Asia, and Africa. PC1 separated Africans from the other populations, PC2 divided Asians from Europeans and Africans, whilst PC3 split Central Asians apart from South Asians.[12]
European population sub-structure
Geneticists agree that Europe is the most genetically homogeneous of all the continents.[13][14] However, some patterns are discernable. Cavalli-Sforza’s principal component analyses revealed five major clinal patterns through out Europe, the meanings of which are included below.[15] Five patterns of genetic relatedness have been identified:
- A cline of genes with highest frequencies in the Middle East, spreading to lowest levels northwest.
- A cline of genes with highest frequencies amongst Finnish and Saami in the extreme north east, and spreading to lowest frequencies in the south west.
- A cline of genes with highest frequencies in the area of the lower Don and Volga rivers in southern Russia, and spreading to lowest frequencies in Iberia, Southern Italy, Greece and the areas inhabited by Saami speakers in the extreme north of Scandinavia.
- A cline of genes with highest frequencies in the Balkans and Southern Italy, spreading to lowest levels in Britain and the Basque country.
- A cline of genes with highest frequencies in the Basque country, and lower levels beyond the area of Iberia and Southern France.
He also created a phylogenetic tree to analysed the internal relationships amongst Europeans. He found four major 'outliers'- Basques, Lapps, Finns and Icelanders; a result he attributed to their relative isolation. Greeks and Yugoslavs represented a second group of less extreme outliers. The remaining populations clustered into several groups : "Celtic", "Germanic", "south-western Europeans", "Scandinavians" and "eastern Europeans".[16]
Semino and Rosser independently performed PCA analyses based on NRY data from several European populations. They found that Y haplogroups showed high degrees of geographic structuring. Semino’s data showed three distinct clusters. A western European group formed a discrete cluster (comprising of Basques, French, Spanish, northern Italians, Germans and Dutch), which was dominated by high frequencies of Hg R1b. An eastern European group (Poles, Hungarians, Macedonians, Croats, Czechs, Ukrainians) was characterised by high frequencies of R1a and I2. A third, middle eastern (Syrians, Lebanese, Turks) group was characterised by high frequencies of haplogroup J lineages.[17] Greeks occupied an intermediate position between European and Middle Eastern populations.[4]
Later studies looking at genetic diversity on a micro–regional scale have revealed that significant internal heterogeneity exists within some countries, cautioning us from assuming that frequencies quoted in pan-continental studies are representative of entire national communities or ethnic groups. This complexity prompts caution in equating similarity in the frequencies of one or more Y chromosomal haplogroups among populations and common descent.[18] In contrast to Y DNA haplogroups, mtDNA haplogroups did not show as much geographical patterning, but were more evenly ubiquitous. Apart from the outlying Saami, all Europeans are characterized by the predominance of haplogroups H, U and T. The lack of observable geographic structuring of mtDNA may be due to socio-cultural factors, namely the phenomena of polygyny and patrilocality.[4]
A later study by Seldin (2006) used over five thousand autosomal SNPs. It showed “a consistent and reproducible distinction between ‘northern’ and ‘southern’ European population groups”. Most individual participants with southern European ancestry (Italian, Greek, Armenian, Portuguese, and Spanish) have >85% membership in the ‘southern’ population; and most northern, western, eastern, and central Europeans have >90% in the ‘northern’ population group. However, many of the participants in this study were actually American citizens who self-identified with different European ethnicities based on familial pedigree.[19]
A similar study in 2007 using samples exclusively from Europe found that the most important genetic differentiation in Europe occurs on a line from the north to the south-east (northern Europe to the Balkans), with another east-west axis of differentiation across Europe. Its findings were consistent with earlier results based on mtDNA and Y-chromosonal DNA that support the theory that modern Iberians (Spanish and Portuguese) hold the most ancient European genetic ancestry, as well as separating Basques and Sami from other European populations. It suggested that the English and Irish cluster with other Northern and Eastern Europeans such as Germans and Poles, while some Basque and Italian individuals also clustered with Northern Europeans. Despite these stratifications, it noted the unusually high degree of European homogeneity: "there is low apparent diversity in Europe with the entire continent-wide samples only marginally more dispersed than single population samples elsewhere in the world."[20]
In 2008, two international research teams published analyses of large-scale genotyping of large samples of Europeans, utilising using over 300, 000 autosomal SNPs. With the exception of usual isolates such as Basques, Finns and Sardinians, the European population lacked sharp discontinuities (clustering) as previous studies have found (see Seldin et al. 2006 and Bauchett et al. 2007), although there was a discernible south to north gradient. Overall, they found only a low level of genetic differentiation between subpopulations, and differences which did exist were characterized by a strong continent-wide correlation between geographic and genetic distance. In addition, they found that diversity was greatest in southern Europe due a larger effective population size and/or population expansion from southern to northern Europe.[21] The researchers take this observation to imply that, genetically speaking, Europeans are not distributed into discrete, populations.[22][23] In fact, according to another European wide study, the main components in the European genomes appear to derive from ancestors whose features were similar to those of modern Basques and Near Easterners, with average values greater than 35% for both these parental populations, regardless of whether or not molecular information is taken into account. The lowest degree of both Basque and Near Eastern admixture is found in Finland, whereas the highest values are, respectively, 70% ("Basque") in Spain and more than 60% ("Near Eastern") in the Balkans.[24][25]
A very recent study in May 2009 [26] that studied 19 populations from Europe using 270,000 SNPs highlighted the genetic diversity or European populations corresponding to the northwest to southeast gradient and distinguished "four several distinct regions" within Europe:
- Finland, showing the greatest distance to the rest of Europeans.
- the Baltic region (Estonia, Latvia and Lithuania), western Russia and eastern Poland.
- Central and Western Europe.
- Italy, "with the southern Italians being more distant".
In this study, barrier analysis revealed "genetic barriers" between Finland, Italy and other countries and interestingly, barriers could also be demonstrated within Finland (between Helsinki and Kuusamo) and Italy (between northern and southern part, Fst= 0.0050). Fst (Fixation index) was found to correlate considerably with geographic distances ranging from ≤0.0010 for neighbouring populations to 0.0200-0.0230 for Southern Italy and Finland. For comparisons, pair-wise Fst of non-European samples were as follows: Europeans – Africans (Yoruba) 0.1530; Europeans – Chinese 0.1100; Africans (Yoruba) – Chinese 0.1900.[27]
Haplogroups in Europe
Human Y-chromosome DNA haplogroups

There are three major Y-chromosome DNA haplogroups which largely account for most of Europe's present-day population.[28]
- Haplogroup R1b is common all over Europe but especially common on the western Atlantic coast of Europe.[29][30] Nearly all of this R1b in Europe is specifically in the form of the R1b1b2 (R-M269) sub-clade, whereas R1b found in Central Asia, Western Asia and Africa tends to be in other clades. Haplogroup R1b frequencies vary from highs in Western Europe in a steadity decreasing cline with growing distance from the Atlantic: 80-90% (Welsh, Basques, Irish, Scots, north-western Spanish and western French); around 50-60% southern Spanish, southern/central Portuguese, eastern French; around 40-50% amongst Germans, Czechs and Slovaks; around 20% amongst Greeks, Sardinians, Slovenians, Swedes, Romanians, Albanians and Bulgarians. It is under 20% in areas such as Poland, Turkey, Mainland Croatia, Hungary, Serbia and Cyprus.[31] Compared to other common Y lineages, R1b remains the most common clade as one moves east to Germany, while further east in Poland, R1a is more common (see below).[32] In Southeastern Europe, R1b drops behind R1a in the area in and around Hungary and Serbia, but it is more common both to the south and north of this specific region.[33]
- Cinnioglu et al. (2004) wrote, concerning R1b1b2 in western Europe that "variance is higher in Iberia than in Western Europe. The decreasing diversity radiating from Turkey towards Southeast Europe, Caucasus and Mesopotamia approximates similar results from Iberia tracing the re-colonization of Northwest Europe by hunter-gatherers during the Holocene as suggested by others (Torroni et al. 1998; Semino et al. 2000a; Wilson et al. 2001)."[34]
- Haplogroup I is found in the form of various distinct sub-clades throughout all of Europe, and is for example relatively common in common in Bosnia and Herzegovina, Croatia, Sweden, Norway, Sardinia and parts of Germany. This clade is only found in Europe, and may have been there since before the LGM.[35]
- Haplogroup R1a is common in Central and Eastern Europe (and is also common in Central Asia and the Indian subcontinent). For example there is a sharp increase in R1a1, and decrease in R1b1b2 presence, as one goes east from Germany to Poland.[36] It also has a substantial presence in Scandinavia (particularly Norway) the Pas Valley[37] and Paris[38] and a small presence in the British Isles.
Putting aside small enclaves there are also several haplogroups apart from the above three, which are most common in certain specific areas of Europe.
- Haplogroup N in the form of its N1c1 sub-clade reaches frequencies of approximately 60% among Finns and approximately 40% among Lithuanians. This clade is found also to the East, as far as Siberia, Japan and China.
- Haplogroup E1b1b1, mainly in the form of its E1b1b1a2 (E-V13) sub-clade reaches frequencies above 40% around the area of Kosovo.[39] This clade is thought to have arrived in Europe from Western Asia either in the later Mesolithic,[40] or the Neolithic.[41]
- Haplogroup J, in various sub-clades, is found in levels of around 15-30% in parts of the Balkans and Italy.[42]
Human mitochondrial DNA haplogroups
There have been a number of studies about the mitochondrial DNA haplogroups (mtDNA) in Europe. According to the University of Oulu Library in Finland:
Classical polymorphic markers (i.e. blood groups, protein electromorphs and HLA antigenes) have suggested that Europe is a genetically homogeneous continent with a few outliers such as the Saami, Sardinians, Icelanders and Basques (Cavalli-Sforza et al. 1993, Piazza 1993). The analysis of mtDNA sequences has also shown a high degree of homogeneity among European populations, and the genetic distances have been found to be much smaller than between populations on other continents, especially Africa (Comas et al. 1997).
The mtDNA haplogroups[43] of Europeans are surveyed by using a combination of data from RFLP analysis of the coding region and sequencing of the hypervariable segment I. About 99% of European mtDNAs fall into one of ten haplogroups: H, I, J, K, M, T, U, V, W or X (Torroni et al. 1996a). Each of these is defined by certain relatively ancient and stable polymorphic sites located in the coding region (Torroni et al. 1996a)... Haplogroup H, which is defined by the absence of a AluI site at bp 7025, is the most prevalent, comprising half of all Europeans (Torroni et al. 1996a, Richards et al. 1998)... Six of the European haplogroups (H, I, J, K, T and W) are essentially confined to European populations (Torroni et al. 1994, 1996a), and probably originated after the ancestral Caucasoids became genetically separated from the ancestors of the modern Africans and Asians.[44]
Apparent Migrations into Europe
The prehistory of the European peoples can be traced by the examination of archaeological sites, linguistic studies, and by the examination of the DNA of the people who live in Europe now, or from recovered ancient DNA. Much of this research is ongoing, and discoveries are still being continually made, so theories rise and fall. Although it is possible to track the various migrations of people across Europe using founder analysis of DNA, most information on these movements comes from archeology. It is important to note that the colonization of Europe did not occur in discrete migrations, as might appear to be suggested. Rather, the settlement process was complex and "likely to have occurred in multiple waves from the east and to have been subsequently obscured by millennia of recurrent gene flow".[45]
Palaeolithic Era
Homo neanderthalensis had inhabited much of Europe and western Asia from as far back as 130, 000 years ago. They continued to exist in Europe as late as 30, 000 years ago. They were replaced by anatomically modern humans (A.M.H.), Cro-Magnoid homo sapiens, who began to appear in Europe c. 40, 000 years ago. Given that the two hominid species likely co-existed in Europe, anthropologists ask whether the two interacted Neanderthals. Before the advent of genetic studies, some anthropologists believed they had discovered skeletons representing Neanderthal/ modern human 'hybrids'. However, these results were deemed 'ambiguous'. Archaeological evidence points to an abrupt transition from Neanderthal artefacts to those related to A.M.H during the Upper Palaeolithic. Y chromosomal and mtDNA data suggest that modern European DNA ultimately derives from Africa, which diverged less than 100, 000 years ago, whereas the last common ancestor between the two species lived between 500, 000 to 600, 000 years ago. Whilst it is conceivable that the autosomes of modern Europeans may retain Neanderthal sequences, Neanderthals were essentially replaced by modern humans. Technological, economical and intellectual advantages not only allowed AMH to better adapt to the Upper Palaeolithic environment of Europe, but probably also resulted in cultural, and therefore biological, barriers between the two species. Neanderhthals were thus probably outcompeted by modern humans, with little or no gene exchange.[46]
That modern humans began to colonize Europe during the Upper Paleolithic about 40,000 years ago is evidenced by the spread of the Aurignacian culture.[47] From a Y-chromosome perspective, Semino proposed that the large haplogroup R1 is an ancient Eurasiatic marker brought in by Homo sapiens who diffused west into Europe ~ 40 ky ago.[48][49] Haplogroup I might represent another putative Palaeolithic marker whose age has been estimated to ~ 22 kYa. Whilst it is 'unique' to Europe, it probably arose in descendants of men arriving from the Middle East c. 20 - 25 kYa, arising from parent haplogroup IJ. At this time, another Upper Palaeolithic culture appears, the Gravettian culture.[50] Thus the genetic data suggests that, from a male perspective, modern humans might have taken two colonizating routes, one from the middle east via the Balkans, and another from Central Asia to the north of the Black Sea.
Martin Richards et al. found that 15- 40% of extant mtDNA lineages trace back to the Palaeolithic migrations (depeding on whether one allows for multiple founder events).[6] MtDNA haplogroup U5, dated to be ~ 40 to 50 kYa, arrived during the first early upper Palaeolithic colonisation. Individually, it accounts for 5-15 % of total mtDNA lineages. Middle U.P. movements are marked by the haplogroups HV, I and U4. HV split into Pre-V (around 26,000 years old) and the larger branch H, both of which spread over all Europe, possibly via Gravettian contacts.[50][51] Haplogroup H accounts for about half the gene lines in Europe, with many subgroups. The above mtDNA lineages, or their precursors, are most likely to have arrived into Europe via the Middle East. This contrasts with Y DNA evidence, whereby some 50%+ of male lineages are characterized by the R1 superfamily, which is of possible central Asian origin. Semino postulates that these differences "may be due in part to the apparent more recent molecular age of Y chromosomes relative to other loci, suggesting more rapid replacement of previous Y chromosomes. Gender-based differential migratory demographic behaviors will also influence the observed patterns of mtDNA and Y variation".
Last Glacial Maximum: Refugia and Re-colonization
About 25,000 years ago began the last very cold period (the Last Glacial Maximum, LGM), rendering much of Europe uninhabitable. Much of northern and central Europe was vacated and people took refuge in climactic sanctuaries (or refugia) as follows:
- Northern Iberia and Southwest France, together making up the so-called "Franco-Cantabrian" refugium.
- The Balkans.
- The Ukraine and more generally the northern coast of the Black Sea.[50]
- Italy[52]
This event decreased the overall genetic diversity in Europe, a "result of drift, consistent with an inferred population bottleneck during the Last Glacial Maximum".[53] As the glaciers receded from about 16 -13 kYa, Europe began to be slowly repopulated by populations within the above-mentioned refugia, leaving traceable genetic signatures.[54]
In particular, some Y haplogroup I clades appear to have diverged from their parental haplogroups sometime during, or shortly after, the LGM.[35] Haplogroup I2 is prevalent in the western Balkans, as well as the rest of southeastern and central-eastern Europe in more moderate frequencies. Its frequency drops rapidly in central Europe, suggesting that the survivors bearing I2 lineages expanded predominantly through south-eastern and central-eastern Europe.[55]
In addition, Cinnioglu sees evidence for the existence of an Anatolian refuge, which also harboured Hg R1b1b2.[56] Today, R1b dominates the y chromosome landscape of western Europe and the British Isles, suggesting that there could have been large populations composition changes based on migrations after the LGM.
Semino, Passarino and Pericic place the origins of haplogroup R1a within the Ukrainian ice-age refuge. Its current distribution throughout eastern Europe and parts of Scandinavia are thus, in part, reflective of a re-peopling of Europe from the southern Russian/ Ukrainian steppes.[57][58][59]
From an mtDNA perspective, Richards et al. found that they majority of mtDNA diversity in Europe is accounted for by post-glacial re-expansions during the late upper Palaeolithic/ Mesolithic. "The regional analyses lend some support to the suggestion that much of western and central Europe was repopulated largely from the southwest when the climate improved. The lineages involved include much of the most common haplogroup, H, as well as much of K, T, W, and X." The study could not elucidate clearly whether there were new migrations of mtDNA lineages from the near east during this period, however, that there was a significant input was deemed unlikely.[45]
Neolithic migrations
The duration of the Neolithic varied from place to place, starting with the introduction of farming and ending with the introduction of bronze implements. In SE Europe it was approximately 7000-3000 BC while in NW Europe 4500-1700 BC. During this era, the so-called Neolithic revolution led to drastic economic as well as socio-cultural changes in Europe. In addition to the introduction of domesticated plants and animals, the Neolithic also saw a dramatic increase in the use of pottery designs as a symbolic expression of group collectiveness. The communis opinio places the origins of Agriculture somewhere in the Fertile Crescent or Near East[60]
An important issue regarding the introduction of neolithic technologies in Europe is the manner by which they were transferred into Europe. Primarily, this question pertains to whether farming was introduced by a significant migratory movement of farmers from the Near East (Cavalli-Sforza's biological demic diffusion model), or a mere "cultural diffusion", or some combination of the two. Secondarily, population genetisist have tried to clarify whether any detectable genetic signatures of Near Eastern origin correspond to the expansion routes postulated by the archaeological evidence.[61]
Martin Richards estimated that 11% of European mtDNA is due to immigration in this period. Gene flow from SE to NW Europe seems to have continued in the Neolithic, the percentage significantly declining towards the British Isles. Classical genetics also suggested that the largest admixture to the European Paleolithic/Mesolithic stock was due to the Neolithic revolution of the 7th to 5th millennia BC.[62] Three main mtDNA gene groups have been identified as contributing Neolithic entrants into Europe: J, T1 and U3 (in that order of importance).[63] With others they amount up to around 20% of the gene pool.[64]
Im 2000, Semino's study on Y DNA revealed the presence of haplotypes belonging to the large clade E1b1b1 (E-M35). These were predominantly found in the souhern Balkans, southern Italy and parts of Iberia. Semino connected this pattern, along with J haplogroup subclades, to be the Y-DNA component of Cavalli-Sforza's Neolithic demic-diffusion.[65]
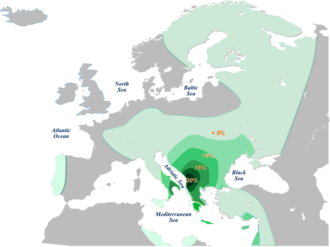
Since then, a lot more information has been discovered about this haplgroup family. The vast majority of European E-M35 clades belong to subhaplogroup E-M78 V-13. In fact, the highest frequencies of V13 are found in Europe - specifically the southern Balkans.[66][41] Whilst the parental E-M78 clade probably arose in northeastern Africa, how and when V-13 came to enjoy significant frequencies in Europe is not entirely clear at present. Cruciani proposed that the mutation which differentiated V-13 from other E-M78 clades first arose in the Near East c. 11 kYa, and at some point experienced a low-grade expansion into Europe. At a later time, it enjoyed a rapid expansion within Europe, accounting for its current frequencies. He dated this to 'no earlier than 5.3 kYa'.[67]. Battaglia et al. (2008) harvcoltxt error: multiple targets (2×): CITEREFBattaglia_et_al.2008 (help) suggest that E-M78 arrived into Europe directly from northern Africa during the humid phase of the Holocene (c. 8.5 kYa). Subsequently, V-13 might have arisen in the region of Macedonia-Peloponesse. They argue that its expansion was due to the 'rapid and robust adoption' of the "Neolithic package" by Balkan Mesolithic peoples, characterised by V-13, who then underwent a rapid demographic expansion. According to either study, the spread of Haplogroup E1b1b into Europe cannot be connected with to large scale migration of 'Neolithic farmers' from the Near East. Other studies, such as those Underhill and Kivisild (2007) and King et al (2008) substantiate the idea that E1b1b clades were already present in Europe prior to the incipient Neolithic.[68] Moreoever, whilst Battaglia connected the expansion phase of V-13 with an expansion of indegenous Balkan "foragers-come-farmers", Cruciani sees little connection between its expansion and that of farming in Europe.
Bronze and Iron Age migrations
The Bronze Age saw the development of long-distance trading networks, particularly along the Atlantic Coast and in the Danube valley. There was migration from Norway to Orkney and Shetland in this period (and to a lesser extent to mainland Scotland and Ireland). There was also migration from Germany to eastern England. Martin Richards estimated that there was about 4% mtDNA immigration to Europe in the Bronze Age. Oppenheimer could find no genetic evidence for any Iron Age migration to Britain.
One theory about the origin of the Indo-European language centres around a hypothetical Proto-Indo-European people, who are traced, in the Kurgan hypothesis, to somewhere north of the Black Sea at about 4500 BCE. They domesticated the horse, and are considered to have spread their culture and genes across Europe. It has been difficult to identify what these "Kurgan" genes might be, though the Y haplogroup R1a is a proposed marker which would indicate that the physical expansion halted in Germany and only the Kurgan culture and language went further. Another hypothesis — the Anatolian hypothesis — suggests an origin in Anatolia with a later expansion from eastern Europe.
To what extent Indo-European migrations replaced the indigenous Mesolithic peoples is debated, but a consensus has been reached that technology and language transfer played a more important role in this process than actual gene-flow.[69]
During the Iron Age, Celts are recorded as having moved from Gaul into northern Italy, Eastern Europe and Anatolia. The relationship between the Celts of Gaul and Spain is unclear as any migration occurred before records exist.
Roman Period
During the period of the Roman Empire, historical sources show that there were many movements of people around Europe, both within and outside the Empire. Historic sources sometimes cite instances of genocide incited by the Romans upon rebellious provincial tribes. If this did in fact occur, it would have been limited given that modern populations show considerable genetic continuity in their respective regions. The process 'Romanization' appeares to have been accomplished by the colonization of provinces by a few Roman-speaking administrators, military personnel and private citizens (merchants, traders) who emanated from the Empire's various regions (and not merely the Italian peninsula). They served as a nucleus for the acculturation of local notables.[70] Given their small numbers and varied origins, Romanization does not appear to have left distinct genetic signatures in Europe. Indeed, Romance-speaking populations in the Balkans have been found to genetically resemble neighbouring Greek and Slavic-speaking peoples rather than modern Italians.[71][72] No direct genetic information on Roman period migrations appears to exist other than a person with a rare Yorkshire surname of African ancestry (Haplogroup A1).[73]
North African Influences
E1b1b is thought to have expanded from Sub-Saharan Africa into North Africa, the Levant and then into Europe. In 2000 Rosser, connected the presence of E1b1b1 (E-M35) clades in southern Europe (the Balkans, Iberia and southern Italy) to a 'northern African influence'. However, the author did not directly connect this with any demographic event or time period.[74] Subsequently, as discussed above, the vast majority of these clades were found to belong to haplogroup E1b1b1a2 (E-V13), whose current frequencies in Europe are due to a demographic expansion originating from within Europe during or after the Neolithic.
Another subclade of E1b1b1 (E-M35) is E1b1b1b (E-M81). This is often considered to be a "Berber marker",[41] or marker of relatively recent northwestern African migrations into Southern Europe. Unlike E1b1b1a2 (E-V13), notable frequencies are limited to specific areas within the Iberian Peninsula, Italy and Sicily. In certain Iberian populations, it is more common than E-M78, at an average frequency of 4-5.6%, with frequencies reaching 9% in Galicia, 10% in Western Andalusia and Northwest Castile and 13 % in Cantabria.[75][76] The highest frequency of this clade found so far in Europe has been observed at 40% the Pasiegos from Cantabria.[77] Flores et al. (2004) propose that the absence of microsatellite variation suggests a very recent arrival from North Africa consistent with historical exchanges across the Mediterranean during the period of Islamic expansion, namely of Berber populations,[42] although in a study of Portuguese Y-chromosome lineages, Gonçalves et al. (2005) revealed that "The mtDNA and Y data indicate that the Berber presence in that region dates prior to the Moorish expansion in 711 AD... Our data indicate that male Berbers, unlike sub-Saharan immigrants, constituted a long-lasting and continuous community in the country".
In Italy, E1b1b1b is typically more common in parts of Sicily, but it is found also in continental Italy[78] (for example near Lucera, a region where Sicilian Moslems are known to have been settled in the time of Frederick II)[76] and France,[77] possibly due to historic migrations during the Islamic, Roman, and Carthaginian empires, as well as the influence of Sephardic Jews.[79] In all, sequences whose origins lay in northern Africa have been "estimated to contribute to the Sicilian gene pool at a rate of 6%". [80]
Apart from E-M81, other related haplogroups within the E-M35 clade are also seen as showing immigration from Northern Africa. Like E-V13, these are mostly subclades of E1b1b1a (E-M78): E-V12, E-V22, and E-V65 all of which have been found in low frequencies in Europe. Cruciani found that these could represent movements emanating from anytime after 15 kYa.[81]
The Y-DNA data has been supported by mtDNA studies. Haplogroups U6 and M1 are present in Iberian populations in ranges of 0 to 7%.[82][83][84][85] However, the presence of U6 in Iberia cannot be equated with certainty to historic Islamic presence given that it happens to be a characteristic genetic marker of the Saami populations of Northern Scandinavia.[83], it is more frequent in the north of the Iberian Peninsula rather than in the south, and is also attested too in the British Islands, again at their northern and western borders. It may be a trace of a prehistoric neolithic/megalithic expansion along the Atlantic coasts from North Africa, perhaps in conjunction with seaborne trade. A trace of these lineages have even made their way to eastern European populations[85]
Sub-Saharan African admixture
In general, Sub-Saharan admixture in Europe is distributed along a South-to-North cline. Peak frequencies of admixture have been detected in the Mediterranean region and Iberia. Gene flow from Africa to Europe is thought to have occurred in both recent and prehistoric times. This has either involved direct contact with Sub-Saharan Africans, or indirect contact through North African or Middle Eastern populations with Sub-Saharan affiliations.
Y-DNA haplogroups A, B and E are the predominant haplogroups in Sub-Saharan Africa. Only Haplogroup E is found at moderate levels(7.2% in one sample[41]) in Europe whereas all others haplogroups are present at low levels, usually less than 1%. Haplogroup E which originated in Sub-Saharan Africa is thought to have entered Europe via the Middle East in the late Pleistocene or Early Holocene.[86][note 1][note 2] The main clade found in Europe is E1b1b. Haplogroup E1b1a dispersed recently in Africa, and therefore its sporadic presence in Europe is usually attributed to recent events, such as the slave trade, the Moorish occupation of Iberia and other historical contacts across the Mediterrenean and Atlantic. Haplogroups A, B and E1* are ancient African lineages that are relatively infrequent in Africa. They have sporadically been detected in Europe but their exact mode of entry into Europe is still not understood. A prehistoric entry of these haplogroups into Europe remains a possibility. [73][87]
MtDNA (haplogroups L1, L2 and L3) are the principal haplogroups found in Sub-Saharan Africa. Haplogroup L lineages are relatively infrequent(less than 1%) throughout Europe with the exception of Iberia, where frequencies as high as 14.8% percent have been reported. These lineages had previously been associated with the African slave trade.[88] However an analysis by Gonzalez et al revealed that most of the L lineages matched Northwest African L lineages rather than contemporary Sub-Saharan L lineages. The authors suggest this pattern indicates that most of the Sub-Saharan L lineages entered Iberia in prehistoric times rather than in more recent periods.[89] [90] [91][41][92]
African autosomal admixture tends to mirror the South-to-North cline profiles of Y-chromosome and MtDNA haplogroups. Halder et al were able to detect West African admixture in Europe wherever haplogroup E was prevalent. Auton et al detected a South-to-North cline of West African haplotypes in Europe with peak frequencies in Iberia. [note 3] [note 4]
Central and East Asian admixture
East Asian mtDNA (haplogroups A, B, C, D, M, N9a and Z) is restricted mainly to Eastern Europe and Scandinavia and is found at generally low frequencies: Lapland (8.5%), Bulgaria/Turkey (6.9%), European Russia (4.2%), Spain/Portugal (2.3%), Czech Republic (1.8%), Western Slavs (1.6%), Eastern Slavs (1.3% to 5.2%), Southern Slavs (1.2%), Scandinavia (0.93%), France/Italy (0.81%), Iceland (0.64%), Germany (0.57%), Finland/Estonia (0.5%), England/Wales (0.23%) and Scotland (0.18%).[93]
Central Asian Y-DNA is much more common. Tat-C (haplogroup N1c) is a Y-chromosome lineage that originated in Siberia[94] and is thought to have spread to Northeastern Europe with male Uralic hunter-gatherer migrations occurring over the last 4000 years.[95] Today it's found in Northern and Northeastern Europe at varying frequencies: Finland (55%), Lithuania (47%), Lapland (42%), Estonia (37%), European Russia (14%), Ukraine (11%), Sweden (8%), Norway (6%), Poland (4%), Germany (3%), Slovakia (3%), Denmark (2%) and Belarus (2%).[96][97]
Though the above rather high frequencies are likely inflated due to genetic drift, which can affect Y-chromosomes,[98] a small but significant Central/East Asian genetic influence in Russians and Finns has been confirmed using population structure analysis.[99]
Inferences from ancient DNA
The genetic history of Europe has mostly been reconstructed from the modern populations of Europe, assuming genetic continuity. This is because of availability of data. However, a small number of ancient mtDNA analyses are available from both the historical and prehistorical periods. These have been summarised by Ellen Levy-Coffman in the Journal of Genetic Genealogy. There are some large differences in the frequencies of occurrence of the various haplogroups compared wth the modern population.
For example, mtDNA Haplogroup N1a, while presently rare (0.18%-0.3%), occurred in as many as 25% of Neolithic Europeans.[100][101] The cause of this reduction is unknown.
She concludes that the genetic profile of Europe has undergone significant transformation over time and that the modern population is not a living fossil of the ancient one. However, the very small sample sizes of the ancient DNA are a problem and more data is needed.
See also
- Archaeogenetics
- Archaeogenetics of the Near East
- Caucasoid
- European ethnic groups
- Human genetic variation
- Population genetics
- White people
- Genetic history of the British Isles
Notes
- ^ Underhill et al, 2001 state" A mesolithic population carrying Group III lineages with the M35/M215 mutation expanded northwards from sub-Saharan to north Africa and the Levant. The Levantine population of farmers that dispersed into Europe during and after the Neolithic carried these African Group III M35/M215 lineages, together with a cluster of Group VI lineages characterized by M172 and M201 mutations."
- ^ Tony Frudakis states "Underhill et al showed that the frequency of the YAP+ haplogroup commonly referred to as haplogroup E or (III) is relatively high (about 25%) in the Middle East and Mediterranean. This haplogroup E is the major haplogroup in Sub-Saharan Africa(75% of all Y chromosomes). Specifically, Europeans contain the E3b subhaplogroup, which was derived from haplogroup E in sub-Saharan Africa and currently is distributed along the North and East of Africa. Thus we refer to the affiliation as African, West African, or North/East African depending on the time frame we wish to refer to and/or our semantic preference. The West African affiliation for continental Europeans and Middle Eastern populations appears to be anthropoligically meaningful information that might help define partitions of substructure in Europe."
- ^ Halder et al state. "We observed patterns of apportionment similar to those described previously using sex and autosomal markers, such as European admixture for African Americans (14.3%) and Mexicans (43.2%), European (65.5%) and East Asian affiliation (27%) for South Asians, and low levels of African admixture (2.8–10.8%) mirroring the distribution of Y E3b haplogroups among various Eurasian populations.
- ^ Auton et al state."These two results therefore suggest that while the initial migrations into Europe came via the Middle East, at least some degree of subsequent gene flow has occurred directly from Africa" "Nonetheless, the haplotype sharing between Europe and the YRI are suggestive of gene flow from Africa, albeit from West Africa and not necessarily North Africa."
Footnotes
- ^ Cavalli-Sforza (1993, p. 39)
- ^ Cavalli-Sforza (1993, p. 51)
- ^ Arredi, Poloni & Tyler-Smith (2007, p. 390)
- ^ a b c d e f Rosser et al. (2000)
- ^ Milisauskas (2002, p. 58)
- ^ a b Richards et al (2000)
- ^ Semino (2000)
- ^ Barbujani & Bertorelle (2001:22-25)
- ^ a b Cavalli-Sforza (1997)
- ^ a b Cavalli-Sforza (1993, p. 90-93)
- ^ a b Bowcock; et al. (1991). "Drift, admixture, and selection in human evolution: A study with DNA polymorphisms".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - ^ Bauchet et al. (2007)
- ^ Cavalli-Sforza (1993)
- ^ Lao et al (2008)
- ^ Cavalli-Sforza (1993, p. 291-296)
- ^ Cavalli-Sforza (1993, p. 268)
- ^ Semino et al (2000)
- ^ F. Di Giacomo et al. (2004), Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe, Human Genetics 115(5):357-71.
- ^ Seldin MF, Shigeta R, Villoslada P; et al. (2006). "European population substructure: clustering of northern and southern populations". PLoS Genet. 2 (9): e143. doi:10.1371/journal.pgen.0020143. PMC 1564423. PMID 17044734.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Measuring European Population Stratification using Microarray Genotype Data
- ^ Lao 2008
- ^ Novembre J, Johnson T, Bryc K; et al. (2008). "Genes mirror geography within Europe". Nature. 456 (7218): 98–101. doi:10.1038/nature07331. PMID 18758442.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Lao O, Lu TT, Nothnagel M; et al. (2008). "Correlation between genetic and geographic structure in Europe". Curr. Biol. 18 (16): 1241–8. doi:10.1016/j.cub.2008.07.049. PMID 18691889. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "Estimating the Impact of Prehistoric Admixture on the Genome of Europeans - Dupanloup et al. 21 (7): 1361 - Molecular Biology and Evolution". Mbe.oxfordjournals.org. Retrieved 2009-07-19.
- ^ "Estimating the Impact of Prehistoric Admixture on the Genome of Europeans - Dupanloup et al. 21 (7): 1361 - Molecular Biology and Evolution". Mbe.oxfordjournals.org. doi:10.1093/molbev/msh135. Retrieved 2009-07-19.
- ^ Genetic Structure of Europeans: A View from the North–East, Nelis et al. 2009
- ^ Pair-wise Fst between European samples
- ^ Semino O, Passarino G, Oefner PJ; et al. (2000). "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective". Science. 290 (5494): 1155–9. PMID 11073453.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) Note: Haplogroup names are different in this article. For example: Haplogroup I is referred as M170 - ^ World haplogroup maps
- ^ Y-chromosome DNA Haplogroups
- ^ Pericic et al. (2005) Table 1 Summarized Percent Frequencies of R1b, R1a, I1b* (xM26), E3b1 and J2e
- ^ Kayser; et al. (2005), "Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis", Human Genetics, 117 (5): 428–443, doi:10.1007/s00439-005-1333-9, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) A copy can be found here [1]. - ^ Pericic et al. (2005)
- ^ Cinnioglu, Cengiz, et al., Excavating Y-Chromosome Haplotype Strata in Anatolia, Human Genetics vol. 114 (2004),
- ^ a b S. Rootsi et al. (2004), Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe, American Journal of Human Genetics 75 128–137
- ^ Kayser; et al. (2005), "Significant genetic differentiation between Poland and Germany follows present-day political borders, as revealed by Y-chromosome analysis", Human Genetics, 117 (5): 428–443, doi:10.1007/s00439-005-1333-9, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) A copy can be found here [2]. - ^ Human Y-Chromosome Variation in the Western Mediterranean Area: Implications for the Peopling of the Region
- ^ page-43
- ^ Peričic et al. (2005), "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations", Mol. Biol. Evol. 22 (10): 1964–75, doi:10.1093/molbev/msi185, PMID 15944443
- ^ Battaglia; et al. (2008), "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe", European Journal of Human Genetics, doi:10.1038/ejhg.2008.249
{{citation}}: Explicit use of et al. in:|author=(help) - ^ a b c d e Cruciani; et al. (2004), "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa", American Journal of Human Genetics, 74: 1014–1022, doi:10.1086/386294, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - ^ a b Semino et al. (2004)
- ^ World mtDNA haplogroup map
- ^ Mitochondrial DNA sequence variation in human populations, Oulu University Library (Finland)
- ^ a b Richards 2000
- ^ Klein RG (2003). "Paleoanthropology. Whither the Neanderthals?". Science. 299 (5612): 1525–7. doi:10.1126/science.1082025. PMID 12624250.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Milisauskas (2002, p. 59)
- ^ Semino 2000. * She refers to it as M 173
- ^ Wells 2001. Eurasian heartland
- ^ a b c Semino 2000
- ^ Torroni A, Bandelt HJ, Macaulay V; et al. (2001). "A signal, from human mtDNA, of postglacial recolonization in Europe". Am. J. Hum. Genet. 69 (4): 844–52. doi:10.1086/323485. PMC 1226069. PMID 11517423. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Pala; et al. (2009), "Mitochondrial Haplogroup U5b3: A Distant Echo of the Epipaleolithic in Italy and the Legacy of the Early Sardinians", The American Journal of Human Genetics, 84 (6): 814:821, doi:10.1016/j.ajhg.2009.05.004, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - ^ R Wells et al. The Eurasian Heartland: A continental perspective on Y-chromosome diversity
- ^ Semino et al. (2000)
- ^ Pericic. 2005
- ^ Cinnioglu et al. Excavating Y-chromosome haplotype strata in Anatolia. 2003
- ^ Pericic et al (2005) For discussion of eastern European dispersal of R1a1
- ^ Passarino et al (2001) For Scandinavian data
- ^ Semino (2000) general introduction
- ^ Milisauskas (2002, p. 1143, 150)
- ^ Milisauskas et al.:146)
- ^ Piazza, Alberto; Cavalli-Sforza, L. L.; Menozzi, Paolo (1994). The history and geography of human genes. Princeton, N.J: Princeton University Press. ISBN 0-691-08750-4.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Oppenheimer
- ^ Richards
- ^ Semino 2000. Here, the clade E-M35 is referred to as "Eu 4".
- ^ Cruciani 2007
- ^ Cruciani 2007
- ^ Underhill's study proposes that E1b1b seems to represent a late-Pleistocene migration from Africa to Europe over the Sinai Peninsula in Egypt, evidence for which does not show up in mitochondrial DNA. Y chromosome data show a signal for a separate late-Pleistocene migration from Africa to Europe via Sinai as evidenced through the distribution of haplogroup E3b lineages, which is not manifested in mtDNA haplogroup distributions.
- ^ See Bryan Sykes, The Seven Daughters of Eve, 1st American ed. (New York: Norton, 2001) for an entertaining account of how this consensus was reached. For historical reasons, in the 1980s mtDNA researchers believed that the Indo-European expansion was overwhelmingly a spread of technology and language, not of genes, while those who studied Y-chromosome lineages believed the opposite. Gradually the mtDNA researchers (Sykes) admitted more physical migration into their scenarios, while the Y folks (Peter Underhill) accepted more technology-copying. Eventually, both groups independently reached a 20% Neolithic - 80% Paleolithic ratio of genetic contribution to today's European population. The mtDNA vs. Y-chromosome discrepancy may be explained by noting that in such conquest-based migrations, a common pattern is of invading foreign males producing offspring with indigenous females, though significant numbers of females of the spreading culture could also arrive with post-conquest settlers. However, where migrations are essentially economic (as most migrations appear to be) it appears equally probable that male family members preceded females into new territory looking for opportunities.
- ^ "Pannonia and Upper Moesia. A History of the Middle Danube Provinces of the Roman Empire. Andras Mocsy. London and Boston, Routledge and Kegan Paul. ISBN 0-7100-7714-9
- ^ Alu insertion polymorphisms in the Balkans and the origins of the Aromuns. David Comas et al. Ann Hum Genet. 2004 Mar;68(Pt 2):120-7.
- ^ Paternal and maternal lineages in the Balkans show a homogeneous landscape over linguistic barriers, except for the isolated Aromuns. Bosh et al., Ann Hum Genet. 2006 Jul;70(Pt 4):459-87.
- ^ a b King; et al. (2007). "Africans in Yorkshire?".
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) Cite error: The named reference "king" was defined multiple times with different content (see the help page). - ^ Rosser et al (2000) Here, it is referred to as Hg 21
- ^ Flores et al. (2005), Beleza et al. (2006), Adams et al. (2008)
- ^ a b Capelli et al. (2009)
- ^ a b Cruciani (2004)
- ^ Gaetano et al. (2008)
- ^ Gonçalves et al. (2005)
- ^ "the contribution of North African populations is estimated to be around 6%. (...) The co-occurrence of the Berber E3b1b-M81 (2.12%) and of the Mid-Eastern J1-M267 (3.81%) Hgs together with the presence of E3b1a1-V12, E3b1a3-V22, E3b1a4-V65 (5.5%) support the hypothesis of intrusion of North African genes. (...) These Hgs are common in northern Africa and are observed only in Mediterranean Europe and together the presence of the E3b1b-M81 highlights the genetic relationships between northern Africa and Sicily. (...) Hg E3b1b-M81 network cluster confirms the genetic affinity between Sicily and North Africa.", Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome, Gaetano et al. 2008
- ^ Cruciani F, La Fratta R, Trombetta B; et al. (2007). "Tracing past human male movements in northern/eastern Africa and western Eurasia: new clues from Y-chromosomal haplogroups E-M78 and J-M12". Mol. Biol. Evol. 24 (6): 1300–11. doi:10.1093/molbev/msm049. PMID 17351267. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) See page 1307 - ^ "Haplogroup U6 is present at frequencies ranging from 0 to 7% in the various Iberian populations, with an average of 1.8%. Given that the frequency of U6 in NW Africa is 10%, the mtDNA contribution of NW Africa to Iberia can be estimated at 18%. This is larger than the contribution estimated with Y-chromosomal lineages (7%) (Bosch et al. 2001)."Joining the Pillars of Hercules: mtDNA Sequences Show Multidirectional Gene Flow in the Western Mediterranean (2003)
- ^ a b Pereira L, Cunha C, Alves C, Amorim A (2005). "African female heritage in Iberia: a reassessment of mtDNA lineage distribution in present times". Hum. Biol. 77 (2): 213–29. PMID 16201138.
Although the absolute value of observed U6 frequency in Iberia is low, it reveals a considerable North African female contribution, if we keep in mind that haplogroup U6 is not very common in North Africa itself and virtually absent in the rest of Europe. Indeed, because the range of variation in western North Africa is 4-28%, the estimated minimum input is 8.54%
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "Our results clearly reinforce, extend, and clarify the preliminary clues of an "important mtDNA contribution from northwest Africa into the Iberian Peninsula" (Côrte-Real et al., 1996; Rando et al., 1998; Flores et al., 2000a; Rocha et al., 1999)(...) Our own data allow us to make minimal estimates of the maternal African pre-Neolithic, Neolithic, and/or recent slave trade input into Iberia. For the former, we consider only the mean value of the U6 frequency in northern African populations, excluding Saharans, Tuareg, and Mauritanians (16%), as the pre-Neolithic frequency in that area, and the present frequency in the whole Iberian Peninsula (2.3%) as the result of the northwest African gene flow at that time. The value obtained (14%) could be as high as 35% using the data of Corte-Real et al. (1996), or 27% with our north Portugal sample." Mitochondrial DNA affinities at the Atlantic fringe of Europe (2003)
- ^ a b Malyarchuk; et al. (2008). "Reconstructing the phylogeny of African mitochondrial DNA lineages in Slavs". doi:10.1038/ejhg.2008.70.
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - ^ Frudakis, Tony (2007). "Apportionment of Autosomal Diversity with Continental Markers". Molecular photofitting. pp. page 326. ISBN 0120884925.
{{cite book}}:|pages=has extra text (help); External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ <cite journal|url=http://www.uma.pt/abrehm/v1.1/docs/downloads/pdfs/Goncalves_Y_Portugal_AnnHumGenet2005.pdf%7Clast=Goncalves%7Ctitle=Y-chromosome Lineages from Portugal, Madeira and A¸cores Record Elements of Sephardim and Berber Ancestry|year=2005}}
- ^ Pereira L, Prata MJ, Amorim A (2000). "Diversity of mtDNA lineages in Portugal: not a genetic edge of European variation". Ann. Hum. Genet. 64 (Pt 6): 491–506. PMID 11281213.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Gonzalez; et al. (2003). "Mitochondrial DNA Affinities at the Atlantic Fringe of Europe" (PDF). doi:10.1002/ajpa.10168.
{{cite journal}}: Cite journal requires|journal=(help); Explicit use of et al. in:|last=(help) - ^ Salas et al. (2004) The African Diaspora: Mitochondrial DNA and the Atlantic Slave Trade. Am J Hum Genet; 74(3): 454-465: "African types in Eurasia, unlike those in America, can therefore be attributed to gene flow from both eastern Africa -- perhaps partly via the Arab slave trade -- and western and southeastern Africa, more likely as a result of the Atlantic slave trade. [...] A contribution from the medieval Arab/Berber conquest of Iberia and Sicily is also possible. [...] However, there are also discernible western and southeastern African components to the (relatively few) mtDNAs of recent African ancestry within Europe, which are likely to be mainly attributable to the more recent Atlantic trade. Portuguese western, southwestern, and southeastern Africa were the main sources for the Atlantic slave trade to Europe."Therefore, it seems unlikely that the impact of mtDNAs of recent African ancestry on Iberia was the result of a single process; indeed, earlier gene flow may also have contributed some of the lineages.
- ^ Achilli et al. 2007, Malyarchuk et al. 2008, Gonzalez et al. (2003)
- ^ Flores et al. 2004, Brion et al. 2005, Brion et al. 2004, Rosser et al. 2000, Semino et al. 2004, DiGiacomo et al. 2003
- ^ Malyarchuk et al. 2008, Helgason et al. 2001
- ^ Lell JT, Sukernik RI, Starikovskaya YB; et al. (2002). "The dual origin and Siberian affinities of Native American Y chromosomes". Am. J. Hum. Genet. 70 (1): 192–206. doi:10.1086/338457. PMC 384887. PMID 11731934.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "The network of Tat-C and DYS7C haplotypes revealed that the ancestral Tat-C haplotype (7C[11-11-10-10]) was found only in southern Middle Siberia, indicating that this Y-chromosome lineage arose in that region. Moreover, the limited microsatellite diversity and resulting compact nature of the network indicates that the Tat-C lineage arose relatively recently (Zerjal et al. 1997). The absence of the Tat-C haplogroup in the Americas, with the exception of a single Navajo (Karafet et al. 1999), along with its high frequency in both northern Europe and northeastern Siberia, indicates that the Tat-C lineage was disseminated from central Asia by both westward and eastward male migrations, the eastward migration reaching Chukotka after the Bering Land Bridge was submerged. Both the M45 and Tat-C haplogroups have been found in Europe, indicating both ancient and recent central Asian influences. However, neither of these major Middle Siberian Y-chromosome lineages appears to have been greatly influenced by the paternal gene pool of Han Chinese or other East Asian populations (Su et al. 1999)."The Dual Origin and Siberian Affinities of Native American Y Chromosomes
- ^ Kittles RA, Perola M, Peltonen L; et al. (1998). "Dual origins of Finns revealed by Y chromosome haplotype variation". Am. J. Hum. Genet. 62 (5): 1171–9. doi:10.1086/301831. PMC 1377088. PMID 9545401. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Passarino G, Cavalleri GL, Lin AA, Cavalli-Sforza LL, Børresen-Dale AL, Underhill PA (2002). "Different genetic components in the Norwegian population revealed by the analysis of mtDNA and Y chromosome polymorphisms". Eur. J. Hum. Genet. 10 (9): 521–9. doi:10.1038/sj.ejhg.5200834. PMID 12173029. Retrieved 2009-07-22.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rosser et al. 2000: "Use of the Y chromosome to investigate human population histories (Jobling and Tyler-Smith 1995) is increasing as convenient polymorphic markers become available. However, the effective population size of this chromosome is one-quarter that of any autosome, and this means that it is particularly influenced by drift. Effective population size may be further reduced through the variance in the number of sons that a father has and perhaps by selective sweeps (Jobling and Tyler-Smith 2000). Conclusions about populations on the basis of this single locus must therefore be made with caution."
- ^ Rosenberg et al. 2002, Bauchet et al. 2007
- ^ Haak W, Forster P, Bramanti B; et al. (2005). "Ancient DNA from the first European farmers in 7500-year-old Neolithic sites". Science. 310 (5750): 1016–8. doi:10.1126/science.1118725. PMID 16284177.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Balter M (2005). "Evolution. Ancient DNA yields clues to the puzzle of European origins". Science. 310 (5750): 964–5. doi:10.1126/science.310.5750.964. PMID 16284157.
{{cite journal}}: Unknown parameter|month=ignored (help)
References
- Adams; et al. (2008), "The Genetic Legacy of Religious Diversity and Intolerance: Paternal Lineages of Christians, Jews, and Muslims in the Iberian Peninsula", The American Journal of Human Genetics, 83: 725, doi:10.1016/j.ajhg.2008.11.007, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Arredi; et al. (2004), "A Predominantly Neolithic Origin for Y-Chromosomal DNA Variation in North Africa", American Journal of Human Genetics, 75: 338–345, doi:10.1086/423147, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - Arredi, B; Poloni, E; Tyler-Smith, C (2007), "The Peopling of Europe", Anthropological Genetics: Theory, Methods and Applications, Cambridge University Press, ISBN 0521546974
- Barbujani, Guido; Bertorelle, Giorgio (2001), "Genetics and the Population History of Europe", PNAS, retrieved 2009-07-22
{{citation}}: Unknown parameter|vol=ignored (|volume=suggested) (help) - Battaglia; et al. (2008), "Y-chromosomal evidence of the cultural diffusion of agriculture in southeast Europe", European Journal of Human Genetics, doi:10.1038/ejhg.2008.249
{{citation}}: Explicit use of et al. in:|author=(help) - Bauchet; et al. (2007), "Measuring European Population Stratification using Microarray Genotype Data", The American Journal of Human Genetics, 80
{{citation}}: Explicit use of et al. in:|author=(help); External link in|title= - Beleza; et al. (2005), "Micro-Phylogeographic and Demographic History of Portuguese Male Lineages", Annals of Human Genetics, 70: 181–194, doi:10.1111/j.1529-8817.2005.00221.x
{{citation}}: Explicit use of et al. in:|last=(help) - Bosch; et al. (2001), "High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between north-western Africa and the Iberian Peninsula", Am J Hum Genet, 68: 1019–1029, doi:10.1086/319521, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - Capelli; et al. (2003), "A Y Chromosome Census of the British Isles", Current Biology, 13 (11): 979–84, doi:10.1016/S0960-9822(03)00373-7, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) also at [3] - Capelli; et al. (2009), "Moors and Saracens in Europe: estimating the medieval North African male legacy in southern Europe", European Journal of Human Genetics, doi:10.1038/ejhg.2008.258
{{citation}}: Explicit use of et al. in:|author=(help) - Menozzi, Paolo; Piazza, Alberto (1993), Cavalli-Sforza, Luigi L. (ed.), The History and Geography of Human Genes, Princeton University Press, ISBN 0691087504, retrieved 2009-07-22
{{citation}}: Missing|author1=(help) - Cavalli-Sforza (1997), "Genes, Peoples and Languages", PNAS, retrieved 2009-07-22
- Cruciani; et al. (2002), "A Back Migration from Asia to Sub-Saharan Africa Is Supported by High-Resolution Analysis of Human Y-Chromosome Haplotypes" (PDF), American Journal of Human Genetics, 70: 1197–1214, doi:10.1086/340257, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - Cruciani; et al. (2004), "Phylogeographic Analysis of Haplogroup E3b (E-M215) Y Chromosomes Reveals Multiple Migratory Events Within and Out Of Africa", American Journal of Human Genetics, 74: 1014–1022, doi:10.1086/386294, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - Cruciani; et al. (2006), "Molecular Dissection of the Y Chromosome Haplogroup E-M78 (E3b1a): A Posteriori Evaluation of a Microsatellite-Network-Based Approach Through Six New Biallelic Markers" (PDF), Human Mutation, 27: 831, doi:10.1002/humu.9445, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) - Cruciani; et al. (2007), "Tracing Past Human Male Movements in Northern/Eastern Africa and Western Eurasia: New Clues from Y-Chromosomal Haplogroups E-M78 and J-M12", Molecular Biology and Evolution, 24: 1300–1311, doi:10.1093/molbev/msm049, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help) Also see Supplementary Data. - Di Gaetano; et al. (2008), "Differential Greek and northern African migrations to Sicily are supported by genetic evidence from the Y chromosome", European Journal of Human Genetics
{{citation}}: Explicit use of et al. in:|author=(help) - Flores; et al. (2004), "Reduced genetic structure of the Iberian peninsula revealed by Y-chromosome analysis: implications for population demography" (PDF), European Journal of Human Genetics, 12: 855–863, doi:10.1038/sj.ejhg.5201225, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Francalacci; et al. (2003), "Peopling of Three Mediterranean Islands (Corsica, Sardinia, and Sicily) Inferred by Y-Chromosome Biallelic Variability", American Journal of Physical Anthropology, 121: 270–279, doi:10.1002/ajpa.10265
{{citation}}: Explicit use of et al. in:|author=(help) - Gonçalves; et al. (2005), "Y-chromosome Lineages from Portugal, Madeira and Açores Record Elements of Sephardim and Berber Ancestry", Annals of Human Genetics, 69: 443–454, doi:10.1046/j.1529-8817.2005.00161.x
{{citation}}: Explicit use of et al. in:|last=(help) Also http://wysinger.homestead.com/portugal.pdf - King and Underhill (2002), "Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages", Antiquity, 76: 707–14
- Lao; et al. (2008), "Correlation between genetic and geographic structure in Europe", Current Biology, 18 (16): 1241–8, doi:10.1016/j.cub.2008.07.049, PMID 18691889, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help); Unknown parameter|month=ignored (help) - Passarino; et al. (2001), "The 49a,f haplotype 11 is a new marker of the EU19 lineage that traces migrations from northern regions of the black sea", Hum. Immunol., vol. 62, no. 9, pp. 922–932, doi:10.1016/S0198-8859(01)00291-9
{{citation}}: Explicit use of et al. in:|author=(help). - Pericic; et al. (2005). "High-resolution phylogenetic analysis of southeastern Europe traces major episodes of paternal gene flow among Slavic populations". Mol. Biol. Evol. 22 (10): 1964–75. doi:10.1093/molbev/msi185. PMID 15944443. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help) - Milisauskas, Sarunas (2002), European Prehistory: a survey, Birkhauser, ISBN 0306467933
- Richards; et al. (2000), "Tracing European founder lineages in the Near Eastern mtDNA pool", American Journal of Human Genetics, 67 (5): 1251–76, PMC 1288566, PMID 11032788, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|last=(help); Unknown parameter|month=ignored (help) - Rosser; et al. (2000), "Y-Chromosomal Diversity in Europe Is Clinal and Influenced Primarily by Geography, Rather than by Language", American Journal of Human Genetics, 67: 1526–1543., doi:10.1086/316890
{{citation}}: Explicit use of et al. in:|last=(help) - Scozzari; et al. (2001), "Human Y-Chromosome Variation in the Western Mediterranean Area: Implications for the Peopling of the Region" (PDF), Human Immunology, 62: 871–884, doi:10.1016/S0198-8859(01)00286-5, retrieved 2009-07-22
{{citation}}: Cite has empty unknown parameter:|unused_data=(help); Explicit use of et al. in:|author=(help) - Semino; et al. (2000). "The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective" (PDF). Science. 290 (5494): 1155–9. doi:10.1126/science.290.5494.1155. PMID 11073453. Retrieved 2009-07-22.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help). - Semino; et al. (2002), "Ethiopians and Khoisan share the deepest clades of the human Y-chromosome phylogeny" (PDF), Am J Hum Genet, vol. 70, pp. 265–8, doi:10.1086/338306, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Semino; et al. (2004), "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area", American Journal of Human Genetics, vol. 74, pp. 1023–1034, doi:10.1086/386295, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Sykes, Bryan (2006), Blood of the Isles: Exploring the Genetic Roots of Our Tribal History, Bantam, ISBN 0593056523, retrieved 2009-07-22
- Underhill; et al. (2000), "Y chromosome sequence variation and the history of human populations", Nat Genet, vol. 26, pp. 358–361, doi:10.1038/81685, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Underhill; et al. (2001), "The phylogeography of Y chromosome binary haplotypes and the origins of modern human populations" (PDF), Ann Hum Genet, vol. 65, pp. 43–62, doi:10.1046/j.1469-1809.2001.6510043.x, retrieved 2009-07-22
{{citation}}: Explicit use of et al. in:|author=(help) - Underhill (2002), Bellwood and Renfrew (ed.), Inference of Neolithic Population Histories using Y-chromosome Haplotypes, Cambridge: McDonald Institute for Archaeological Research, ISBN 1-902937-20-1
{{citation}}: Unknown parameter|booktitle=ignored (help) - Underhill and Kivisild (2007), "Use of Y Chromosome and Mitochondrial DNA Population Structure in Tracing Human Migrations", Annu. Rev. Genet., 41: 539–64, doi:10.1146/annurev.genet.41.110306.130407
- Perlès C, Monthel G ( 2001) The Early Neolithic in Greece: The First Farming Communities in Europe. Cambridge University Press, Cambridge.
- Runnels C (2003) The origins of the Greek Neolithic: a personal view, in Ammerman and Biagi (2003 eds).
External links
- International Society of Genetic Genealogy - 2009 tree of haplogroup R
- Journal of Genetic Genealogy - We Are Not Our Ancestors: Evidence for Discontinuity between Prehistoric and Modern Europeans
- Atlas of the Human Journey
- World Haplogroups Maps
- Origins of Europeans
- Genetic Structure of Human Populations.
- Haplotype R1b
- Haplogroup R1b
