Haplogroup L3: Difference between revisions
Skllagyook (talk | contribs) |
Skllagyook (talk | contribs) |
||
| Line 20: | Line 20: | ||
An [[Asia]]n center of origin and dispersal for haplogroup L3 has also been hypothesized based on the similar coalescence dates of L3 and its Eurasian-distributed [[Haplogroup M (mtDNA)|M]] and [[Haplogroup N (mtDNA)|N]] derivative clades (~71 kya), the distant location in [[Southeast Asia]] of the oldest subclades of M and N, and the comparable age of the paternal haplogroup [[Haplogroup DE (Y-DNA)|DE]]. According to this hypothesis, after an initial Out-of-Africa migration of early [[anatomically modern human]]s around 125 kya, fully modern human L3-carrying females are thus proposed to have back-migrated from the maternal haplogroup's place of origin in Eurasia around 70 kya along with males bearing the paternal haplogroup [[Haplogroup E-M96|E]], which is also proposed to have originated in Eurasia. These new Eurasian lineages are then suggested to have largely replaced the old autochthonous male and female North-East African lineages.<ref name="AsiaThesis1">{{cite journal|vauthors=Cabrera VM, Marrero P, Abu-Amero KK, Larruga JM|date=June 2018|title=Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago|journal=BMC Evolutionary Biology|volume=18|issue=1|pages=98|doi=10.1186/s12862-018-1211-4|pmc=6009813|pmid=29921229}}</ref> |
An [[Asia]]n center of origin and dispersal for haplogroup L3 has also been hypothesized based on the similar coalescence dates of L3 and its Eurasian-distributed [[Haplogroup M (mtDNA)|M]] and [[Haplogroup N (mtDNA)|N]] derivative clades (~71 kya), the distant location in [[Southeast Asia]] of the oldest subclades of M and N, and the comparable age of the paternal haplogroup [[Haplogroup DE (Y-DNA)|DE]]. According to this hypothesis, after an initial Out-of-Africa migration of early [[anatomically modern human]]s around 125 kya, fully modern human L3-carrying females are thus proposed to have back-migrated from the maternal haplogroup's place of origin in Eurasia around 70 kya along with males bearing the paternal haplogroup [[Haplogroup E-M96|E]], which is also proposed to have originated in Eurasia. These new Eurasian lineages are then suggested to have largely replaced the old autochthonous male and female North-East African lineages.<ref name="AsiaThesis1">{{cite journal|vauthors=Cabrera VM, Marrero P, Abu-Amero KK, Larruga JM|date=June 2018|title=Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago|journal=BMC Evolutionary Biology|volume=18|issue=1|pages=98|doi=10.1186/s12862-018-1211-4|pmc=6009813|pmid=29921229}}</ref> |
||
According to other research, though earlier migrations out of Africa of anatomically modern humans occurred, current Eurasian populations descend from a later migration from Africa dated between 50,000 and 70,000 years ago.<ref name="Vai">{{cite journal|vauthors=Vai S, Sarno S, Lari M,Luiselli D, Manzi G, Gallinaro M, Mataich S, Hübner A, Modi A, Pilli E, Tafuri MA, Caramelli D, di Lernia S|date=March 2019|title=Ancestral mitochondrial N lineage from the Neolithic ‘green’ Sahara|journal=Sci Rep|doi=10.1038/s41598-019-39802-1|pmc= 6401177|pmid=30837540}}</ref><ref name="ReferenceC">{{cite journal | vauthors = Haber M, Jones AL, Connel BA, Asan, Arciero E, Huanming Y, Thomas MG, Xue Y, Tyler-Smith C | title = A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa | journal = Genetics | pages = genetics.302368.2019 | date = June 2019 | pmid = 31196864 | doi = 10.1534/genetics.119.302368 }}</ref> |
According to other research, though earlier migrations out of Africa of anatomically modern humans occurred, current Eurasian populations descend from a later migration from Africa dated between 50,000 and 70,000 years ago.<ref name=Posth2016>{{harvp|Posth et al.|2016}}.<br>See also [http://dienekes.blogspot.nl/2016/02/mtdna-from-55-hunter-gatherers-across.html ''mtDNA from 55 hunter-gatherers across 35,000 years in Europe''], Dienekes' Anthroplogy Bog.</ref><ref name="Vai">{{cite journal|vauthors=Vai S, Sarno S, Lari M,Luiselli D, Manzi G, Gallinaro M, Mataich S, Hübner A, Modi A, Pilli E, Tafuri MA, Caramelli D, di Lernia S|date=March 2019|title=Ancestral mitochondrial N lineage from the Neolithic ‘green’ Sahara|journal=Sci Rep|doi=10.1038/s41598-019-39802-1|pmc= 6401177|pmid=30837540}}</ref><ref name="ReferenceC">{{cite journal | vauthors = Haber M, Jones AL, Connel BA, Asan, Arciero E, Huanming Y, Thomas MG, Xue Y, Tyler-Smith C | title = A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa | journal = Genetics | pages = genetics.302368.2019 | date = June 2019 | pmid = 31196864 | doi = 10.1534/genetics.119.302368 }}</ref> |
||
A study (by Vai et al. 2019) on a newly discovered basal branch of maternal haplogroup N found in early Neolithic North African remains, suggests that haplogroup L3 may have originated in Africa between 60-70,000 years ago, with haplogroup N diverging from it soon after (between about 50,000-65,000 years ago) either in Arabia or possibly North Africa, and haplogroup M originating in the Middle East.<ref name="Vai">{{cite journal|vauthors=Vai S, Sarno S, Lari M,Luiselli D, Manzi G, Gallinaro M, Mataich S, Hübner A, Modi A, Pilli E, Tafuri MA, Caramelli D, di Lernia S|date=March 2019|title=Ancestral mitochondrial N lineage from the Neolithic ‘green’ Sahara|journal=Sci Rep|doi=10.1038/s41598-019-39802-1|pmc= 6401177|pmid=30837540}}</ref> |
A study (by Vai et al. 2019) on a newly discovered basal branch of maternal haplogroup N found in early Neolithic North African remains, suggests that haplogroup L3 may have originated in Africa between 60-70,000 years ago, with haplogroup N diverging from it soon after (between about 50,000-65,000 years ago) either in Arabia or possibly North Africa, and haplogroup M originating in the Middle East.<ref name="Vai">{{cite journal|vauthors=Vai S, Sarno S, Lari M,Luiselli D, Manzi G, Gallinaro M, Mataich S, Hübner A, Modi A, Pilli E, Tafuri MA, Caramelli D, di Lernia S|date=March 2019|title=Ancestral mitochondrial N lineage from the Neolithic ‘green’ Sahara|journal=Sci Rep|doi=10.1038/s41598-019-39802-1|pmc= 6401177|pmid=30837540}}</ref> |
||
Revision as of 04:33, 4 September 2019
| Haplogroup L3 | |
|---|---|
| Possible time of origin | 60,000–70,000 YBP[1] |
| Possible place of origin | East Africa[1][2] or Asia[3] |
| Ancestor | L3'4 |
| Descendants | L3a, L3b'f, L3c'd, L3e'i'k'x, L3h, M, N |
| Defining mutations | 769, 1018, 16311[4] |
Haplogroup L3 is a human mitochondrial DNA (mtDNA) haplogroup. The clade has played a pivotal role in the prehistory of the human species. It represents the most common parent maternal lineage of all people outside Africa, and for many people within the Africa continent as well.[5][6]
Origin

Haplogroup L3's exact place of origin is uncertain. According to the Recent African origin of modern humans (Out-of-Africa) theory, the clade is believed to have arisen and dispersed from East Africa, initially thought to have occurred between 84,000 and 104,000 years ago.[7] An analysis of 369 complete African L3 sequences placed the maximal date of the clade's expansion at around 70,000 years ago. This virtually rules out a successful exit out of Africa before 74,000, the date of the Toba volcanic super-eruption in Sumatra, thus making an origin and expansion around 70,000 years ago most likely.[1] The Time to Most Recent Common Ancestor for the L3 lineage has also recently been estimated to date to between 58,900 and 70,200 years ago, around the time of and associated with the Out-of-Africa expansion of the ancestors of non-African modern humans from Eastern Africa into Eurasia around 70,000 years ago, and also with a similar expansion within Africa around that time also from the East of the continent.[1]
Phylogenetically, haplogroup L6 and L4 are the closest to L3 out of the L lineages. Both L6 and L4 are primarily distributed and have their greatest diversity in Eastern Africa. L4'6 (L3'4'6) has a TMRCA of 114,288 years before present while L3'4 link at 95,240 ybp in the middle paleolithic.[5]

a: Exit of the L3 precursor to Eurasia. b: Return to Africa and expansion to Asia of basal L3 lineages with subsequent differentiation in both continents.
An Asian center of origin and dispersal for haplogroup L3 has also been hypothesized based on the similar coalescence dates of L3 and its Eurasian-distributed M and N derivative clades (~71 kya), the distant location in Southeast Asia of the oldest subclades of M and N, and the comparable age of the paternal haplogroup DE. According to this hypothesis, after an initial Out-of-Africa migration of early anatomically modern humans around 125 kya, fully modern human L3-carrying females are thus proposed to have back-migrated from the maternal haplogroup's place of origin in Eurasia around 70 kya along with males bearing the paternal haplogroup E, which is also proposed to have originated in Eurasia. These new Eurasian lineages are then suggested to have largely replaced the old autochthonous male and female North-East African lineages.[3]
According to other research, though earlier migrations out of Africa of anatomically modern humans occurred, current Eurasian populations descend from a later migration from Africa dated between 50,000 and 70,000 years ago.[8][2][9] A study (by Vai et al. 2019) on a newly discovered basal branch of maternal haplogroup N found in early Neolithic North African remains, suggests that haplogroup L3 may have originated in Africa between 60-70,000 years ago, with haplogroup N diverging from it soon after (between about 50,000-65,000 years ago) either in Arabia or possibly North Africa, and haplogroup M originating in the Middle East.[2]
Distribution
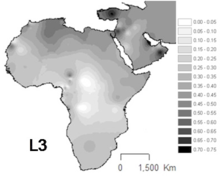
L3 is common in Northeast Africa and some other parts of East Africa,[10] in contrast to others parts of Africa where the haplogroups L1 and L2 represent around two thirds of mtDNA lineages.[11] L3 sublineages are also frequent in the Arabian peninsula.
L3 is subdivided into several clades, two of which spawned the macrohaplogroups M and N that are today carried by most people outside Africa.[11] There is at least one relatively deep non-M, non-N clade of L3 outside Africa, L3f1b6, which is found at a frequency of 1% in Asturias, Spain. It diverged from African L3 lineages at least 10,000 years ago.[12]
According to Maca-Meyer et al. (2001), "L3 is more related to Eurasian haplogroups than to the most divergent African clusters L1 and L2".[13] L3 is the haplogroup from which all modern humans outside Africa derive.[14] However, there is a greater diversity of major L3 branches within Africa than outside of it, the two major non-African branches being the L3 offshoots M and N.
Subclade distribution

L3 has seven equidistant descendants: L3a, L3b'f, L3c'd, L3e'i'k'x, L3h, M, N. Five are African, while two are associated with the Out of Africa event.
- N – Eurasia and parts of Africa due to back-migration.[6][15]
- M – Asia, the Mediterranean Basin, and parts of Africa due to back-migration.[6][15]
- L3a – East Africa.[5][6] Moderate to high frequencies found among the Sanye, Samburu, Iraqw, Yaaku, El-Molo and other minor indigenous populations from the East African Rift Valley. It is infrequent to nonexistent in Sudan and the Sahel zone.[16]
- L3b'f
- L3b – Spread from East Africa in the upper paleolithic to West-Central Africa. Some subclades spread from Central Africa to East Africa with the Bantu migration.[6]
- L3f – Northeast Africa, Sahel, Arabian peninsula, Iberia. Gaalien,[19] Beja[19]
- L3f1
- L3f2 – Primarily distributed in East Africa.[6] Also found in North Africa and Central Africa.[18]
- L3f3 – Spread from Eastern Africa to Chad and the Sahel around 8-9 ka.[6] Found in the Chad Basin.[18][20]
- L3c'd
- L3c – Extremely rare lineage with only two samples found so far in Eastern Africa and the Near East.[6]
- L3d – Spread from East Africa in the upper paleolithic to Central Africa. Some subclades spread to East Africa with the Bantu migration.[6] Found among the Fulani,[5] Chadians,[5] Ethiopians,[21] Akan people,[22] Mozambique,[21] Yemenites,[21] Egyptians, Berbers[23]
- L3e'i'k'x
- L3e – Spread from East Africa in the upper paleolithic to West-Central Africa. It is the most common L3 sub-clade in Bantu-speaking populations.[24] L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, Afro-Brazilians and Caribbeans[25]
- L3e1 – Spread from West-Central Africa to Southwest Africa with the Bantu migration. Found in Angola (6.8%).[26] Mozambique, Sudanese and Kikuyu from Kenya as well as in Yemen and among the Akan people[22]
- L3e5 – Originated in the Chad Basin. Found in Algeria,[27] as well as Burkina Faso, Nigeria, South Tunisia, South Morocco and Egypt[28]
- L3i Almost exclusively found in East Africa.[6]
- L3k – Rare haplogroup primarily found in North Africa and the Sahel.[6][18]
- L3x – Almost exclusively found in East Africa.[6] Found among Ethiopian Oromos,[21] Egyptians[Note 2][29]
- L3e – Spread from East Africa in the upper paleolithic to West-Central Africa. It is the most common L3 sub-clade in Bantu-speaking populations.[24] L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, Afro-Brazilians and Caribbeans[25]
- L3h – Almost exclusively found in East Africa.[6]
- L3h1 – Primarily found in East Africa with branches of L3h1b1 sporadically found in the Sahel and North Africa.[17][18]
- L3h2 – Found in Northeast Africa and Socotra. Split from other L3h branches as early as 65-69 ka during the middle paleolithic.[17][18]
Ancient and historic samples
Haplogroup L3 has been observed in an ancient fossil belonging to the Pre-Pottery Neolithic B culture.[30] L3x2a was observed in a 4,500 year old hunter-gather excavated in Mota, Ethiopia, with the ancient fossil found to be most closely related to modern Southwest Ethiopian populations.[31][32] Haplogroup L3 has also been found among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the Pre-Ptolemaic/late New Kingdom and Ptolemaic periods.[33] Additionally, haplogroup L3 has been observed in ancient Guanche fossils excavated in Gran Canaria and Tenerife on the Canary Islands, which have been radiocarbon-dated to between the 7th and 11th centuries CE. All of the clade-bearing individuals were inhumed at the Gran Canaria site, with most of these specimens found to belong to the L3b1a subclade (3/4; 75%). The Guanche skeletons also bore an autochthonous Maghrebi genomic component that peaks among modern Berbers, which suggests that they originated from ancestral Berber populations inhabiting northwestern Affoundnat a high ncy[34]
A variety of L3 have been uncovered in ancient remains associated with the Pastoral Neolithic and Pastoral Iron Age of East Africa.[35]
| Culture | Genetic cluster or affinity | Country | Site | Date | Maternal Haplogroup | Paternal Haplogroup | Source |
| Early pastoral | PN | Kenya | Prettejohn's Gully (GsJi11) | 4060-3860 | L3f1b | - | Prendergast 2019 |
| Pastoral Neolithic | PN | Kenya | Cole’s Burial (GrJj5a) | 3350-3180 | L3i2 | E-V32 | Prendergast 2019 |
| Pastoral Neolithic or Elmenteitan | PN | Kenya | Rigo Cave (GrJh3) | 2710-2380 | L3f | E-M293 | Prendergast 2019 |
| Pastoral Neolithic | PN | Kenya | Naishi Rockshelter | 2750-2500 | L3x1a | E-V1515 (prob. E-M293) | Prendergast 2019 |
| Pastoral Neolithic | PN | Tanzania | Gishimangeda Cave | 2490-2350 | L3x1 | - | Prendergast 2019 |
| Pastoral Neolithic | PN | Kenya | Naivasha Burial Site | 2350-2210 | L3h1a1 | E-M293 | Prendergast 2019 |
| Pastoral Neolithic | PN | Kenya | Naivasha Burial Site | 2320-2150 | L3x1a | E-M293 | Prendergast 2019 |
| Pastoral Neolithic | PN | Tanzania | Gishimangeda Cave | 2150-2020 | L3i2 | E-M293 | Prendergast 2019 |
| Pastoral Neolithic or Elmenteitan | PN | Kenya | Njoro River Cave II | 2110-1930 | L3h1a2a1 | - | Prendergast 2019 |
| Pastoral Neolithic | N/A | Tanzania | Gishimangeda Cave | 2000-1900 | L3h1a2a1 | - | Prendergast 2019 |
| Pastoral Neolithic | PN | Kenya | Ol Kalou | 1810-1620 | L3d1d | E-M293 | Prendergast 2019 |
| Pastoral Iron Age | PIA | Kenya | Kisima Farm, C4 | 1060-940 | L3h1a1 | E-M75 (excl. M98) | Prendergast 2019 |
| Pastoral Iron Age | PIA | Kenya | Emurua Ole Polos (GvJh122) | 420-160 | L3h1a1 | E-M293 | Prendergast 2019 |
| Pastoral Iron Age | PN outlier | Kenya | Kokurmatakore | N/A | L3a2a | E-M35 (not E-M293) | Prendergast 2019 |
Tree
This phylogenetic tree of haplogroup L3 subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[4] and subsequent published research.[36]
Most Recent Common Ancestor (MRCA)
- L1-6
- L2-6
- L2'3'4'6
- L3'4'6
- L3'4
- L3
- L3a
- L3a1
- L3a1a
- L3a1b
- L3a2
- L3a2a
- L3a1
- L3b'f
- L3b
- L3b1
- L3b1a
- L3b1a1
- L3b1a2
- L3b1a3
- L3b1a4
- L3b1a5
- L3b1a5a
- L3b1a6
- L3b1a7
- L3b1a7
- L3b1a8
- L3b1a9
- L3b1a9a
- L3b1a10
- L3b1a11
- L3b1b
- L3b1b1
- L3b1a
- L3b2
- L3b2a
- L3b2a
- L3b3
- L3b1
- L3f
- L3f1
- L3f1a
- L3f1a1
- L3f1b
- L3f1b1
- L3f1b2
- L3f1b2a
- L3f1b3
- L3f1b4
- L3f1b4a
- L3f1b4a1
- L3f1b4b
- L3f1b4c
- L3f1b4a
- L3f1b5
- L3f1a
- L3f2
- L3f2a
- L3f2b
- L3f3
- L3f3a
- L3f3b
- L3f1
- L3b
- L3c'd
- L3c
- L3d
- L3d1-5
- L3d1
- L3d1a
- L3d1a1
- L3d1a1a
- L3d1a1
- L3d1b
- L3d1b1
- L3d1c
- L3d1d
- L3d1a
- 199
- L3d2
- L3d5
- L3d3
- L3d3a
- L3d4
- L3d5
- L3d1
- L3d1-5
- L3e'i'k'x
- L3e
- L3e1
- L3e1a
- L3e1a1
- L3e1a1a
- 152
- L3e1a2
- L3e1a3
- L3e1a1
- L3e1b
- L3e1c
- L3e1d
- L3e1e
- L3e1a
- L3e2
- L3e2a
- L3e2a1
- L3e2a1a
- L3e2a1b
- L3e2a1b1
- L3e2a1
- L3e2b
- L3e2b1
- L3e2b1a
- L3e2b2
- L3e2b3
- L3e2b1
- L3e2a
- L3e3'4'5
- L3e3'4
- L3e3
- L3e3a
- L3e3b
- L3e3b1
- L3e4
- L3e3
- L3e5
- L3e3'4
- L3e1
- L3i
- L3i1
- L3i1a
- L3i1b
- L3i2
- L3i1
- L3k
- L3k1
- L3x
- L3x1
- L3x1a
- L3x1a1
- L3x1a2
- L3x1b
- L3x1a
- L3x2
- L3x2a
- L3x2a1
- L3x2a1a
- L3x2a1
- L3x2b
- L3x2a
- L3x1
- L3e
- L3h
- L3h1
- L3h1a
- L3h1a1
- L3h1a2
- L3h1a2a
- L3h1a2b
- L3h1b
- L3h1b1
- L3h1b1a
- L3h1b1a1
- L3h1b1a
- L3h1b2
- L3h1b1
- L3h1a
- L3h2
- L3h1
- M
- N
- L3a
- L3
- L3'4
- L3'4'6
- L2'3'4'6
- L2-6
See also
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- ^ a b c d Soares, P; Alshamali, F; Pereira, J. B; Fernandes, V; Silva, N. M; Afonso, C; Costa, M. D; Musilova, E; MacAulay, V; Richards, M. B; Cerny, V; Pereira, L (2011). "The Expansion of mtDNA Haplogroup L3 within and out of Africa". Molecular Biology and Evolution. 29 (3): 915–927. doi:10.1093/molbev/msr245. PMID 22096215.
- ^ a b c Vai S, Sarno S, Lari M, Luiselli D, Manzi G, Gallinaro M, Mataich S, Hübner A, Modi A, Pilli E, Tafuri MA, Caramelli D, di Lernia S (March 2019). "Ancestral mitochondrial N lineage from the Neolithic 'green' Sahara". Sci Rep. doi:10.1038/s41598-019-39802-1. PMC 6401177. PMID 30837540.
- ^ a b c Cabrera VM, Marrero P, Abu-Amero KK, Larruga JM (June 2018). "Carriers of mitochondrial DNA macrohaplogroup L3 basal lineages migrated back to Africa from Asia around 70,000 years ago". BMC Evolutionary Biology. 18 (1): 98. doi:10.1186/s12862-018-1211-4. PMC 6009813. PMID 29921229.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Van Oven, Mannis; Kayser, Manfred (2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–94. doi:10.1002/humu.20921. PMID 18853457.
- ^ a b c d e Behar, Doron M.; Villems, Richard; Soodyall, Himla; Blue-Smith, Jason; Pereira, Luisa; Metspalu, Ene; Scozzari, Rosaria; Makkan, Heeran; et al. (2008). "The Dawn of Human Matrilineal Diversity" (PDF). The American Journal of Human Genetics. 82 (5): 1130–40. doi:10.1016/j.ajhg.2008.04.002. PMC 2427203. PMID 18439549.
- ^ a b c d e f g h i j k l m n o p q r s Soares, P.; Alshamali, F.; Pereira, J. B.; Fernandes, V.; Silva, N. M.; Afonso, C.; Costa, M. D.; Musilova, E.; Macaulay, V. (2011-11-16). "The Expansion of mtDNA Haplogroup L3 within and out of Africa". Molecular Biology and Evolution. 29 (3): 915–927. CiteSeerX 10.1.1.923.345. doi:10.1093/molbev/msr245. ISSN 0737-4038. PMID 22096215.
- ^ Gonder, M. K.; Mortensen, H. M.; Reed, F. A.; De Sousa, A.; Tishkoff, S. A. (2006). "Whole-mtDNA Genome Sequence Analysis of Ancient African Lineages". Molecular Biology and Evolution. 24 (3): 757–68. doi:10.1093/molbev/msl209. PMID 17194802.
- ^ Posth et al. (2016).
See also mtDNA from 55 hunter-gatherers across 35,000 years in Europe, Dienekes' Anthroplogy Bog. - ^ Haber M, Jones AL, Connel BA, Asan, Arciero E, Huanming Y, Thomas MG, Xue Y, Tyler-Smith C (June 2019). "A Rare Deep-Rooting D0 African Y-chromosomal Haplogroup and its Implications for the Expansion of Modern Humans Out of Africa". Genetics: genetics.302368.2019. doi:10.1534/genetics.119.302368. PMID 31196864.
- ^ Martina Kujanova; Luisa Pereira; Veronica Fernandes; Joana B. Pereira; Viktor Cerny (2009). "Near Eastern Neolithic Genetic Input in a Small Oasis of the Egyptian Western Desert". American Journal of Physical Anthropology. 140 (2): 336–46. doi:10.1002/ajpa.21078. PMID 19425100.
- ^ a b Wallace, D; Brown, MD; Lott, MT (1999). "Mitochondrial DNA variation in human evolution and disease". Gene. 238 (1): 211–30. doi:10.1016/S0378-1119(99)00295-4. PMID 10570998.
- ^ a b Pardiñas, AF; Martínez, JL; Roca, A; García-Vazquez, E; López, B (2014). "Over the sands and far away: Interpreting an Iberian mitochondrial lineage with ancient Western African origins". Am. J. Hum. Biol. 26 (6): 777–83. doi:10.1002/ajhb.22601. PMID 25130626.
- ^ Maca-Meyer, Nicole; González, Ana M; Larruga, José M; Flores, Carlos; Cabrera, Vicente M (2001). "Major genomic mitochondrial lineages delineate early human expansions". BMC Genetics. 2: 13. doi:10.1186/1471-2156-2-13. PMC 55343. PMID 11553319.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Archived copy". Archived from the original on 2011-07-08. Retrieved 2009-03-09.
{{cite web}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help)CS1 maint: archived copy as title (link) - ^ a b "Carriers of mitochondrial DNA macrohaplogroup L3 basic lineages migrated back to Africa from Asia around 70,000 years ago". 2017. bioRxiv 233502.
{{cite bioRxiv}}: Check|biorxiv=value (help); Cite uses deprecated parameter|authors=(help) - ^ Boru, Hirbo, Jibril (2011). "Complex Genetic History of East African Human Populations" (PDF): 118.
{{cite journal}}: Cite journal requires|journal=(help)CS1 maint: multiple names: authors list (link) - ^ a b c d e Soares, P.; Alshamali, F.; Pereira, J. B.; Fernandes, V.; Silva, N. M.; Afonso, C.; Costa, M. D.; Musilova, E.; Macaulay, V.; Richards, M. B.; Cerny, V.; Pereira, L. (16 November 2011). "The Expansion of mtDNA Haplogroup L3 within and out of Africa". Molecular Biology and Evolution. 29 (3): 915–927. doi:10.1093/molbev/msr245. PMID 22096215.
{{cite journal}}: Invalid|ref=harv(help) Supplementary data at [1] - ^ a b c d e f g h i Hernández, Candela L; Soares, Pedro; Dugoujon, Jean M; Novelletto, Andrea; Rodríguez, Juan N; Rito, Teresa; Oliveira, Marisa; Melhaoui, Mohammed; Baali, Abdellatif; Pereira, Luisa; Calderón, Rosario (2015). "Early Holocenic and Historic mtDNA African Signatures in the Iberian Peninsula: The Andalusian Region as a Paradigm". PLOS ONE. 10 (10): e0139784. Bibcode:2015PLoSO..1039784H. doi:10.1371/journal.pone.0139784. PMC 4624789. PMID 26509580.
{{cite journal}}: CS1 maint: unflagged free DOI (link) Supplementary data doi:10.1371/journal.pone.0139784.s006. - ^ a b Mohamed, Hisham Yousif Hassan. "Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan" (PDF). University of Khartoum. Retrieved 14 June 2016.
- ^ Černý, Viktor; Fernandes, Verónica; Costa, Marta D; Hájek, Martin; Mulligan, Connie J; Pereira, Luísa (2009). "Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup". BMC Evolutionary Biology. 9: 63. doi:10.1186/1471-2148-9-63. PMC 2680838. PMID 19309521.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d Kivisild, T; Reidla, M; Metspalu, E; Rosa, A; Brehm, A; Pennarun, E; Parik, J; Geberhiwot, T; et al. (2004). "Ethiopian Mitochondrial DNA Heritage: Tracking Gene Flow Across and Around the Gate of Tears". The American Journal of Human Genetics. 75 (5): 752–70. doi:10.1086/425161. PMC 1182106. PMID 15457403.
- ^ a b Fendt, Liane; Röck, Alexander; Zimmermann, Bettina; Bodner, Martin; Thye, Thorsten; Tschentscher, Frank; Owusu-Dabo, Ellis; Göbel, Tanja M.K.; Schneider, Peter M.; Parson, Walther (2012). "MtDNA diversity of Ghana: a forensic and phylogeographic view". Forensic Science International: Genetics. 6 (2): 244–49. doi:10.1016/j.fsigen.2011.05.011. PMC 3314991. PMID 21723214.
- ^ Sheet1 – PLOS Pathogens
- ^ Anderson, S. 2006, Phylogenetic and phylogeographic analysis of African mitochondrial DNA variation. Archived 2011-09-10 at the Wayback Machine
- ^ Bandelt, HJ; Alves-Silva, J; Guimarães, PE; Santos, MS; Brehm, A; Pereira, L; Coppa, A; Larruga, JM; et al. (2001). "Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade". Annals of Human Genetics. 65 (Pt 6): 549–63. doi:10.1046/j.1469-1809.2001.6560549.x. PMID 11851985.
- ^ Plaza, Stéphanie; Salas, Antonio; Calafell, Francesc; Corte-Real, Francisco; Bertranpetit, Jaume; Carracedo, Ángel; Comas, David (2004). "Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola". Human Genetics. 115 (5): 439–47. doi:10.1007/s00439-004-1164-0. PMID 15340834.
- ^ Asmahan Bekada; Lara R. Arauna; Tahria Deba; Francesc Calafell; Soraya Benhamamouch; David Comas (September 24, 2015). "Genetic Heterogeneity in Algerian Human Populations". PLoS ONE. 10 (9): e0138453. Bibcode:2015PLoSO..1038453B. doi:10.1371/journal.pone.0138453. PMC 4581715. PMID 26402429.
{{cite journal}}: CS1 maint: unflagged free DOI (link); S5 Table - ^ Fadhlaoui-Zid, K.; Plaza, S.; Calafell, F.; Ben Amor, M.; Comas, D.; Bennamar, A.; Gaaied, El (2004). "Mitochondrial DNA Heterogeneity in Tunisian Berbers". Annals of Human Genetics. 68 (Pt 3): 222–33. doi:10.1046/j.1529-8817.2004.00096.x. PMID 15180702.
- ^ Stevanovitch, A.; Gilles, A.; Bouzaid, E.; Kefi, R.; Paris, F.; Gayraud, R. P.; Spadoni, J. L.; El-Chenawi, F.; Beraud-Colomb, E. (2004). "Mitochondrial DNA Sequence Diversity in a Sedentary Population from Egypt". Annals of Human Genetics. 68 (Pt 1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
- ^ Fernández, Eva; et al. (2014). "Ancient DNA analysis of 8000 BC near eastern farmers supports an early neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands". PLoS Genetics. 10 (6): e1004401. doi:10.1371/journal.pgen.1004401. PMC 4046922. PMID 24901650.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ See supplementary materials from Llorente, M. Gallego; Jones, E. R.; Eriksson, A.; Siska, V.; Arthur, K. W.; Arthur, J. W.; Curtis, M. C.; Stock, J. T.; Coltorti, M.; Pieruccini, P.; Stretton, S.; Brock, F.; Higham, T.; Park, Y.; Hofreiter, M.; Bradley, D. G.; Bhak, J.; Pinhasi, R.; Manica, A. (13 November 2015). "Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa". Science. 350 (6262): 820–822. doi:10.1126/science.aad2879. PMID 26449472.
- ^ Llorente, M. Gallego; Jones, E. R.; Eriksson, A.; Siska, V.; Arthur, K. W.; Arthur, J. W.; Curtis, M. C.; Stock, J. T.; Coltorti, M. (2015-11-13). "Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa". Science. 350 (6262): 820–822. Bibcode:2015Sci...350..820L. doi:10.1126/science.aad2879. PMID 26449472.
- ^ Schuenemann, Verena J.; et al. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications. 8: 15694. Bibcode:2017NatCo...815694S. doi:10.1038/ncomms15694. PMC 5459999. PMID 28556824.
- ^ Rodrı́guez-Varela; et al. (2017). "Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans". Current Biology. 27 (1–7): 3396–3402.e5. Bibcode:1996CBio....6.1213A. doi:10.1016/j.cub.2017.09.059. PMID 29107554. Retrieved 27 October 2017.
- ^ Prendergast, Mary E.; Lipson, Mark; Sawchuk, Elizabeth A.; Olalde, Iñigo; Ogola, Christine A.; Rohland, Nadin; Sirak, Kendra A.; Adamski, Nicole; Bernardos, Rebecca (2019-05-30). "Ancient DNA reveals a multistep spread of the first herders into sub-Saharan Africa". Science: eaaw6275. doi:10.1126/science.aaw6275. ISSN 0036-8075.
- ^ "PhyloTree.org | tree | L3". phylotree.org. Retrieved 2018-06-25.
Notes
- ^ See Supplemental_TreeUpdatedOctober.xls found under the Supplementary data of Soares et al. 2011 harvnb error: multiple targets (3×): CITEREFSoaresAlshamaliPereiraFernandes2011 (help)
- ^ GUR46 on table 1. is a mtDNA haplogroup L3x2a.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- Haplogroup L3
- Mannis van Oven's PhyloTree.org – mtDNA subtree L3
- Spread of Haplogroup L3, from National Geographic
