Retroviral ribonuclease H
| Retroviral ribonuclease H | |||||||||
|---|---|---|---|---|---|---|---|---|---|
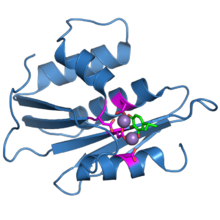 The ribonuclease H domain from the HIV-1 reverse transcriptase protein. The four active-site carboxylate residues are shown in magenta. Two bound manganese ions are shown as purple spheres. A bound inhibitor molecule, beta-thujaplicinol, is shown in green.[1] | |||||||||
| Identifiers | |||||||||
| EC no. | 3.1.26.13 | ||||||||
| CAS no. | 9050-76-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
The retroviral ribonuclease H (retroviral RNase H) is a catalytic domain of the retroviral reverse transcriptase (RT) enzyme. The RT enzyme is used to generate complementary DNA (cDNA) from the retroviral RNA genome. This process is called reverse transcription. To complete this complex process, the retroviral RT enzymes need to adopt a multifunctional nature. They therefore possess 3 of the following biochemical activities: RNA-dependent DNA polymerase, ribonuclease H, and DNA-dependent DNA polymerase activities.[2] Like all RNase H enzymes, the retroviral RNase H domain cleaves DNA/RNA duplexes and will not degrade DNA or unhybridized RNA.
Structure
[edit]

Although the RT structures from human, murine and avian retroviruses display different subunits, the relative sizes, orientation and connection of the DNA polymerase and RNase H domains are strikingly similar. The RNase H domain occupies ~25% of the RT protein C-terminal. The DNA polymerase domain occupies ~55% of the RT protein N-terminal.[5] The RNase H domains of MMLV and HIV-1 RT enzymes are structural very similar to the Escherichia coli and Bacillus halodurans RNases H as well as to human RNaseH1.[6][7][8][9][10] In general, the folded structures of retroviral RNase H domains take the form of 5-stranded mixed beta sheets flanked by four alpha helices in an asymmetric distribution. A notable difference between the various RNase H proteins is the presence or absence of the C-helix (present in E. coli, MLV and human RNases H, absent in HIV-1, B. halodurans and ASLV RNases H), a positively charged alpha helix also referred to as the basic loop or protrusion.[10] It is believed to have a role in substrate binding.[10]

Function
[edit]During reverse transcription of the viral genomic RNA into cDNA, an RNA/DNA hybrid is created. The RNA strand is then hydrolyzed by the RNase H domain to enable synthesis of the second DNA strand by the DNA polymerase function of the RT enzyme.[5] In addition, retroviral virions package a single tRNA molecule that they use as a primer during reverse transcription of the viral genomic RNA.[11] The retroviral RNase H is needed to digest the tRNA molecule when it is no longer needed. These processes happen in a Mg2+ dependent fashion.[12][13]
Retroviral RNases H cleave their substrates through 3 different modes:
- sequence-specific internal cleavage of RNA [1-4]. Human immunodeficiency virus type 1 and Moloney murine leukemia virus enzymes prefer to cleave the RNA strand one nucleotide away from the RNA-DNA junction.
- RNA 5'-end directed cleavage 13-19 nucleotides from the RNA end.
- DNA 3'-end directed cleavage 15-20 nucleotides away from the primer terminus.
The two end-directed modes are unique to the retroviral RNases H because of a number of effects of the associated polymerase domain of retroviral RT.[6] In the more universal internal cleavage mode, the RNases H behave as typical endonucleases and cleave the RNA along the length of a DNA / RNA hybrid substrate in the absence of any ‘end’ effects.[14][15][16][17]
References
[edit]- ^ a b Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C, Clark AD, Ryan K, Hickey MJ, Love RA, Hughes SH, Bergqvist S, Arnold E (December 2009). "Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site". Structure. 17 (12): 1625–1635. doi:10.1016/j.str.2009.09.016. PMC 3365588. PMID 20004166.
- ^ Worthington, Von (1993). Worthington Enzyme Manual. Worthington. p. 280.
- ^ Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Nakamura H, Ikehara M, Matsuzaki T, Morikawa K (February 1992). "Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution". Journal of Molecular Biology. 223 (4): 1029–52. doi:10.1016/0022-2836(92)90260-q. PMID 1311386.
- ^ Sarafianos SG, Das K, Tantillo C, Clark AD, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E (March 2001). "Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA". The EMBO Journal. 20 (6): 1449–61. doi:10.1093/emboj/20.6.1449. PMC 145536. PMID 11250910.
- ^ a b Beilhartz GL, Götte M (April 2010). "HIV-1 Ribonuclease H: Structure, Catalytic Mechanism and Inhibitors". Viruses. 2 (4): 900–26. doi:10.3390/v2040900. PMC 3185654. PMID 21994660.
- ^ a b Lim D, Gregorio GG, Bingman C, Martinez-Hackert E, Hendrickson WA, Goff SP (September 2006). "Crystal structure of the moloney murine leukemia virus RNase H domain". Journal of Virology. 80 (17): 8379–89. doi:10.1128/jvi.00750-06. PMC 1563865. PMID 16912289.
- ^ Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K (September 1990). "Three-dimensional structure of ribonuclease H from E. coli". Nature. 347 (6290): 306–9. Bibcode:1990Natur.347..306K. doi:10.1038/347306a0. PMID 1698262. S2CID 4234320.
- ^ Yang W, Hendrickson WA, Crouch RJ, Satow Y (September 1990). "Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein". Science. 249 (4975): 1398–405. doi:10.1126/science.2169648. PMID 2169648.
- ^ Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (July 2005). "Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis". Cell. 121 (7): 1005–16. doi:10.1016/j.cell.2005.04.024. PMID 15989951.
- ^ a b c Leo B, Schweimer K, Rösch P, Hartl MJ, Wöhrl BM (September 2012). "The solution structure of the prototype foamy virus RNase H domain indicates an important role of the basic loop in substrate binding". Retrovirology. 9 (73): 73. doi:10.1186/1742-4690-9-73. PMC 3443672. PMID 22962864.
- ^ Fu, Tie-Bo; John taylor (27 March 1992). "When retroviral reverse transcriptases reach the end of their RNA templates". Journal of Virology. 66 (7): 4271–4278. doi:10.1128/JVI.66.7.4271-4278.1992. PMC 241232. PMID 1376369.
- ^ Taylor JM (March 1977). "An analysis of the role of tRNA species as primers for the transcription into DNA of RNA tumor virus genomes". Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 473 (1): 57–71. doi:10.1016/0304-419x(77)90007-5. PMID 66067.
- ^ Talele TT, Upadhyay A, Pandey VN (October 2009). "Influence of the RNase H domain of retroviral reverse transcriptases on the metal specificity and substrate selection of their polymerase domains". Virology Journal. 6 (159): 159. doi:10.1186/1743-422x-6-159. PMC 2765437. PMID 19814799.
- ^ Schultz SJ, Zhang M, Champoux JJ (November 2004). "Recognition of internal cleavage sites by retroviral RNases H". Journal of Molecular Biology. 344 (3): 635–52. doi:10.1016/j.jmb.2004.09.081. PMID 15533434.
- ^ Krug MS, Berger SL (May 1989). "Ribonuclease H activities associated with viral reverse transcriptases are endonucleases". Proceedings of the National Academy of Sciences of the United States of America. 86 (10): 3539–43. Bibcode:1989PNAS...86.3539K. doi:10.1073/pnas.86.10.3539. PMC 287173. PMID 2471188.
- ^ Champoux JJ, Schultz SJ (March 2009). "Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription". The FEBS Journal. 276 (6): 1506–16. doi:10.1111/j.1742-4658.2009.06909.x. PMC 2742777. PMID 19228195.
- ^ Schultz SJ, Champoux JJ (June 2008). "RNase H activity: structure, specificity, and function in reverse transcription". Virus Research. 134 (1–2): 86–103. doi:10.1016/j.virusres.2007.12.007. PMC 2464458. PMID 18261820.
External links
[edit]- Retroviral+ribonuclease+H at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
