Human evolutionary genetics
Human evolutionary genetics studies how one human genome differs from another human genome, the evolutionary past that gave rise to it, and its current effects. Differences between genomes have anthropological, medical and forensic implications and applications. Genetic data can provide important insight into human evolution.
Origin of apes
−10 — – −9 — – −8 — – −7 — – −6 — – −5 — – −4 — – −3 — – −2 — – −1 — – 0 — |
| |||||||||||||||||||||||||||||
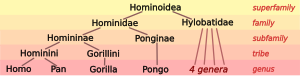
Biologists classify humans, along with only a few other species, as great apes (species in the family Hominidae). The living Hominidae include two distinct species of chimpanzee (the bonobo, Pan paniscus, and the common chimpanzee, Pan troglodytes), two species of gorilla (the western gorilla, Gorilla gorilla, and the eastern gorilla, Gorilla graueri), and two species of orangutan (the Bornean orangutan, Pongo pygmaeus, and the Sumatran orangutan, Pongo abelii). The great apes with the family Hylobatidae of gibbons form the superfamily Hominoidea of apes.
Apes, in turn, belong to the primate order (>400 species), along with the Old World monkeys, the New World monkeys, and others. Data from both mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) indicate that primates belong to the group of Euarchontoglires, together with Rodentia, Lagomorpha, Dermoptera, and Scandentia.[1] This is further supported by Alu-like short interspersed nuclear elements (SINEs) which have been found only in members of the Euarchontoglires.[2]
Cladistics
A phylogenetic tree is usually derived from DNA or protein sequences from populations. Often, mitochondrial DNA or Y chromosome sequences are used to study ancient human demographics. These single-locus sources of DNA do not recombine and are almost always inherited from a single parent, with only one known exception in mtDNA.[3] Individuals from closer geographic regions generally tend to be more similar than individuals from regions farther away. Distance on a phylogenetic tree can be used approximately to indicate:
- Genetic distance. The genetic difference between humans and chimps is less than 2%,[4] or twenty times larger than the variation among modern humans.
- Temporal remoteness of the most recent common ancestor. The mitochondrial most recent common ancestor of modern humans lived roughly 200,000 years ago[citation needed], latest common ancestors of humans and chimps between four and seven million years ago.
Speciation of humans and the African apes
The separation of humans from their closest relatives, the non-human apes (chimpanzees and gorillas), has been studied extensively for more than a century. Five major questions have been addressed:
- Which apes are our closest ancestors?
- When did the separations occur?
- What was the effective population size of the common ancestor before the split?
- Are there traces of population structure (subpopulations) preceding the speciation or partial admixture succeeding it?
- What were the specific events (including fusion of chromosomes 2a and 2b) prior to and subsequent to the separation?
General observations
As discussed before, different parts of the genome show different sequence divergence between different hominoids. It has also been shown that the sequence divergence between DNA from humans and chimpanzees varies greatly. For example, the sequence divergence varies between 0% to 2.66% between non-coding, non-repetitive genomic regions of humans and chimpanzees.[5] Additionally gene trees, generated by comparative analysis of DNA segments, do not always fit the species tree. Summing up:
- The sequence divergence varies significantly between humans, chimpanzees and gorillas.
- For most DNA sequences, humans and chimpanzees appear to be most closely related, but some point to a human-gorilla or chimpanzee-gorilla clade.
- The human genome has been sequenced, as well as the chimpanzee genome. Humans have 23 pairs of chromosomes, while chimpanzees, gorillas, and orangutans have 24. Human chromosome 2 is a fusion of two chromosomes 2a and 2b that remained separate in the other primates.[6]
Divergence times
The divergence time of humans from other apes is of great interest. One of the first molecular studies, published in 1967 measured immunological distances (IDs) between different primates.[7] Basically the study measured the strength of immunological response that an antigen from one species (human albumin) induces in the immune system of another species (human, chimpanzee, gorilla and Old World monkeys). Closely related species should have similar antigens and therefore weaker immunological response to each other's antigens. The immunological response of a species to its own antigens (e.g. human to human) was set to be 1.
The ID between humans and gorillas was determined to be 1.09, that between humans and chimpanzees was determined as 1.14. However the distance to six different Old World monkeys was on average 2.46, indicating that the African apes are more closely related to humans than to monkeys. The authors consider the divergence time between Old World monkeys and hominoids to be 30 million years ago (MYA), based on fossil data, and the immunological distance was considered to grow at a constant rate. They concluded that divergence time of humans and the African apes to be roughly ~5 MYA. That was a surprising result. Most scientists at that time thought that humans and great apes diverged much earlier (>15 MYA).
The gorilla was, in ID terms, closer to human than to chimpanzees; however, the difference was so slight that the trichotomy could not be resolved with certainty. Later studies based on molecular genetics were able to resolve the trichotomy: chimpanzees are phylogenetically closer to humans than to gorillas. However, some divergence times estimated later (using much more sophisticated methods in molecular genetics) do not substantially differ from the very first estimate in 1967, but a recent paper[8] puts it at 11-14 MYA.
Divergence times and ancestral effective population size

Current methods to determine divergence times use DNA sequence alignments and molecular clocks. Usually the molecular clock is calibrated assuming that the orangutan split from the African apes (including humans) 12-16 MYA. Some studies also include some old world monkeys and set the divergence time of them from hominoids to 25-30 MYA. Both calibration points are based on very little fossil data and have been criticized.[9]
If these dates are revised, the divergence times estimated from molecular data will change as well. However, the relative divergence times are unlikely to change. Even if we can't tell absolute divergence times exactly, we can be pretty sure that the divergence time between chimpanzees and humans is about sixfold shorter than between chimpanzees (or humans) and monkeys.
One study (Takahata et al., 1995) used 15 DNA sequences from different regions of the genome from human and chimpanzee and 7 DNA sequences from human, chimpanzee and gorilla.[10] They determined that chimpanzees are more closely related to humans than gorillas. Using various statistical methods, they estimated the divergence time human-chimp to be 4.7 MYA and the divergence time between gorillas and humans (and chimps) to be 7.2 MYA.
Additionally they estimated the effective population size of the common ancestor of humans and chimpanzees to be ~100,000. This was somewhat surprising since the present day effective population size of humans is estimated to be only ~10,000. If true that means that the human lineage would have experienced an immense decrease of its effective population size (and thus genetic diversity) in its evolution. (see Toba catastrophe theory)

Another study (Chen & Li, 2001) sequenced 53 non-repetitive, intergenic DNA segments from a human, a chimpanzee, a gorilla, and orangutan.[5] When the DNA sequences were concatenated to a single long sequence, the generated neighbor-joining tree supported the Homo-Pan clade with 100% bootstrap (that is that humans and chimpanzees are the closest related species of the four). When three species are fairly closely related to each other (like human, chimpanzee and gorilla), the trees obtained from DNA sequence data may not be congruent with the tree that represents the speciation (species tree).
The shorter internodal time span (TIN) the more common are incongruent gene trees. The effective population size (Ne) of the internodal population determines how long genetic lineages are preserved in the population. A higher effective population size causes more incongruent gene trees. Therefore, if the internodal time span is known, the ancestral effective population size of the common ancestor of humans and chimpanzees can be calculated.
When each segment was analyzed individually, 31 supported the Homo-Pan clade, 10 supported the Homo-Gorilla clade, and 12 supported the Pan-Gorilla clade. Using the molecular clock the authors estimated that gorillas split up first 6.2-8.4 MYA and chimpanzees and humans split up 1.6-2.2 million years later (internodal time span) 4.6-6.2 MYA. The internodal time span is useful to estimate the ancestral effective population size of the common ancestor of humans and chimpanzees.
A parsimonious analysis revealed that 24 loci supported the Homo-Pan clade, 7 supported the Homo-Gorilla clade, 2 supported the Pan-Gorilla clade and 20 gave no resolution. Additionally they took 35 protein coding loci from databases. Of these 12 supported the Homo-Pan clade, 3 the Homo-Gorilla clade, 4 the Pan-Gorilla clade and 16 gave no resolution. Therefore, only ~70% of the 52 loci that gave a resolution (33 intergenic, 19 protein coding) support the 'correct' species tree. From the fraction of loci which did not support the species tree and the internodal time span they estimated previously, the effective population of the common ancestor of humans and chimpanzees was estimated to be ~52 000 to 96 000. This value is not as high as that from the first study (Takahata), but still much higher than present day effective population size of humans.[dubious – discuss]
A third study (Yang, 2002) used the same dataset that Chen and Li used but estimated the ancestral effective population of 'only' ~12,000 to 21,000, using a different statistical method.[11]
Genetic differences between humans and other great apes
The alignable sequences within genomes of humans and chimpanzees differ by about 35 million single-nucleotide substitutions. Additionally about 3% of the complete genomes differ by deletions, insertions and duplications.[12]
Since mutation rate is relatively constant, roughly one half of these changes occurred in the human lineage. Only a very tiny fraction of those fixed differences gave rise to the different phenotypes of humans and chimpanzees and finding those is a great challenge. The vast majority of the differences are neutral and do not affect the phenotype.[citation needed]
Molecular evolution may act in different ways, through protein evolution, gene loss, differential gene regulation and RNA evolution. All are thought to have played some part in human evolution.
Gene loss
Many different mutations can inactivate a gene, but few will change its function in a specific way. Inactivation mutations will therefore be readily available for selection to act on. Gene loss could thus be a common mechanism of evolutionary adaptation (the "less-is-more" hypothesis).[13]
80 genes were lost in the human lineage after separation from the last common ancestor with the chimpanzee. 36 of those were for olfactory receptors. Genes involved in chemoreception and immune response are overrepresented.[14] Another study estimated that 86 genes had been lost.[15]
Hair keratin gene KRTHAP1
A gene for type I hair keratin was lost in the human lineage. Keratins are a major component of hairs. Humans still have nine functional type I hair keratin genes, but the loss of that particular gene may have caused the thinning of human body hair. The gene loss occurred relatively recently in human evolution—less than 240,000 years ago.[16]
Myosin gene MYH16
Stedman et al. (2004) stated that the loss of the sarcomeric myosin gene MYH16 in the human lineage led to smaller masticatory muscles. They estimated that the mutation that led to the inactivation (a two base pair deletion) occurred 2.4 million years ago, predating the appearance of Homo ergaster/erectus in Africa. The period that followed was marked by a strong increase in cranial capacity, promoting speculation that the loss of the gene may have removed an evolutionary constraint on brain size in the genus Homo.[17]
Another estimate for the loss of the MYH16 gene is 5.3 million years ago, long before Homo appeared.[18]
Other
- CASPASE12, a cysteinyl aspartate proteinase. The loss of this gene is speculated to have reduced the lethality of bacterial infection in humans.[14]
Gene addition
Segmental duplications (SDs or LCRs) have had roles in creating new primate genes and shaping human genetic variation.
Human-specific DNA insertions
When the human genome was compared to the genomes of five comparison primate species, including the chimpanzee, gorilla, orangutan, gibbon, and macaque, it was found that there are approximately 20,000 human-specific insertions believed to be regulatory. While most insertions appear to be fitness neutral, a small amount have been identified in positively selected genes showing associations to neural phenotypes and some relating to dental and sensory perception-related phenotypes. These findings hint at the seemingly important role of human-specific insertions in the recent evolution of humans.[19]
Selection pressures
Human accelerated regions are areas of the genome that differ between humans and chimpanzees to a greater extent than can be explained by genetic drift over the time since the two species shared a common ancestor. These regions show signs of being subject to natural selection, leading to the evolution of distinctly human traits. Two examples are HAR1F, which is believed to be related to brain development and HAR2 (a.k.a. HACNS1) that may have played a role in the development of the opposable thumb.
It has also been hypothesized that much of the difference between humans and chimpanzees is attributable to the regulation of gene expression rather than differences in the genes themselves. Analyses of conserved non-coding sequences, which often contain functional and thus positively selected regulatory regions, address this possibility.[20]
Sequence divergence between humans and apes
When the draft sequence of the common chimpanzee (Pan troglodytes) genome was published in the summer 2005, 2400 million bases (of ~3160 million bases) were sequenced and assembled well enough to be compared to the human genome.[12] 1.23% of this sequenced differed by single-base substitutions. Of this, 1.06% or less was thought to represent fixed differences between the species, with the rest being variant sites in humans or chimpanzees. Another type of difference, called indels (insertions/deletions) accounted for many fewer differences (15% as many), but contributed ~1.5% of unique sequence to each genome, since each insertion or deletion can involve anywhere from one base to millions of bases.[12]
A companion paper examined segmental duplications in the two genomes,[21] whose insertion and deletion into the genome account for much of the indel sequence. They found that a total of 2.7% of euchromatic sequence had been differentially duplicated in one or the other lineage.
| Locus | Human-Chimp | Human-Gorilla | Human-Orangutan |
|---|---|---|---|
| Alu elements | 2 | - | - |
| Non-coding (Chr. Y) | 1.68 ± 0.19 | 2.33 ± 0.2 | 5.63 ± 0.35 |
| Pseudogenes (autosomal) | 1.64 ± 0.10 | 1.87 ± 0.11 | - |
| Pseudogenes (Chr. X) | 1.47 ± 0.17 | - | - |
| Noncoding (autosomal) | 1.24 ± 0.07 | 1.62 ± 0.08 | 3.08 ± 0.11 |
| Genes (Ks) | 1.11 | 1.48 | 2.98 |
| Introns | 0.93 ± 0.08 | 1.23 ± 0.09 | - |
| Xq13.3 | 0.92 ± 0.10 | 1.42 ± 0.12 | 3.00 ± 0.18 |
| Subtotal for X chromosome | 1.16 ± 0.07 | 1.47 ± 0.08 | - |
| Genes (Ka) | 0.8 | 0.93 | 1.96 |
The sequence divergence has generally the following pattern: Human-Chimp < Human-Gorilla << Human-Orangutan, highlighting the close kinship between humans and the African apes. Alu elements diverge quickly due to their high frequency of CpG dinucleotides which mutate roughly 10 times more often than the average nucleotide in the genome. The mutation rate is higher in the male germ line, therefore the divergence in the Y chromosome—which is inherited solely from the father—is higher than in autosomes. The X chromosome is inherited twice as often through the female germ line as through the male germ line and therefore shows slightly lower sequence divergence. The sequence divergence of the Xq13.3 region is surprisingly low between humans and chimpanzees.[22]
Mutations altering the amino acid sequence of proteins (Ka) are the least common. In fact ~29% of all orthologous proteins are identical between human and chimpanzee. The typical protein differs by only two amino acids.[12] The measures of sequence divergence shown in the table only take the substitutional differences, for example from an A (adenine) to a G (guanine), into account. DNA sequences may however also differ by insertions and deletions (indels) of bases. These are usually stripped from the alignments before the calculation of sequence divergence is performed.
Genetic differences between modern humans and Neanderthals
An international group of scientists completed a draft sequence of the Neanderthal genome in May 2010. The results indicate some breeding between modern humans (Homo sapiens) and Neanderthals (Homo neanderthalensis), as the genomes of non-African humans have 1-4% more in common with Neanderthals than do the genomes of subsaharan Africans. Neanderthals and most modern humans share a lactose-intolerant variant of the lactase gene that encodes an enzyme that is unable to break down lactose in milk after weaning. Modern humans and Neanderthals also share the FOXP2 gene variant associated with brain development and with speech in modern humans, indicating that Neanderthals may have been able to speak. Chimps have two amino acid differences in FOXP2 compared with human and Neanderthal FOXP2.[23][24][25]
Modern humans
Molecular biologists starting with Wesley Brown[26] on mtDNA and Allan Wilson[26] on mtDNA have produced observations relevant to human evolution.
Age of the common ancestor
By estimating the rate at which mutations occur in mtDNA, the age of the common ancestral mtDNA type can be estimated: "the common ancestral mtDNA (type a) links mtDNA types that have diverged by an average of nearly 0.57%. Assuming a rate of 2%-4% per million years, this implies that the common ancestor of all surviving mtDNA types existed 140,000-290,000 years ago."[26]
Most, but not all (see fr. Multiregional origin of modern humans), scientists in the relevant fields consider this observation robust. This common direct ancestor in the line of mother to daughter (or mitochondrial most recent common ancestor (mtMRCA)) of all extant humans has become known as Mitochondrial Eve. (Mitochondria are inherited from the mother only.) The observation that the mtMRCA is the direct matrilineal ancestor of all living humans does not mean either that she was the first anatomically modern woman, nor that no other women lived concurrently with her, nor even excluding the existence of other women being ancestors of today's people. Other women would have lived at the same time and passed nuclear genes down to living humans, but their mitochondrial lineages were lost over time. This could be due to events such as producing only male children.
African origin for modern humans
There is evidence that modern human mtDNA has an African origin: "We infer from the tree of minimum length... that Africa is a likely source of the human mitochondrial gene pool. This inference comes from the observation that one of the two primary branches leads exclusively to African mtDNAs... while the second primary branch also leads to African mtDNAs... By postulating that the common ancestral mtDNA... was African, we minimize the number of intercontinental migrations needed to account for the geographic distribution of mtDNA types."[26]
The broad study of African genetic diversity headed by Sarah Tishkoff found the San people to express the greatest genetic diversity among the 113 distinct populations sampled, making them one of 14 "ancestral population clusters". The research also located the origin of modern human migration in south-western Africa, near the coastal border of Namibia and Angola.[27][28]
Y chromosome lineages

The Y chromosome is much larger than mtDNA, and is relatively homogeneous; therefore it has taken much longer to find distinct lineages and to analyse them. Conversely, because the Y chromosome is so large by comparison, it holds more genetic information. Y chromosome studies show similar findings to those made with mtDNA. The estimate for the age of the ancestral Y chromosome for all extant Y chromosomes is given at about 70,000 years ago and is also placed in Africa; the individual who contributed this Y chromosomal heritage is sometimes referred to as Y chromosome Adam. The difference in dates between Y chromosome Adam and mitochondrial Eve is usually attributed to a higher extinction rate for Y chromosomes due to greater differential reproductive success between individual men, which means that a small number of very successful men may produce many children, while a larger number of less successful men will produce far fewer children.
At a recent conference at the American Society of Human Genetics, there was a study presented by Melissa Wilson Sayres from the University of California Berkeley which called into question the amount to which people had more female ancestors than male. While it didn't deny that men have historically produced at a higher rate than females, a polygamous mating system doesn't explain how the y-chromosome is so uniform. "Other people have suggested it's as few as four females for one male. We find that it's probably skewed, but it's more like four females to three males" said Sayres.[29][30]
Ancestral components
Modern DNA sequencing has identified various ancestral components in contemporary populations. A number of these genetic elements have West Eurasian origins. They include the following ancestral components, with their geographical hubs and main associated populations:
| # | West Eurasian component | Geographical hub | Peak population | Notes |
|---|---|---|---|---|
| 1 | Ancestral North Indian | North India, Pakistan | North Indians, Pakistanis | Main West Eurasian component in the Indian subcontinent. Peaks among Indo-European-speaking caste populations in the northern areas, but also found at significant frequencies among some Dravidian-speaking caste groups. Associated with either the arrival of Indo-European speakers from West Asia or Central Asia between 3,000 and 4,000 years before present, or with the spread of agriculture and West Asian crops beginning around 8,000-9,000 ybp, or with migrations from West Asia in the pre-agricultural period. Contrasted with the indigenous Ancestral South Indian component, which peaks among the Onge Andamanese inhabiting the Andaman Islands.[31][32] |
| 2 | Arabian | Arabian peninsula | Yemenis, Saudis, Qataris, Bedouins | Main West Eurasian component in the Gulf region. Most closely associated with local Arabic, Semitic-speaking populations.[33] Also found at significant frequencies in parts of the Levant, Egypt and Libya.[33][34] |
| 3 | Coptic | Nile Valley | Copts, Beja, Afro-Asiatic Ethiopians, Sudanese Arabs, Nubians | Main West Eurasian component in Northeast Africa.[35] Roughly equivalent with the Ethio-Somali component.[35][36] Peaks among Egyptian Copts in Sudan. Also found at high frequencies among other Afro-Asiatic (Hamito-Semitic) speakers in Ethiopia and Sudan, as well as among many Nubians. Associated with Ancient Egyptian ancestry, without the later Arabian influence present among modern Egyptians. Contrasted with the indigenous Nilo-Saharan component, which peaks among Nilo-Saharan- and Kordofanian-speaking populations inhabiting the southern part of the Nile Valley.[35] |
| 4 | Ethio-Somali | Horn of Africa | Somalis, Afars, Amhara, Oromos, Tigrinya | Main West Eurasian component in the Horn.[36] Roughly equivalent with the Coptic component.[35][36] Associated with the arrival of Afro-Asiatic speakers in the region during antiquity. Peaks among Cushitic- and Ethiopian Semitic-speaking populations in the northern areas. Diverged from the Maghrebi component around 23,000 ybp, and from the Arabian component about 25,000 ybp. Contrasted with the indigenous Omotic component, which peaks among the Omotic-speaking Ari ironworkers inhabiting southern Ethiopia.[36] |
| 5 | European | Europe | Europeans | Main West Eurasian component in Europe. Also found at significant frequencies in adjacent geographical areas outside of the continent, in Anatolia, the Caucasus, the Iranian plateau, and parts of the Levant.[33] |
| 6 | Levantine | Near East, Caucasus | Druze, Lebanese, Cypriots, Syrians, Jordanians, Palestinians, Armenians, Georgians, Sephardic Jews, Ashkenazi Jews, Iranians, Turks, Sardinians, Adygei | Main West Eurasian component in the Near East and Caucasus. Peaks among Druze populations in the Levant. Found amongst local Afro-Asiatic, Indo-European, Caucasus and Turkish speakers alike. Diverged from the European component around 9,100-15,900 ybp, and from the Arabian component about 15,500-23,700 ypb. Also found at significant frequencies in Southern Europe as well as parts of the Arabian peninsula.[33] |
| 7 | Maghrebi | Northwest Africa | Berbers, Maghrebis, Sahrawis, Tuareg | Main West Eurasian component in the Maghreb. Peaks among the Berber (non-Arabized) populations in the region.[34] Diverged from the Ethio-Somali/Coptic, Arabian, Levantine and European components prior to the Holocene.[34][36] |
See also
- Archaeogenetics of the Near East
- Chimpanzee genome project
- FOXP2 and human evolution
- Genetics and archaeogenetics of South Asia
- Genetic history of Europe
- Genetic history of indigenous peoples of the Americas
- Genetic history of Italy
- Genetic history of the British Isles
- Homininae
- List of haplogroups of historical and famous figures
- The Journey of Man: A Genetic Odyssey
- Y-DNA haplogroups by ethnic groups
- Race and genetics
References
- ^ Murphy, W.J.; Eizirik, E.; O'Brien, S.J.; Madsen, O.; Scally, M.; Douady, C.J.; Teeling, E.; Ryder, O.A.; Stanhope, M.J.; de Jong, W.W. & Springer, M.S.; Eizirik; O'Brien; Madsen; Scally; Douady; Teeling; Ryder; Stanhope; De Jong; Springer (2001). "Resolution of the early placental mammal radiation using Bayesian phylogenetics". Science. 294 (5550): 2348–2351. Bibcode:2001Sci...294.2348M. doi:10.1126/science.1067179. PMID 11743200.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Kriegs, J.O.; Churakov, G.; Kiefmann, M.; Jordan, U.; Brosius, J.; Schmitz, J. (2006). "Retroposed elements as archives for the evolutionary history of placental mammals". PLoS Biol. 4 (4): e91. doi:10.1371/journal.pbio.0040091. PMC 1395351. PMID 16515367.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Schwartz M, Vissing J; Vissing (2002). "Paternal Inheritance of Mitochondrial DNA". N Engl J Med. 347 (8): 576–580. doi:10.1056/NEJMoa020350. PMID 12192017.
- ^ "Human Chromosome 2." PBS.
- ^ a b c Chen, F.C. & Li, W.H.; Li (2001). "Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees". Am J Hum Genet. 68 (2): 444–456. doi:10.1086/318206. PMC 1235277. PMID 11170892.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ken Miller in the Kitzmiller v. Dover trial transcripts.
- ^ Sarich, V.M. & Wilson, A.C.; Wilson (1967). "Immunological time scale for hominid evolution". Science. 158 (3805): 1200–1203. Bibcode:1967Sci...158.1200S. doi:10.1126/science.158.3805.1200. PMID 4964406.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Venn, Oliver; Turner, Isaac; Mathieson, Iain; de Groot, Natasja; Bontrop, Ronald; McVean, Gil (June 2014). "Strong male bias drives germline mutation in chimpanzees". Science. 33 (6189): 1272–1275. doi:10.1126/science.344.6189.1272.
- ^ Yoder, A.D. & Yang, Z.; Yang (1 July 2000). "Estimation of primate speciation dates using local molecular clocks". Mol Biol Evol. 17 (7): 1081–1090. doi:10.1093/oxfordjournals.molbev.a026389. PMID 10889221.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Takahata, N.; Satta, Y.; Klein, J. (1995). "Divergence time and population size in the lineage leading to modern humans". Theor Popul Biol. 48 (2): 198–221. doi:10.1006/tpbi.1995.1026. PMID 7482371.
- ^ Yang, Z. (1 December 2002). "Likelihood and Bayes estimation of ancestral population sizes in hominoids using data from multiple loci" (abstract page). Genetics. 162 (4): 1811–1823. PMC 1462394. PMID 12524351.
- ^ a b c d Chimpanzee Sequencing; Analysis Consortium (2005). "Initial sequence of the chimpanzee genome and comparison with the human genome". Nature. 437 (7055): 69–87. Bibcode:2005Natur.437...69.. doi:10.1038/nature04072. PMID 16136131.
- ^ Olson, M.V. (1999). "When less is more: gene loss as an engine of evolutionary change". Am J Hum Genet. 64 (1): 18–23. doi:10.1086/302219. PMC 1377697. PMID 9915938.
- ^ a b Wang, X.; Grus, W.E.; Zhang, J. (2006). "Gene losses during human origins". PLoS Biol. 4 (3): e52. doi:10.1371/journal.pbio.0040052. PMC 1361800. PMID 16464126.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Demuth, Jeffery P.; Bie, Tijl De; Stajich, Jason E.; Cristianini, Nello; Hahn, Matthew W. (December 2006). Borevitz, Justin (ed.). "The Evolution of Mammalian Gene Families". PLoS ONE. 1: e85. Bibcode:2006PLoSO...1...85D. doi:10.1371/journal.pone.0000085. PMC 1762380. PMID 17183716.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Winter, H.; Langbein, L.; Krawczak, M.; Cooper, D.N.; Suarez, L.F.J.; Rogers, M.A.; Praetzel, S.; Heidt, P.J.; Schweizer, J. (2001). "Human type I hair keratin pseudogene phihHaA has functional orthologs in the chimpanzee and gorilla: evidence for recent inactivation of the human gene after the Pan-Homo divergence". Hum Genet. 108 (1): 37–42. doi:10.1007/s004390000439. PMID 11214905.
- ^ Stedman, H.H.; Kozyak, B.W.; Nelson, A.; Thesier, D.M.; Su, L.T.; Low, D.W.; Bridges, C.R.; Shrager, J.B.; Purvis, N.M. & Mitchell, M.A.; Kozyak; Nelson; Thesier; Su; Low; Bridges; Shrager; Minugh-Purvis; Mitchell (2004). "Myosin gene mutation correlates with anatomical changes in the human lineage". Nature. 428 (6981): 415–418. Bibcode:2004Natur.428..415S. doi:10.1038/nature02358. PMID 15042088.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Perry, G.H.; Verrelli, B.C.; Stone, A.C. (2005). "Comparative analyses reveal a complex history of molecular evolution for human MYH16". Mol Biol Evol. 22 (3): 379–382. doi:10.1093/molbev/msi004. PMID 15470226.
- ^ Hellen, Elizabeth H. B.; Kern, Andrew D. (2015-04-01). "The Role of DNA Insertions in Phenotypic Differentiation between Humans and Other Primates". Genome Biology and Evolution. 7 (4): 1168–1178. doi:10.1093/gbe/evv012. ISSN 1759-6653. PMC 4419785. PMID 25635043.
- ^ Bird, Christine P.; Liu, Maureen; Thomas, Daryl J; Ingle, Catherine E; Beazley, Claude; Miller, Webb; Hurles, Matthew E; Dermitzakis, Emmanouil T (2007). "Fast-evolving noncoding sequences in the human genome". Genome Biology. 8 (6): R118. doi:10.1186/gb-2007-8-6-r118. PMC 2394770. PMID 17578567.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Cheng, Z.; Ventura, M.; She, X.; Khaitovich, P.; Graves, T.; Osoegawa, K.; Church, D; Pieter DeJong, P.; Wilson, R. K.; Paabo, S.; Rocchi, M; Eichler, E. E. (2005). "A genome-wide comparison of recent chimpanzee and human segmental duplications". Nature. 437 (1 September 2005): 88–93. Bibcode:2005Natur.437...88C. doi:10.1038/nature04000. PMID 16136132.
- ^ Kaessmann, H.; Heissig, F.; von Haeseler, A.; Pääbo, S. (1999). "DNA sequence variation in a non-coding region of low recombination on the human X chromosome". Nat Genet. 22 (1): 78–81. doi:10.1038/8785. PMID 10319866.
- ^ Saey, Tina Hesman (2009). "Story one: Team decodes neandertal DNA: Genome draft may reveal secrets of human evolution". Science News. 175 (6): 5–7. doi:10.1002/scin.2009.5591750604.
- ^ Green, Richard E.; Krause; Briggs; Maricic; Stenzel; Kircher; Patterson; Li; Zhai; Fritz; Hansen; Durand; Malaspinas; Jensen; Marques-Bonet; Alkan; Prüfer; Meyer; Burbano; Good; Schultz; Aximu-Petri; Butthof; Höber; Höffner; Siegemund; Weihmann; Nusbaum; Lander; et al. (2010-05-07). "A Draft Sequence of the Neandertal Genome". Science. 328 (5979): 710–722. Bibcode:2010Sci...328..710G. doi:10.1126/science.1188021. PMID 20448178.
- ^ "NEANDERTALS LIVE!". john hawks weblog. Retrieved 2010-12-31.
- ^ a b c d Cann, RL; Stoneking, M; Wilson, AC (1987). "Mitochondrial DNA and human evolution". Nature. 325 (6099): 31–6. Bibcode:1987Natur.325...31C. doi:10.1038/325031a0. PMID 3025745.
- ^ Tishkoff, SA.; Reed, FA.; Friedlaender, FR.; Ehret, C.; Ranciaro, A.; Froment, A.; Hirbo, JB.; Awomoyi, AA.; Bodo, JM. (May 2009). "The genetic structure and history of Africans and African Americans". Science. 324 (5930): 1035–44. doi:10.1126/science.1172257. PMC 2947357. PMID 19407144.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ BBC World News "Africa's genetic secrets unlocked", 1 May 2009; the results were published in the online edition of the journal Science.
- ^ Sayres, Melissa A. Wilson; Venditti, Chris; Pagel, Mark; Makova, Kateryna D. (2011). "Do variations in substitution rates and male mutation bias correlate with life history traits? A study of 32 mammalian genomes". Evolution. 65 (10): 2800–2815. doi:10.1111/j.1558-5646.2011.01337.x. ISSN 0014-3820.
- ^ Ghose, T. (13 November 2012). "Y chromosome study casts doubt on theories regarding junk DNA, sexually dominant men". Huffington Post. Retrieved 5 December 2012.
- ^ Priya Moorjani, Kumarasamy Thangaraj, Nick Patterson, Mark Lipson, Po-Ru Loh, Periyasamy Govindaraj, Bonnie Berger, David Reich, Lalji Singh (5 September 2013). "Genetic Evidence for Recent Population Mixture in India" (PDF). American Journal of Human Genetics. 93 (3): 422–438. doi:10.1016/j.ajhg.2013.07.006. PMC 3769933. PMID 23932107. Retrieved 17 May 2015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Rakesh Tamang, Lalji Singh, Kumarasamy Thangaraj (November 2012). "Complex genetic origin of Indian populations and its implications" (PDF). Journal of Biosciences. 37 (5): 911–919. doi:10.1007/s12038-012-9256-9. Retrieved 17 May 2015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Marc Haber, Dominique Gauguier, Sonia Youhanna, Nick Patterson, Priya Moorjani, Laura R. Botigué, Daniel E. Platt, Elizabeth Matisoo-Smith, David F. Soria-Hernanz, R. Spencer Wells, Jaume Bertranpetit, Chris Tyler-Smith, David Comas, Pierre A. Zalloua (February 28, 2013). "Genome-Wide Diversity in the Levant Reveals Recent Structuring by Culture". PLOS Genetics. 9 (2): e1003316. doi:10.1371/journal.pgen.1003316. PMC 3585000. PMID 23468648. Retrieved 17 May 2015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c Brenna M. Henn, Laura R. Botigué, Simon Gravel, Wei Wang, Abra Brisbin, Jake K. Byrnes, Karima Fadhlaoui-Zid, Pierre A. Zalloua, Andres Moreno-Estrada, Jaume Bertranpetit, Carlos D. Bustamante, David Comas (January 12, 2012). "Genomic Ancestry of North Africans Supports Back-to-Africa Migrations". PLOS Genetics. 8 (1): e1002397. doi:10.1371/journal.pgen.1002397. PMC 3257290. PMID 22253600. Retrieved 17 May 2015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c d Begoña Dobon, Hisham Y. Hassan, Hafid Laayouni, Pierre Luisi, Isis Ricaño-Ponce, Alexandra Zhernakova, Cisca Wijmenga, Hanan Tahir, David Comas, Mihai G. Netea, Jaume Bertranpetit (28 May 2015). "The genetics of East African populations: a Nilo-Saharan component in the African genetic landscape". Scientific Reports. 5: 9996. doi:10.1038/srep09996. Retrieved 13 June 2015.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e Jason A. Hodgson, Connie J. Mulligan, Ali Al-Meeri, Ryan L. Raaum (June 12, 2014). "Early Back-to-Africa Migration into the Horn of Africa". PLOS Genetics. 10 (6): e1004393. doi:10.1371/journal.pgen.1004393.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link); "Supplementary Text S1: Affinities of the Ethio-Somali ancestry component". doi:10.1371/journal.pgen.1004393.s017. Retrieved 17 May 2015.{{cite journal}}: Cite journal requires|journal=(help)CS1 maint: unflagged free DOI (link)
Further reading
- Jobling, Mark A.; Hollox, Edward; Hurles, Matthew; Kivisild, Toomas; Tyler-Smith, Chris (2013). Human Evolutionary Genetics. New York: Garland Science. ISBN 978-0-8153-4148-2. OCLC 829099073.
- Rannala B, Yang Z; Yang (August 2003). "Bayes estimation of species divergence times and ancestral population sizes using DNA sequences from multiple loci". Genetics. 164 (4): 1645–56. PMC 1462670. PMID 12930768.
