PETase
| PETase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
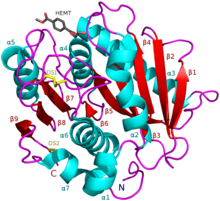 I. sakaiensis PETase (A0A0K8P6T7) in complex with HEMT, a PET analogue (PDB: 5XH3). | |||||||||
| Identifiers | |||||||||
| EC no. | 3.1.1.101 | ||||||||
| Alt. names | PET hydrolase, poly(ethylene terephthalate) hydrolase | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
PETases are an esterase class of enzymes that catalyze the hydrolysis of polyethylene terephthalate (PET) plastic to monomeric mono-2-hydroxyethyl terephthalate (MHET). The idealized chemical reaction is (where n is the number of monomers in the polymer chain):[1]
- (ethylene terephthalate)n + H2O → (ethylene terephthalate)n-1 + MHET
Trace amount of the PET breaks down to bis(2-hydroxyethyl) terephthalate (BHET). PETases can also break down PEF-plastic (polyethylene-2,5-furandicarboxylate), which is a bioderived PET replacement, into the analogous MHEF. PETases can't catalyze the hydrolysis of aliphatic polyesters like polybutylene succinate or polylactic acid.[2]
Non-enzymatic natural degradation of PET will take hundreds of years, but PETases can degrade PET in matter of days.[3]
History
The first PETase was discovered in 2016 from Ideonella sakaiensis strain 201-F6 bacteria found from sludge samples collected close to a Japanese PET bottle recycling site.[1][4] Other types of PET degrading hydrolases have been known before this discovery.[2] These include hydrolases such as lipases, esterases, and cutinases.[5] Discoveries of polyester degrading enzymes date at least as far back as 1975 (α-chymotrypsin)[6] and 1977 (lipase) for example.[7]
PET plastic was put into widespread use in the 1970s and it has been suggested that PETases in bacteria evolved only recently.[2] PETase may have had past enzymatic activity associated with degradation of a waxy coating on plants.[8]
Structure
As of April 2019, there were 17 known three-dimensional crystal structures of PETases: 6QGC, 6ILX, 6ILW, 5YFE, 6EQD, 6EQE, 6EQF, 6EQG, 6EQH, 6ANE, 5XJH, 5YNS, 5XFY, 5XFZ, 5XG0, 5XH2 and 5XH3.
PETase exhibits shared qualities with both lipases and cutinases in that it possesses an α/β-hydrolase fold; although, the active-site cleft observed in PETase is more open than in cutinases.[2] The Ideonella sakaiensis PETase is similar to dienelactone hydrolase, according to Pfam. According to ESTHER, it falls into the Polyesterase-lipase-cutinase family.
There are approximately 69 PETase-like enzymes comprising a variety of diverse organisms, and there are two classifications of these enzymes including type I and type II. It is suggested that 57 enzymes fall into the type I category whereas the rest fall into the type II group, including the PETase enzyme found in the Ideonella sakaiensis. Within all 69 PETase-like enzymes, there exists the same three residues within the active site, suggesting that the catalytic mechanism is the same in all forms of PETase-like enzymes.[9]
-
Surface of the PETase double mutant (R103G and S131A) with HEMT (1-(2-hydroxyethyl) 4-methyl terephthalate) bound to its active site. HEMT is an analogue of MHET, and has an additional methanol esterified to it. PDBID: 5XH3.
-
Ribbon diagram of PETase with three residues Ser160, Asp206, and His237. The catalytic triad is represented by cyan-colored sticks. The active site is shown in orange to represent stimulation by a 2-HE(MHET)4 molecule.[9]
Mutations
In 2018 scientists from the University of Portsmouth with the collaboration of the National Renewable Energy Laboratory of the United States Department of Energy developed a mutant of this PETase that degrades PET faster than the one in its natural state. In this study it was also shown that PETases can degrade polyethylene 2,5-furandicarboxylate (PEF).[2][10]
Biological pathway

In I. sakaiensis, the resultant MHET is further broken down by the action of MHETase enzyme to terephthalic acid and ethylene glycol.[1] Laboratory experiments showed that chimeric proteins that artificially link a MHETase and a PETase outperform similar mixtures of free enzymes.[12]
See also
- Organisms breaking down plastic
- Galleria mellonella, a caterpillar that can digest polyethylene.
- Aspergillus tubingensis, a fungus that can digest polyurethane.
- Pestalotiopsis microspora, an endophytic fungus species able to break down polyurethane.
- cutinase, an esterase enzyme of similar geometric shape
References
- ^ a b c Yoshida S, Hiraga K, Takehana T, Taniguchi I, Yamaji H, Maeda Y, et al. (March 2016). "A bacterium that degrades and assimilates poly(ethylene terephthalate)". Science. 351 (6278): 1196–9. doi:10.1126/science.aad6359. PMID 26965627. S2CID 31146235.
- "Discovery of a Bacterium that Degrades and Assimilates Poly(ethylene terephthalate) could Serve as a Degradation and/or Fermentation Platform for Biological Recycling of PET Waste Products" (PDF). Kyoto Institute of Technology (Press release). 2016-03-30.
- ^ a b c d e Austin HP, Allen MD, Donohoe BS, Rorrer NA, Kearns FL, Silveira RL, et al. (May 2018). "Characterization and engineering of a plastic-degrading aromatic polyesterase". Proceedings of the National Academy of Sciences of the United States of America. 115 (19): E4350–E4357. doi:10.1073/pnas.1718804115. PMC 5948967. PMID 29666242.
- ^ Dockrill, Peter. "Scientists Have Accidentally Created a Mutant Enzyme That Eats Plastic Waste". ScienceAlert. Retrieved 2018-11-27.
- ^ Tanasupawat S, Takehana T, Yoshida S, Hiraga K, Oda K (August 2016). "Ideonella sakaiensis sp. nov., isolated from a microbial consortium that degrades poly(ethylene terephthalate)". International Journal of Systematic and Evolutionary Microbiology. 66 (8): 2813–8. doi:10.1099/ijsem.0.001058. PMID 27045688.
- ^ Han X, Liu W, Huang JW, Ma J, Zheng Y, Ko TP, et al. (December 2017). "Structural insight into catalytic mechanism of PET hydrolase". Nature Communications. 8 (1): 2106. doi:10.1038/s41467-017-02255-z. PMC 5727383. PMID 29235460.
- ^ Tabushi I, Yamada H, Matsuzaki H, Furukawa J (August 1975). "Polyester readily hydrolyzable by chymotrypsin". Journal of Polymer Science: Polymer Letters Edition. 13 (8): 447–450. doi:10.1002/pol.1975.130130801.
- ^ Tokiwa Y, Suzuki T (November 1977). "Hydrolysis of polyesters by lipases". Nature. 270 (5632): 76–8. doi:10.1038/270076a0. PMID 927523. S2CID 4145159.
- ^ "Lab 'Accident' Becomes Mutant Enzyme That Devours Plastic". Live Science. Retrieved 2018-11-27.
- ^ a b Joo S, Cho IJ, Seo H, Son HF, Sagong HY, Shin TJ, et al. (January 2018). "Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation". Nature Communications. 9 (1): 382. doi:10.1038/s41467-018-02881-1. PMC 5785972. PMID 29374183.
- ^ Carrington, Damian. "New super-enzyme eats plastic bottles six times faster". The Guardian.
- ^ Allison Chan (2016). "The Future of Bacteria Cleaning Our Plastic Waste" (PDF).
- ^ Knott BC, Erickson E, Allen MD, Gado JE, Graham R, Kearns FL, et al. (Oct 2020). "Characterization and engineering of a two-enzyme system for plastics depolymerization". Proc Natl Acad Sci U S A. 117 (41): 25476–25485. doi:10.1073/pnas.2006753117. PMC 7568301. PMID 32989159.


![Ribbon diagram of PETase with three residues Ser160, Asp206, and His237. The catalytic triad is represented by cyan-colored sticks. The active site is shown in orange to represent stimulation by a 2-HE(MHET)4 molecule.[9]](http://upload.wikimedia.org/wikipedia/commons/thumb/d/db/PETase_active_site.png/739px-PETase_active_site.png)