Haplogroup L3: Difference between revisions
Soupforone (talk | contribs) No edit summary |
Rescuing 5 sources and tagging 0 as dead. #IABot (v1.6) |
||
| Line 20: | Line 20: | ||
L3 is sub-divided into several clades, two of which spawned the macro-haplogroups [[Haplogroup M (mtDNA)|M]] and [[Haplogroup N (mtDNA)|N]] that are today carried by most people outside of Africa.<ref>{{cite journal |last1=Wallace |first1=D |last2=Brown |first2=MD |last3=Lott |first3=MT |title=Mitochondrial DNA variation in human evolution and disease |journal=Gene |volume=238 |issue=1 |pages=211–30 |year=1999 |pmid=10570998 |doi=10.1016/S0378-1119(99)00295-4}}</ref> There is at least one relatively deep non-M, non-N clade of L3 outside Africa, L3f1b6, found at 1% in Asturias Spain, which diverged from African L3 lineages at least 10,000 years ago.<ref name="Pardiñas2014">{{cite journal|last1=Pardiñas|first1=AF|last2=Martínez|first2=JL|last3=Roca|first3= A|last4=García-Vazquez|first4= E|last5=López|first5=B|title=Over the sands and far away: Interpreting an Iberian mitochondrial lineage with ancient Western African origins|journal=Am J Hum Biol.|volume=26|issue=6|pages=777–83|year=2014|pmid= 25130626|doi=10.1002/ajhb.22601}}</ref> |
L3 is sub-divided into several clades, two of which spawned the macro-haplogroups [[Haplogroup M (mtDNA)|M]] and [[Haplogroup N (mtDNA)|N]] that are today carried by most people outside of Africa.<ref>{{cite journal |last1=Wallace |first1=D |last2=Brown |first2=MD |last3=Lott |first3=MT |title=Mitochondrial DNA variation in human evolution and disease |journal=Gene |volume=238 |issue=1 |pages=211–30 |year=1999 |pmid=10570998 |doi=10.1016/S0378-1119(99)00295-4}}</ref> There is at least one relatively deep non-M, non-N clade of L3 outside Africa, L3f1b6, found at 1% in Asturias Spain, which diverged from African L3 lineages at least 10,000 years ago.<ref name="Pardiñas2014">{{cite journal|last1=Pardiñas|first1=AF|last2=Martínez|first2=JL|last3=Roca|first3= A|last4=García-Vazquez|first4= E|last5=López|first5=B|title=Over the sands and far away: Interpreting an Iberian mitochondrial lineage with ancient Western African origins|journal=Am J Hum Biol.|volume=26|issue=6|pages=777–83|year=2014|pmid= 25130626|doi=10.1002/ajhb.22601}}</ref> |
||
According to Maca-Meyer et al. (2001), "L3 is more related to [[Eurasia]]n haplogroups than to the most divergent [[Ethnic groups of Africa|African]] clusters [[Haplogroup L1 (mtDNA)|L1]] and [[Haplogroup L2 (mtDNA)|L2]]".<ref>{{cite journal |last1=Maca-Meyer |first1=Nicole |last2=González |first2=Ana M |last3=Larruga |first3=José M |last4=Flores |first4=Carlos |last5=Cabrera |first5=Vicente M |title=Major genomic mitochondrial lineages delineate early human expansions |journal=BMC Genetics |volume=2 |page=13 |year=2001 |pmid=11553319 |pmc=55343 |doi=10.1186/1471-2156-2-13}}</ref> L3 is the haplogroup from which all modern humans outside of Africa derive.<ref>https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step3</ref> |
According to Maca-Meyer et al. (2001), "L3 is more related to [[Eurasia]]n haplogroups than to the most divergent [[Ethnic groups of Africa|African]] clusters [[Haplogroup L1 (mtDNA)|L1]] and [[Haplogroup L2 (mtDNA)|L2]]".<ref>{{cite journal |last1=Maca-Meyer |first1=Nicole |last2=González |first2=Ana M |last3=Larruga |first3=José M |last4=Flores |first4=Carlos |last5=Cabrera |first5=Vicente M |title=Major genomic mitochondrial lineages delineate early human expansions |journal=BMC Genetics |volume=2 |page=13 |year=2001 |pmid=11553319 |pmc=55343 |doi=10.1186/1471-2156-2-13}}</ref> L3 is the haplogroup from which all modern humans outside of Africa derive.<ref>{{cite web |url=https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step3 |title=Archived copy |accessdate=2009-03-09 |deadurl=yes |archiveurl=https://web.archive.org/web/20110708112911/https://www.cambridgedna.com/genealogy-dna-ancient-migrations-slideshow.php?view=step3 |archivedate=2011-07-08 |df= }}</ref> |
||
Basal haplogroup L3* is found among [[Nubian people|Nubians]] (13.8%),<ref name="Hassan2009">{{cite web|last1=Mohamed|first1=Hisham Yousif Hassan|title=Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan|url=http://khartoumspace.uofk.edu/bitstream/handle/123456789/6376/Genetic%20Patterns%20of%20Y-chromosome%20and%20Mitochondrial.pdf?sequence=1|publisher=University of Khartoum|accessdate=14 June 2016}}</ref> as well as [[Socotra|Socotri]] (4.3%).<ref name="Cerny2009">{{cite journal|last1=Černý, Viktor|title=Out of Arabia – the settlement of island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity|journal=American journal of Physical Anthropology|date=2009|volume=138|issue=4|pages=439–47|url=http://ychrom.invint.net/upload/iblock/f30/Cerny%202009%20Out%20of%20ArabiarusThe%20Settlement%20of%20Island%20Soqotra%20as%20Revealed%20by%20Mitochondrial%20and%20Y.pdf|accessdate=14 June 2016|display-authors=etal|doi=10.1002/ajpa.20960|pmid=19012329}}</ref> |
Basal haplogroup L3* is found among [[Nubian people|Nubians]] (13.8%),<ref name="Hassan2009">{{cite web|last1=Mohamed|first1=Hisham Yousif Hassan|title=Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan|url=http://khartoumspace.uofk.edu/bitstream/handle/123456789/6376/Genetic%20Patterns%20of%20Y-chromosome%20and%20Mitochondrial.pdf?sequence=1|publisher=University of Khartoum|accessdate=14 June 2016}}</ref> as well as [[Socotra|Socotri]] (4.3%).<ref name="Cerny2009">{{cite journal|last1=Černý, Viktor|title=Out of Arabia – the settlement of island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity|journal=American journal of Physical Anthropology|date=2009|volume=138|issue=4|pages=439–47|url=http://ychrom.invint.net/upload/iblock/f30/Cerny%202009%20Out%20of%20ArabiarusThe%20Settlement%20of%20Island%20Soqotra%20as%20Revealed%20by%20Mitochondrial%20and%20Y.pdf|accessdate=14 June 2016|display-authors=etal|doi=10.1002/ajpa.20960|pmid=19012329|deadurl=yes|archiveurl=https://web.archive.org/web/20161006125303/http://ychrom.invint.net/upload/iblock/f30/Cerny%202009%20Out%20of%20ArabiarusThe%20Settlement%20of%20Island%20Soqotra%20as%20Revealed%20by%20Mitochondrial%20and%20Y.pdf|archivedate=6 October 2016|df=}}</ref> |
||
Haplogroup L3 has been observed in an ancient fossil belonging to the [[Pre-Pottery Neolithic B]] culture.<ref>{{cite journal|last1=Fernández, Eva, et al.|title=Ancient DNA analysis of 8000 BC near eastern farmers supports an early neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands|journal=PLoS Genetics|date=2014|volume=10|issue=6|pmc=4046922|pmid=24901650|doi=10.1371/journal.pgen.1004401|page=e1004401}}</ref> The clade has also been found among [[ancient Egypt]]ian mummies excavated at the [[Abusir|Abusir el-Meleq]] archaeological site in Middle Egypt, which date from the Pre-[[Ptolemaic Kingdom|Ptolemaic]]/late [[New Kingdom of Egypt|New Kingdom]] and Ptolemaic periods.<ref>{{cite journal|last1=Schuenemann, Verena J., et al.|title=Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods|journal=Nature Communications|date=2017|volume=8|page=15694|pmid=28556824|pmc=5459999|doi=10.1038/ncomms15694|bibcode=2017NatCo...815694S}}</ref> Additionally, haplogroup L3 has been observed in ancient [[Guanches|Guanche]] fossils excavated in [[Gran Canaria]] and [[Tenerife]] on the [[Canary Islands]], which have been radiocarbon-dated to between the 7th and 11th centuries CE. All of the clade-bearing individuals were inhumed at the Gran Canaria site, with most of these specimens found to belong to the L3b1a subclade (3/4; 75%).<ref>{{cite journal|last1=Rodrı́guez-Varela et al.|title=Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans|journal=Current Biology|date=2017|volume=27|issue=1–7|url=http://www.cell.com/current-biology/fulltext/S0960-9822(17)31257-5|accessdate=27 October 2017}}</ref> |
Haplogroup L3 has been observed in an ancient fossil belonging to the [[Pre-Pottery Neolithic B]] culture.<ref>{{cite journal|last1=Fernández, Eva, et al.|title=Ancient DNA analysis of 8000 BC near eastern farmers supports an early neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands|journal=PLoS Genetics|date=2014|volume=10|issue=6|pmc=4046922|pmid=24901650|doi=10.1371/journal.pgen.1004401|page=e1004401}}</ref> The clade has also been found among [[ancient Egypt]]ian mummies excavated at the [[Abusir|Abusir el-Meleq]] archaeological site in Middle Egypt, which date from the Pre-[[Ptolemaic Kingdom|Ptolemaic]]/late [[New Kingdom of Egypt|New Kingdom]] and Ptolemaic periods.<ref>{{cite journal|last1=Schuenemann, Verena J., et al.|title=Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods|journal=Nature Communications|date=2017|volume=8|page=15694|pmid=28556824|pmc=5459999|doi=10.1038/ncomms15694|bibcode=2017NatCo...815694S}}</ref> Additionally, haplogroup L3 has been observed in ancient [[Guanches|Guanche]] fossils excavated in [[Gran Canaria]] and [[Tenerife]] on the [[Canary Islands]], which have been radiocarbon-dated to between the 7th and 11th centuries CE. All of the clade-bearing individuals were inhumed at the Gran Canaria site, with most of these specimens found to belong to the L3b1a subclade (3/4; 75%).<ref>{{cite journal|last1=Rodrı́guez-Varela et al.|title=Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans|journal=Current Biology|date=2017|volume=27|issue=1–7|url=http://www.cell.com/current-biology/fulltext/S0960-9822(17)31257-5|accessdate=27 October 2017}}</ref> |
||
| Line 39: | Line 39: | ||
** '''L3d''' – Wide distribution in Africa. Among the [[Fulani]],<ref name="Behar" /> [[Chad]]ians,<ref name="Behar" /> [[Ethiopian]]s,<ref name="Kivisild">{{cite journal |last1=Kivisild |first1=T |last2=Reidla |first2=M |last3=Metspalu |first3=E |last4=Rosa |first4=A |last5=Brehm |first5=A |last6=Pennarun |first6=E |last7=Parik |first7=J |last8=Geberhiwot |first8=T |last9=Usanga |first9=E |last10=Villems |first10=Richard |title=Ethiopian Mitochondrial DNA Heritage: Tracking Gene Flow Across and Around the Gate of Tears |journal=The American Journal of Human Genetics |volume=75 |issue=5 |pages=752–70 |year=2004 |doi=10.1086/425161 |pmid=15457403 |pmc=1182106 |display-authors=8 }}</ref> [[Akan people]],<ref name="Fendt">{{cite journal|url = http://www.sciencedirect.com/science/article/pii/S1872497311001268#sec0085 | doi=10.1016/j.fsigen.2011.05.011 | volume=6 | issue=2 | title=MtDNA diversity of Ghana: a forensic and phylogeographic view | year=2012 | journal=Forensic Science International: Genetics | pages=244–49 | last1 = Fendt | first1 = Liane | last2 = Röck | first2 = Alexander | last3 = Zimmermann | first3 = Bettina | last4 = Bodner | first4 = Martin | last5 = Thye | first5 = Thorsten | last6 = Tschentscher | first6 = Frank | last7 = Owusu-Dabo | first7 = Ellis | last8 = Göbel | first8 = Tanja M.K. | last9 = Schneider | first9 = Peter M. | last10 = Parson | first10 = Walther}}</ref> [[Mozambique]],<ref name="Kivisild"/> [[Yemen]]<ref name="Kivisild" /> [[Egyptians]], [[Berbers]]<ref>Sheet1 – PLOS Pathogens</ref> |
** '''L3d''' – Wide distribution in Africa. Among the [[Fulani]],<ref name="Behar" /> [[Chad]]ians,<ref name="Behar" /> [[Ethiopian]]s,<ref name="Kivisild">{{cite journal |last1=Kivisild |first1=T |last2=Reidla |first2=M |last3=Metspalu |first3=E |last4=Rosa |first4=A |last5=Brehm |first5=A |last6=Pennarun |first6=E |last7=Parik |first7=J |last8=Geberhiwot |first8=T |last9=Usanga |first9=E |last10=Villems |first10=Richard |title=Ethiopian Mitochondrial DNA Heritage: Tracking Gene Flow Across and Around the Gate of Tears |journal=The American Journal of Human Genetics |volume=75 |issue=5 |pages=752–70 |year=2004 |doi=10.1086/425161 |pmid=15457403 |pmc=1182106 |display-authors=8 }}</ref> [[Akan people]],<ref name="Fendt">{{cite journal|url = http://www.sciencedirect.com/science/article/pii/S1872497311001268#sec0085 | doi=10.1016/j.fsigen.2011.05.011 | volume=6 | issue=2 | title=MtDNA diversity of Ghana: a forensic and phylogeographic view | year=2012 | journal=Forensic Science International: Genetics | pages=244–49 | last1 = Fendt | first1 = Liane | last2 = Röck | first2 = Alexander | last3 = Zimmermann | first3 = Bettina | last4 = Bodner | first4 = Martin | last5 = Thye | first5 = Thorsten | last6 = Tschentscher | first6 = Frank | last7 = Owusu-Dabo | first7 = Ellis | last8 = Göbel | first8 = Tanja M.K. | last9 = Schneider | first9 = Peter M. | last10 = Parson | first10 = Walther}}</ref> [[Mozambique]],<ref name="Kivisild"/> [[Yemen]]<ref name="Kivisild" /> [[Egyptians]], [[Berbers]]<ref>Sheet1 – PLOS Pathogens</ref> |
||
* L3e'i'k'x |
* L3e'i'k'x |
||
** '''L3e''' – West-Central Africa. It is the most common L3 sub-clade in [[Bantu languages|Bantu]]-speaking populations.<ref>Anderson, S. 2006, [http://www.biolog-e.leeds.ac.uk/Biolog-e/uploads/SadieAnderson-Mann.pdf Phylogenetic and phylogeographic analysis of African mitochondrial DNA variation.]</ref> L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, [[Afro-Brazilian]]s and [[Caribbean]]s<ref name="Bandelt">{{cite journal |last1=Bandelt |first1=HJ |last2=Alves-Silva |first2=J |last3=Guimarães |first3=PE |last4=Santos |first4=MS |last5=Brehm |first5=A |last6=Pereira |first6=L |last7=Coppa |first7=A |last8=Larruga |first8=JM |last9=Rengo |first9=C |last10=Scozzari |first10=R |last11=Torroni |first11=A |last12=Prata |first12=M. J. |last13=Amorim |first13=A |last14=Prado |first14=V. F. |last15=Pena |first15=S. D. |title=Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade |journal=Annals of Human Genetics |volume=65 |issue=Pt 6 |pages=549–63 |year=2001 |pmid=11851985 |doi=10.1017/S0003480001008892 |display-authors=8 |doi-broken-date=2017-01-16 }}</ref> |
** '''L3e''' – West-Central Africa. It is the most common L3 sub-clade in [[Bantu languages|Bantu]]-speaking populations.<ref>Anderson, S. 2006, [http://www.biolog-e.leeds.ac.uk/Biolog-e/uploads/SadieAnderson-Mann.pdf Phylogenetic and phylogeographic analysis of African mitochondrial DNA variation.] {{webarchive|url=https://web.archive.org/web/20110910155424/http://www.biolog-e.leeds.ac.uk/Biolog-e/uploads/SadieAnderson-Mann.pdf |date=2011-09-10 }}</ref> L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, [[Afro-Brazilian]]s and [[Caribbean]]s<ref name="Bandelt">{{cite journal |last1=Bandelt |first1=HJ |last2=Alves-Silva |first2=J |last3=Guimarães |first3=PE |last4=Santos |first4=MS |last5=Brehm |first5=A |last6=Pereira |first6=L |last7=Coppa |first7=A |last8=Larruga |first8=JM |last9=Rengo |first9=C |last10=Scozzari |first10=R |last11=Torroni |first11=A |last12=Prata |first12=M. J. |last13=Amorim |first13=A |last14=Prado |first14=V. F. |last15=Pena |first15=S. D. |title=Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade |journal=Annals of Human Genetics |volume=65 |issue=Pt 6 |pages=549–63 |year=2001 |pmid=11851985 |doi=10.1017/S0003480001008892 |display-authors=8 |doi-broken-date=2017-01-16 }}</ref> |
||
*** L3e1 – Central Africa origin and is found in [[Algeria]], [[Cameroon]], [[Angola]],<ref name="Plaza2004">{{cite journal |last1=Plaza |first1=Stéphanie |last2=Salas |first2=Antonio |last3=Calafell |first3=Francesc |last4=Corte-Real |first4=Francisco |last5=Bertranpetit |first5=Jaume |last6=Carracedo |first6=Ángel |last7=Comas |first7=David |title=Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola |journal=Human Genetics |volume=115 |issue=5 |pages=439–47 |year=2004 |pmid=15340834 |doi=10.1007/s00439-004-1164-0}}</ref> Mozambique, [[Sudanese]] and [[Kikuyu people|Kikuyu]] from [[Kenya]] as well as in Yemen and among the [[Akan people]]<ref name="Fendt"/> |
*** L3e1 – Central Africa origin and is found in [[Algeria]], [[Cameroon]], [[Angola]],<ref name="Plaza2004">{{cite journal |last1=Plaza |first1=Stéphanie |last2=Salas |first2=Antonio |last3=Calafell |first3=Francesc |last4=Corte-Real |first4=Francisco |last5=Bertranpetit |first5=Jaume |last6=Carracedo |first6=Ángel |last7=Comas |first7=David |title=Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola |journal=Human Genetics |volume=115 |issue=5 |pages=439–47 |year=2004 |pmid=15340834 |doi=10.1007/s00439-004-1164-0}}</ref> Mozambique, [[Sudanese]] and [[Kikuyu people|Kikuyu]] from [[Kenya]] as well as in Yemen and among the [[Akan people]]<ref name="Fendt"/> |
||
*** L3e5 – Originated in the Chad Basin. Found in [[Algeria]],<ref name="Bekada2015">{{cite journal|author1=Asmahan Bekada |author2=Lara R. Arauna |author3=Tahria Deba |author4=Francesc Calafell |author5=Soraya Benhamamouch |author6=David Comas |title=Genetic Heterogeneity in Algerian Human Populations|journal=PLoS ONE|date=September 24, 2015|volume=10|issue=9|doi=10.1371/journal.pone.0138453|url=http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0138453|accessdate=13 May 2016 |pmid=26402429 |pmc=4581715 |pages=e0138453|bibcode=2015PLoSO..1038453B }}; [http://journals.plos.org/plosone/article/asset?unique&id=info:doi/10.1371/journal.pone.0138453.s007 S5 Table]</ref> as well as [[Burkina Faso]], [[Nigeria]], South [[Tunisia]], South [[Morocco]] and [[Egypt]]<ref name= "Fadhlaoui-Zid">{{cite journal |last1=Fadhlaoui-Zid |first1=K. |last2=Plaza |first2=S. |last3=Calafell |first3=F. |last4=Ben Amor |first4=M. |last5=Comas |first5=D. |last6=Bennamar |first6=A. |last7=Gaaied |first7=El |title=Mitochondrial DNA Heterogeneity in Tunisian Berbers |journal=Annals of Human Genetics |volume=68 |issue=Pt 3 |pages=222–33 |year=2004 |pmid=15180702 |doi=10.1046/j.1529-8817.2004.00096.x}}</ref> |
*** L3e5 – Originated in the Chad Basin. Found in [[Algeria]],<ref name="Bekada2015">{{cite journal|author1=Asmahan Bekada |author2=Lara R. Arauna |author3=Tahria Deba |author4=Francesc Calafell |author5=Soraya Benhamamouch |author6=David Comas |title=Genetic Heterogeneity in Algerian Human Populations|journal=PLoS ONE|date=September 24, 2015|volume=10|issue=9|doi=10.1371/journal.pone.0138453|url=http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0138453|accessdate=13 May 2016 |pmid=26402429 |pmc=4581715 |pages=e0138453|bibcode=2015PLoSO..1038453B }}; [http://journals.plos.org/plosone/article/asset?unique&id=info:doi/10.1371/journal.pone.0138453.s007 S5 Table]</ref> as well as [[Burkina Faso]], [[Nigeria]], South [[Tunisia]], South [[Morocco]] and [[Egypt]]<ref name= "Fadhlaoui-Zid">{{cite journal |last1=Fadhlaoui-Zid |first1=K. |last2=Plaza |first2=S. |last3=Calafell |first3=F. |last4=Ben Amor |first4=M. |last5=Comas |first5=D. |last6=Bennamar |first6=A. |last7=Gaaied |first7=El |title=Mitochondrial DNA Heterogeneity in Tunisian Berbers |journal=Annals of Human Genetics |volume=68 |issue=Pt 3 |pages=222–33 |year=2004 |pmid=15180702 |doi=10.1046/j.1529-8817.2004.00096.x}}</ref> |
||
| Line 182: | Line 182: | ||
** Ian Logan's [http://www.ianlogan.co.uk/mtDNA.htm Mitochondrial DNA Site] |
** Ian Logan's [http://www.ianlogan.co.uk/mtDNA.htm Mitochondrial DNA Site] |
||
* Haplogroup L3 |
* Haplogroup L3 |
||
** Mannis van Oven's [http://www.phylotree.org/tree/subtree_L3.htm PhyloTree.org – mtDNA subtree L3] |
** Mannis van Oven's [https://web.archive.org/web/20091117111251/http://www.phylotree.org/tree/subtree_L3.htm PhyloTree.org – mtDNA subtree L3] |
||
** [https://www3.nationalgeographic.com/genographic/atlas.html?card=mm003 Spread of Haplogroup L3], from ''[[National Geographic (magazine)|National Geographic]]'' |
** [https://web.archive.org/web/20060320203146/https://www3.nationalgeographic.com/genographic/atlas.html?card=mm003 Spread of Haplogroup L3], from ''[[National Geographic (magazine)|National Geographic]]'' |
||
[[Category:Human mtDNA haplogroups|L3]] |
[[Category:Human mtDNA haplogroups|L3]] |
||
Revision as of 21:10, 29 October 2017
| Haplogroup L3 | |
|---|---|
| Possible time of origin | 80,000–104,000 YBP[1] or 60,000–70,000 YBP[2] |
| Possible place of origin | East Africa[3] |
| Ancestor | L3'4 |
| Descendants | L3a, L3b'f, L3c'd, L3e'i'k'x, L3h, M, N |
| Defining mutations | 769, 1018, 16311[4] |
Haplogroup L3 is a human mitochondrial DNA (mtDNA) haplogroup. The clade has played a pivotal role in the prehistory of the human species. It represents the most common parent maternal lineage of all people outside of Africa, and for many individuals within the continent as well.[5]
Origin

Haplogroup L3's exact place of origin is uncertain. According to the Recent African origin of modern humans (Out-of-Africa) theory, the clade is believed to have arisen in Africa and dispersed from East Africa between 84,000 and 104,000 years ago.[1] An analysis of 369 complete African L3 sequences placed the maximal date of the clade's expansion at ∼70 ka. This virtually rules out a successful exit out of Africa before 74 ka, the date of the Toba volcanic super-eruption in Sumatra.[2] The Time to Most Recent Common Ancestor for the L3 lineage has also recently been estimated to be between 58,900 and 70,200 years ago.[2]
Distribution
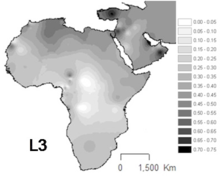
L3 is common in Northeast Africa, in contrast to others parts of Africa where the haplogroups L1 and L2 represent two thirds of mtDNAs.[6][7] L3 sublineages are also frequent in the Arabian peninsula.
L3 is sub-divided into several clades, two of which spawned the macro-haplogroups M and N that are today carried by most people outside of Africa.[8] There is at least one relatively deep non-M, non-N clade of L3 outside Africa, L3f1b6, found at 1% in Asturias Spain, which diverged from African L3 lineages at least 10,000 years ago.[9]
According to Maca-Meyer et al. (2001), "L3 is more related to Eurasian haplogroups than to the most divergent African clusters L1 and L2".[10] L3 is the haplogroup from which all modern humans outside of Africa derive.[11]
Basal haplogroup L3* is found among Nubians (13.8%),[12] as well as Socotri (4.3%).[13]
Haplogroup L3 has been observed in an ancient fossil belonging to the Pre-Pottery Neolithic B culture.[14] The clade has also been found among ancient Egyptian mummies excavated at the Abusir el-Meleq archaeological site in Middle Egypt, which date from the Pre-Ptolemaic/late New Kingdom and Ptolemaic periods.[15] Additionally, haplogroup L3 has been observed in ancient Guanche fossils excavated in Gran Canaria and Tenerife on the Canary Islands, which have been radiocarbon-dated to between the 7th and 11th centuries CE. All of the clade-bearing individuals were inhumed at the Gran Canaria site, with most of these specimens found to belong to the L3b1a subclade (3/4; 75%).[16]
Subclade distribution

Undifferentiated haplogroup L3 is widely distributed, particularly in the Chad Basin.[17] It is also found among Egyptians inhabiting El-Hayez oasis (11.4%).[7]
- L3a – East Africa.[5]
- L3b'f
- L3c'd
- L3c – Northeast Africa, South Arabia. Ethiopian Jews,[5] Yemeni Jews[5]
- L3d – Wide distribution in Africa. Among the Fulani,[5] Chadians,[5] Ethiopians,[19] Akan people,[20] Mozambique,[19] Yemen[19] Egyptians, Berbers[21]
- L3e'i'k'x
- L3e – West-Central Africa. It is the most common L3 sub-clade in Bantu-speaking populations.[22] L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, Afro-Brazilians and Caribbeans[23]
- L3e1 – Central Africa origin and is found in Algeria, Cameroon, Angola,[24] Mozambique, Sudanese and Kikuyu from Kenya as well as in Yemen and among the Akan people[20]
- L3e5 – Originated in the Chad Basin. Found in Algeria,[25] as well as Burkina Faso, Nigeria, South Tunisia, South Morocco and Egypt[26]
- L3i
- L3k – North Africa. Libyans,[5] Tunisians.[5]
- L3x – Northeast Africa. Ethiopian Oromos,[19] Egyptians[Note 1][27]
- L3e – West-Central Africa. It is the most common L3 sub-clade in Bantu-speaking populations.[22] L3e is suggested to be associated with a Central African origin and is also the most common L3 subclade amongst African Americans, Afro-Brazilians and Caribbeans[23]
- L3h – East Africa. Ethiopians.[19]
Tree
This phylogenetic tree of haplogroup L3 subclades is based on the paper by Mannis van Oven and Manfred Kayser Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation[4] and subsequent published research.
Most Recent Common Ancestor (MRCA)
- L1-6
- L2-6
- L2'3'4'6
- L3'4'6
- L3'4
- L3
- L3a
- L3b'f
- L3b
- L3b1
- L3b1a
- L3b1a1
- L3b1a2
- L3b1b
- L3b1b1
- L3b1a
- L3b2
- L3b1
- L3f
- L3f1
- L3f1a
- L3f1b
- L3f1b1
- L3f1b2
- L3f1b2a
- 150
- L3f1b3
- L3f1b4
- L3f1b4a
- L3f1b4a1
- L3f1b4a
- L3f2
- L3f2b
- L3f3
- L3f1
- L3b
- L3c'd
- L3c
- L3d
- L3d1-5
- L3d1
- L3d1a
- L3d1a1
- L3d1a1a
- L3d1a1
- L3d1b
- L3d1b1
- L3d1c
- L3d1d
- L3d1a
- 199
- L3d2
- L3d5
- L3d3
- L3d3a
- L3d4
- L3d5
- L3d1
- L3d1-5
- L3e'i'k'x
- L3e
- L3e1
- L3e1a
- L3e1a1
- L3e1a1a
- 152
- L3e1a2
- L3e1a3
- L3e1a1
- L3e1b
- L3e1c
- L3e1d
- L3e1e
- L3e1a
- L3e2
- L3e2a
- L3e2a1
- L3e2a1a
- L3e2a1b
- L3e2a1b1
- L3e2a1
- L3e2b
- L3e2b1
- L3e2b1a
- L3e2b2
- L3e2b3
- L3e2b1
- L3e2a
- L3e3'4'5
- L3e3'4
- L3e3
- L3e3a
- L3e3b
- L3e3b1
- L3e4
- L3e3
- L3e5
- L3e3'4
- L3e1
- L3i
- L3i1
- L3i1a
- L3i1b
- L3i2
- L3i1
- L3k
- L3x
- L3x1
- L3x2
- L3x2a
- L3x2a1
- L3x2a1a
- L3x2a1
- L3x2b
- L3x2a
- L3e
- L3h
- L3h1
- L3h1a
- L3h1a1
- L3h1a2
- L3h1a2a
- L3h1a2b
- L3h1b
- L3h1b1
- L3h1b1a
- L3h1b1a1
- L3h1b1a
- L3h1b2
- L3h1b1
- L3h1a
- L3h2
- L3h1
- M
- N
- L3
- L3'4
- L3'4'6
- L2'3'4'6
- L2-6
See also
|
Phylogenetic tree of human mitochondrial DNA (mtDNA) haplogroups | |||||||||||||||||||||||||||||||||||||||
| Mitochondrial Eve (L) | |||||||||||||||||||||||||||||||||||||||
| L0 | L1–6 | ||||||||||||||||||||||||||||||||||||||
| L1 | L2 | L3 | L4 | L5 | L6 | ||||||||||||||||||||||||||||||||||
| M | N | ||||||||||||||||||||||||||||||||||||||
| CZ | D | E | G | Q | O | A | S | R | I | W | X | Y | |||||||||||||||||||||||||||
| C | Z | B | F | R0 | pre-JT | P | U | ||||||||||||||||||||||||||||||||
| HV | JT | K | |||||||||||||||||||||||||||||||||||||
| H | V | J | T | ||||||||||||||||||||||||||||||||||||
References
- ^ a b Gonder, M. K.; Mortensen, H. M.; Reed, F. A.; De Sousa, A.; Tishkoff, S. A. (2006). "Whole-mtDNA Genome Sequence Analysis of Ancient African Lineages". Molecular Biology and Evolution. 24 (3): 757–68. doi:10.1093/molbev/msl209. PMID 17194802.
- ^ a b c http://mbe.oxfordjournals.org/content/early/2011/11/16/molbev.msr245.short?rss=1
- ^ Salas, A; Richards, Martin; de la Fe, Tomás; Lareu, María-Victoria; Sobrino, Beatriz; Sánchez-Diz, Paula; MacAulay, Vincent; Carracedo, Ángel (2002). "The Making of the African mtDNA Landscape". The American Journal of Human Genetics. 71 (5): 1082–111. doi:10.1086/344348. PMC 385086. PMID 12395296.
- ^ a b Van Oven, Mannis; Kayser, Manfred (2009). "Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation". Human Mutation. 30 (2): E386–94. doi:10.1002/humu.20921. PMID 18853457.
- ^ a b c d e f g h Behar, Doron M.; Villems, Richard; Soodyall, Himla; Blue-Smith, Jason; Pereira, Luisa; Metspalu, Ene; Scozzari, Rosaria; Makkan, Heeran; et al. (2008). "The Dawn of Human Matrilineal Diversity". The American Journal of Human Genetics. 82 (5): 1130–40. doi:10.1016/j.ajhg.2008.04.002. PMC 2427203. PMID 18439549.
- ^ Wallace DC et al. (2000), Origin of haplogroup M in Ethiopia, Am J Hum Genet 67(Suppl):217[verification needed]
- ^ a b Martina Kujanova; Luisa Pereira; Veronica Fernandes; Joana B. Pereira; Viktor Cerny (2009). "Near Eastern Neolithic Genetic Input in a Small Oasis of the Egyptian Western Desert". American Journal of Physical Anthropology. 140 (2): 336–46. doi:10.1002/ajpa.21078. PMID 19425100.
- ^ Wallace, D; Brown, MD; Lott, MT (1999). "Mitochondrial DNA variation in human evolution and disease". Gene. 238 (1): 211–30. doi:10.1016/S0378-1119(99)00295-4. PMID 10570998.
- ^ a b Pardiñas, AF; Martínez, JL; Roca, A; García-Vazquez, E; López, B (2014). "Over the sands and far away: Interpreting an Iberian mitochondrial lineage with ancient Western African origins". Am J Hum Biol. 26 (6): 777–83. doi:10.1002/ajhb.22601. PMID 25130626.
- ^ Maca-Meyer, Nicole; González, Ana M; Larruga, José M; Flores, Carlos; Cabrera, Vicente M (2001). "Major genomic mitochondrial lineages delineate early human expansions". BMC Genetics. 2: 13. doi:10.1186/1471-2156-2-13. PMC 55343. PMID 11553319.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Archived copy". Archived from the original on 2011-07-08. Retrieved 2009-03-09.
{{cite web}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help)CS1 maint: archived copy as title (link) - ^ a b c Mohamed, Hisham Yousif Hassan. "Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan" (PDF). University of Khartoum. Retrieved 14 June 2016.
- ^ Černý, Viktor; et al. (2009). "Out of Arabia – the settlement of island Soqotra as revealed by mitochondrial and Y chromosome genetic diversity" (PDF). American journal of Physical Anthropology. 138 (4): 439–47. doi:10.1002/ajpa.20960. PMID 19012329. Archived from the original (PDF) on 6 October 2016. Retrieved 14 June 2016.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help) - ^ Fernández, Eva; et al. (2014). "Ancient DNA analysis of 8000 BC near eastern farmers supports an early neolithic pioneer maritime colonization of Mainland Europe through Cyprus and the Aegean Islands". PLoS Genetics. 10 (6): e1004401. doi:10.1371/journal.pgen.1004401. PMC 4046922. PMID 24901650.
{{cite journal}}: Explicit use of et al. in:|last1=(help)CS1 maint: unflagged free DOI (link) - ^ Schuenemann, Verena J.; et al. (2017). "Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods". Nature Communications. 8: 15694. Bibcode:2017NatCo...815694S. doi:10.1038/ncomms15694. PMC 5459999. PMID 28556824.
{{cite journal}}: Explicit use of et al. in:|last1=(help) - ^ Rodrı́guez-Varela; et al. (2017). "Genomic Analyses of Pre-European Conquest Human Remains from the Canary Islands Reveal Close Affinity to Modern North Africans". Current Biology. 27 (1–7). Retrieved 27 October 2017.
{{cite journal}}: Explicit use of et al. in:|last1=(help) - ^ Cerezo, María; et al. (2011). "New insights into the Lake Chad Basin population structure revealed by high-throughput genotyping of mitochondrial DNA coding SNPs". PLoS ONE. 6 (4): e18682. Bibcode:2011PLoSO...618682C. doi:10.1371/journal.pone.0018682.
{{cite journal}}: Explicit use of et al. in:|last1=(help)CS1 maint: unflagged free DOI (link) - ^ Černý, Viktor; Fernandes, Verónica; Costa, Marta D; Hájek, Martin; Mulligan, Connie J; Pereira, Luísa (2009). "Migration of Chadic speaking pastoralists within Africa based on population structure of Chad Basin and phylogeography of mitochondrial L3f haplogroup". BMC Evolutionary Biology. 9: 63. doi:10.1186/1471-2148-9-63. PMC 2680838. PMID 19309521.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e f g Kivisild, T; Reidla, M; Metspalu, E; Rosa, A; Brehm, A; Pennarun, E; Parik, J; Geberhiwot, T; et al. (2004). "Ethiopian Mitochondrial DNA Heritage: Tracking Gene Flow Across and Around the Gate of Tears". The American Journal of Human Genetics. 75 (5): 752–70. doi:10.1086/425161. PMC 1182106. PMID 15457403.
- ^ a b Fendt, Liane; Röck, Alexander; Zimmermann, Bettina; Bodner, Martin; Thye, Thorsten; Tschentscher, Frank; Owusu-Dabo, Ellis; Göbel, Tanja M.K.; Schneider, Peter M.; Parson, Walther (2012). "MtDNA diversity of Ghana: a forensic and phylogeographic view". Forensic Science International: Genetics. 6 (2): 244–49. doi:10.1016/j.fsigen.2011.05.011.
- ^ Sheet1 – PLOS Pathogens
- ^ Anderson, S. 2006, Phylogenetic and phylogeographic analysis of African mitochondrial DNA variation. Archived 2011-09-10 at the Wayback Machine
- ^ Bandelt, HJ; Alves-Silva, J; Guimarães, PE; Santos, MS; Brehm, A; Pereira, L; Coppa, A; Larruga, JM; et al. (2001). "Phylogeography of the human mitochondrial haplogroup L3e: a snapshot of African prehistory and Atlantic slave trade". Annals of Human Genetics. 65 (Pt 6): 549–63. doi:10.1017/S0003480001008892 (inactive 2017-01-16). PMID 11851985.
{{cite journal}}: CS1 maint: DOI inactive as of January 2017 (link) - ^ Plaza, Stéphanie; Salas, Antonio; Calafell, Francesc; Corte-Real, Francisco; Bertranpetit, Jaume; Carracedo, Ángel; Comas, David (2004). "Insights into the western Bantu dispersal: mtDNA lineage analysis in Angola". Human Genetics. 115 (5): 439–47. doi:10.1007/s00439-004-1164-0. PMID 15340834.
- ^ Asmahan Bekada; Lara R. Arauna; Tahria Deba; Francesc Calafell; Soraya Benhamamouch; David Comas (September 24, 2015). "Genetic Heterogeneity in Algerian Human Populations". PLoS ONE. 10 (9): e0138453. Bibcode:2015PLoSO..1038453B. doi:10.1371/journal.pone.0138453. PMC 4581715. PMID 26402429. Retrieved 13 May 2016.
{{cite journal}}: CS1 maint: unflagged free DOI (link); S5 Table - ^ Fadhlaoui-Zid, K.; Plaza, S.; Calafell, F.; Ben Amor, M.; Comas, D.; Bennamar, A.; Gaaied, El (2004). "Mitochondrial DNA Heterogeneity in Tunisian Berbers". Annals of Human Genetics. 68 (Pt 3): 222–33. doi:10.1046/j.1529-8817.2004.00096.x. PMID 15180702.
- ^ Stevanovitch, A.; Gilles, A.; Bouzaid, E.; Kefi, R.; Paris, F.; Gayraud, R. P.; Spadoni, J. L.; El-Chenawi, F.; Beraud-Colomb, E. (2004). "Mitochondrial DNA Sequence Diversity in a Sedentary Population from Egypt". Annals of Human Genetics. 68 (Pt 1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
Notes
- ^ GUR46 on table 1. is a mtDNA haplogroup L3x2a.
External links
- General
- Ian Logan's Mitochondrial DNA Site
- Haplogroup L3
- Mannis van Oven's PhyloTree.org – mtDNA subtree L3
- Spread of Haplogroup L3, from National Geographic
