RNA interference
RNA interference (RNAi) is a biological process in which RNA molecules inhibit gene expression, typically by causing the destruction of specific mRNA molecules. Historically, it was known by other names, including co-suppression, post-transcriptional gene silencing (PTGS), and quelling. Only after these apparently unrelated processes were fully understood did it become clear that they all described the RNAi phenomenon. Andrew Fire and Craig C. Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNA interference in the nematode worm Caenorhabditis elegans, which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired gene. RNAi is now known as precise, efficient, stable and better than antisense technology for gene suppression.[1]
Two types of small ribonucleic acid (RNA) molecules – microRNA (miRNA) and small interfering RNA (siRNA) – are central to RNA interference. RNAs are the direct products of genes, and these small RNAs can bind to other specific messenger RNA (mRNA) molecules and either increase or decrease their activity, for example by preventing an mRNA from producing a protein. RNA interference has an important role in defending cells against parasitic nucleotide sequences – viruses and transposons. It also influences development.
The RNAi pathway is found in many eukaryotes, including animals, and is initiated by the enzyme Dicer, which cleaves long double-stranded RNA (dsRNA) molecules into short double-stranded fragments of ~20 nucleotide siRNAs. Each siRNA is unwound into two single-stranded RNAs (ssRNAs), the passenger strand and the guide strand. The passenger strand is degraded and the guide strand is incorporated into the RNA-induced silencing complex (RISC). The most well-studied outcome is post-transcriptional gene silencing, which occurs when the guide strand pairs with a complementary sequence in a messenger RNA molecule and induces cleavage by Argonaute, the catalytic component of the RISC complex. In some organisms, this process spreads systemically, despite the initially limited molar concentrations of siRNA.
RNAi is a valuable research tool, both in cell culture and in living organisms, because synthetic dsRNA introduced into cells can selectively and robustly induce suppression of specific genes of interest. RNAi may be used for large-scale screens that systematically shut down each gene in the cell, which can help to identify the components necessary for a particular cellular process or an event such as cell division. The pathway is also used as a practical tool in biotechnology, medicine and insecticides.[2]
Cellular mechanism

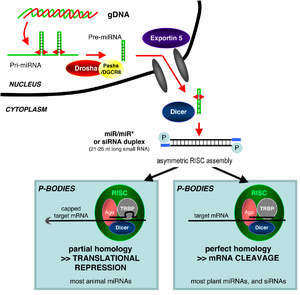
RNAi is an RNA-dependent gene silencing process that is controlled by the RNA-induced silencing complex (RISC) and is initiated by short double-stranded RNA molecules in a cell's cytoplasm, where they interact with the catalytic RISC component argonaute.[4] When the dsRNA is exogenous (coming from infection by a virus with an RNA genome or laboratory manipulations), the RNA is imported directly into the cytoplasm and cleaved to short fragments by Dicer. The initiating dsRNA can also be endogenous (originating in the cell), as in pre-microRNAs expressed from RNA-coding genes in the genome. The primary transcripts from such genes are first processed to form the characteristic stem-loop structure of pre-miRNA in the nucleus, then exported to the cytoplasm. Thus, the two dsRNA pathways, exogenous and endogenous, converge at the RISC.[5]
dsRNA cleavage
Endogenous dsRNA initiates RNAi by activating the ribonuclease protein Dicer,[6] which binds and cleaves double-stranded RNAs (dsRNAs) to produce double-stranded fragments of 20–25 base pairs with a 2-nucleotide overhang at the 3' end.[7] Bioinformatics studies on the genomes of multiple organisms suggest this length maximizes target-gene specificity and minimizes non-specific effects.[8] These short double-stranded fragments are called small interfering RNAs (siRNAs). These siRNAs are then separated into single strands and integrated into an active RISC complex. After integration into the RISC, siRNAs base-pair to their target mRNA and cleave it, thereby preventing it from being used as a translation template.[9]
Exogenous dsRNA is detected and bound by an effector protein, known as RDE-4 in C. elegans and R2D2 in Drosophila, that stimulates dicer activity.[10] This protein only binds long dsRNAs, but the mechanism producing this length specificity is unknown.[10] This RNA-binding protein then facilitates the transfer of cleaved siRNAs to the RISC complex.[11]
In C. elegans this initiation response is amplified through the synthesis of a population of 'secondary' siRNAs during which the dicer-produced initiating or 'primary' siRNAs are used as templates.[12] These 'secondary' siRNAs are structurally distinct from dicer-produced siRNAs and appear to be produced by an RNA-dependent RNA polymerase (RdRP).[13][14]
MicroRNA

MicroRNAs (miRNAs) are genomically encoded non-coding RNAs that help regulate gene expression, particularly during development.[15] The phenomenon of RNA interference, broadly defined, includes the endogenously induced gene silencing effects of miRNAs as well as silencing triggered by foreign dsRNA. Mature miRNAs are structurally similar to siRNAs produced from exogenous dsRNA, but before reaching maturity, miRNAs must first undergo extensive post-transcriptional modification. A miRNA is expressed from a much longer RNA-coding gene as a primary transcript known as a pri-miRNA which is processed, in the cell nucleus, to a 70-nucleotide stem-loop structure called a pre-miRNA by the microprocessor complex. This complex consists of an RNase III enzyme called Drosha and a dsRNA-binding protein DGCR8. The dsRNA portion of this pre-miRNA is bound and cleaved by Dicer to produce the mature miRNA molecule that can be integrated into the RISC complex; thus, miRNA and siRNA share the same downstream cellular machinery.[16] First, viral encoded miRNA was described in EBV.[17] Thereafter, an increasing number of microRNAs have been described in viruses. VIRmiRNA is a comprehensive catalogue covering viral microRNA, their targets and anti-viral miRNAs [18] (see also VIRmiRNA resource: http://crdd.osdd.net/servers/virmirna/).
siRNAs derived from long dsRNA precursors differ from miRNAs in that miRNAs, especially those in animals, typically have incomplete base pairing to a target and inhibit the translation of many different mRNAs with similar sequences. In contrast, siRNAs typically base-pair perfectly and induce mRNA cleavage only in a single, specific target.[19] In Drosophila and C. elegans, miRNA and siRNA are processed by distinct argonaute proteins and dicer enzymes.[20][21]
Three prime untranslated regions and microRNAs
Three prime untranslated regions (3'UTRs) of messenger RNAs (mRNAs) often contain regulatory sequences that post-transcriptionally cause RNA interference. Such 3'-UTRs often contain both binding sites for microRNAs (miRNAs) as well as for regulatory proteins. By binding to specific sites within the 3'-UTR, miRNAs can decrease gene expression of various mRNAs by either inhibiting translation or directly causing degradation of the transcript. The 3'-UTR also may have silencer regions that bind repressor proteins that inhibit the expression of a mRNA.
The 3'-UTR often contains microRNA response elements (MREs). MREs are sequences to which miRNAs bind. These are prevalent motifs within 3'-UTRs. Among all regulatory motifs within the 3'-UTRs (e.g. including silencer regions), MREs make up about half of the motifs.
As of 2014, the miRBase web site,[22] an archive of miRNA sequences and annotations, listed 28,645 entries in 233 biologic species. Of these, 1,881 miRNAs were in annotated human miRNA loci. miRNAs were predicted to have an average of about four hundred target mRNAs (affecting expression of several hundred genes).[23] Friedman et al.[23] estimate that >45,000 miRNA target sites within human mRNA 3'UTRs are conserved above background levels, and >60% of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs.
Direct experiments show that a single miRNA can reduce the stability of hundreds of unique mRNAs.[24] Other experiments show that a single miRNA may repress the production of hundreds of proteins, but that this repression often is relatively mild (less than 2-fold).[25][26]
The effects of miRNA dysregulation of gene expression seem to be important in cancer.[27] For instance, in gastrointestinal cancers, nine miRNAs have been identified as epigenetically altered and effective in down regulating DNA repair enzymes.[28]
The effects of miRNA dysregulation of gene expression also seem to be important in neuropsychiatric disorders, such as schizophrenia, bipolar disorder, major depression, Parkinson's disease, Alzheimer's disease and autism spectrum disorders.[29][30][31]
RISC activation and catalysis

The active components of an RNA-induced silencing complex (RISC) are endonucleases called argonaute proteins, which cleave the target mRNA strand complementary to their bound siRNA.[4] As the fragments produced by dicer are double-stranded, they could each in theory produce a functional siRNA. However, only one of the two strands, which is known as the guide strand, binds the argonaute protein and directs gene silencing. The other anti-guide strand or passenger strand is degraded during RISC activation.[32] Although it was first believed that an ATP-dependent helicase separated these two strands,[33] the process proved to be ATP-independent and performed directly by the protein components of RISC.[34][35] However, an in vitro kinetic analysis of RNAi in the presence and absence of ATP showed that ATP may be required to unwind and remove the cleaved mRNA strand from the RISC complex after catalysis.[36] The guide strand tends to be the one whose 5' end is less stably paired to its complement,[37] but strand selection is unaffected by the direction in which dicer cleaves the dsRNA before RISC incorporation.[38] Instead, the R2D2 protein may serve as the differentiating factor by binding the more-stable 5' end of the passenger strand.[39]
The structural basis for binding of RNA to the argonaute protein was examined by X-ray crystallography of the binding domain of an RNA-bound argonaute protein. Here, the phosphorylated 5' end of the RNA strand enters a conserved basic surface pocket and makes contacts through a divalent cation (an atom with two positive charges) such as magnesium and by aromatic stacking (a process that allows more than one atom to share an electron by passing it back and forth) between the 5' nucleotide in the siRNA and a conserved tyrosine residue. This site is thought to form a nucleation site for the binding of the siRNA to its mRNA target.[40] Analysis of the inhibitory effect of mismatches in either the 5’ or 3’ end of the guide strand has demonstrated that the 5’ end of the guide strand is likely responsible for matching and binding the target mRNA, while the 3’ end is responsible for physically arranging target mRNA into a cleavage-favorable RISC region.[36]
It is not understood how the activated RISC complex locates complementary mRNAs within the cell. Although the cleavage process has been proposed to be linked to translation, translation of the mRNA target is not essential for RNAi-mediated degradation.[41] Indeed, RNAi may be more effective against mRNA targets that are not translated.[42] Argonaute proteins are localized to specific regions in the cytoplasm called P-bodies (also cytoplasmic bodies or GW bodies), which are regions with high rates of mRNA decay;[43] miRNA activity is also clustered in P-bodies.[44] Disruption of P-bodies decreases the efficiency of RNA interference, suggesting that they are a critical site in the RNAi process.[45]
Transcriptional silencing
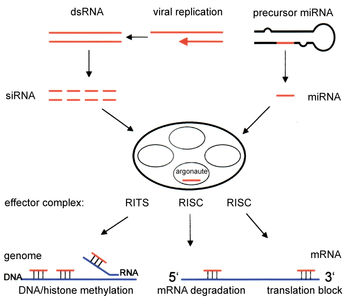
Components of the RNAi pathway are used in many eukaryotes in the maintenance of the organization and structure of their genomes. Modification of histones and associated induction of heterochromatin formation serves to downregulate genes pre-transcriptionally;[47] this process is referred to as RNA-induced transcriptional silencing (RITS), and is carried out by a complex of proteins called the RITS complex. In fission yeast this complex contains argonaute, a chromodomain protein Chp1, and a protein called Tas3 of unknown function.[48] As a consequence, the induction and spread of heterochromatic regions requires the argonaute and RdRP proteins.[49] Indeed, deletion of these genes in the fission yeast S. pombe disrupts histone methylation and centromere formation,[50] causing slow or stalled anaphase during cell division.[51] In some cases, similar processes associated with histone modification have been observed to transcriptionally upregulate genes.[52]
The mechanism by which the RITS complex induces heterochromatin formation and organization is not well understood. Most studies have focused on the mating-type region in fission yeast, which may not be representative of activities in other genomic regions/organisms. In maintenance of existing heterochromatin regions, RITS forms a complex with siRNAs complementary to the local genes and stably binds local methylated histones, acting co-transcriptionally to degrade any nascent pre-mRNA transcripts that are initiated by RNA polymerase. The formation of such a heterochromatin region, though not its maintenance, is dicer-dependent, presumably because dicer is required to generate the initial complement of siRNAs that target subsequent transcripts.[53] Heterochromatin maintenance has been suggested to function as a self-reinforcing feedback loop, as new siRNAs are formed from the occasional nascent transcripts by RdRP for incorporation into local RITS complexes.[54] The relevance of observations from fission yeast mating-type regions and centromeres to mammals is not clear, as heterochromatin maintenance in mammalian cells may be independent of the components of the RNAi pathway.[55]
Crosstalk with RNA editing
The type of RNA editing that is most prevalent in higher eukaryotes converts adenosine nucleotides into inosine in dsRNAs via the enzyme adenosine deaminase (ADAR).[56] It was originally proposed in 2000 that the RNAi and A→I RNA editing pathways might compete for a common dsRNA substrate.[57] Some pre-miRNAs do undergo A→I RNA editing[58][59] and this mechanism may regulate the processing and expression of mature miRNAs.[59] Furthermore, at least one mammalian ADAR can sequester siRNAs from RNAi pathway components.[60] Further support for this model comes from studies on ADAR-null C. elegans strains indicating that A→I RNA editing may counteract RNAi silencing of endogenous genes and transgenes.[61]
Variation among organisms
Organisms vary in their ability to take up foreign dsRNA and use it in the RNAi pathway. The effects of RNA interference can be both systemic and heritable in plants and C. elegans, although not in Drosophila or mammals. In plants, RNAi is thought to propagate by the transfer of siRNAs between cells through plasmodesmata (channels in the cell walls that enable communication and transport).[33] Heritability comes from methylation of promoters targeted by RNAi; the new methylation pattern is copied in each new generation of the cell.[63] A broad general distinction between plants and animals lies in the targeting of endogenously produced miRNAs; in plants, miRNAs are usually perfectly or nearly perfectly complementary to their target genes and induce direct mRNA cleavage by RISC, while animals' miRNAs tend to be more divergent in sequence and induce translational repression.[62] This translational effect may be produced by inhibiting the interactions of translation initiation factors with the messenger RNA's polyadenine tail.[64]
Some eukaryotic protozoa such as Leishmania major and Trypanosoma cruzi lack the RNAi pathway entirely.[65][66] Most or all of the components are also missing in some fungi, most notably the model organism Saccharomyces cerevisiae.[67] The presence of RNAi in other budding yeast species such as Saccharomyces castellii and Candida albicans, further demonstrates that inducing two RNAi-related proteins from S. castellii facilitates RNAi in S. cerevisiae.[68] That certain ascomycetes and basidiomycetes are missing RNA interference pathways indicates that proteins required for RNA silencing have been lost independently from many fungal lineages, possibly due to the evolution of a novel pathway with similar function, or to the lack of selective advantage in certain niches.[69]
Related prokaryotic systems
Gene expression in prokaryotes is influenced by an RNA-based system similar in some respects to RNAi. Here, RNA-encoding genes control mRNA abundance or translation by producing a complementary RNA that anneals to an mRNA. However these regulatory RNAs are not generally considered to be analogous to miRNAs because the dicer enzyme is not involved.[70] It has been suggested that CRISPR interference systems in prokaryotes are analogous to eukaryotic RNA interference systems, although none of the protein components are orthologous.[71]
Biological functions
Immunity
RNA interference is a vital part of the immune response to viruses and other foreign genetic material, especially in plants where it may also prevent the self-propagation of transposons.[72] Plants such as Arabidopsis thaliana express multiple dicer homologs that are specialized to react differently when the plant is exposed to different viruses.[73] Even before the RNAi pathway was fully understood, it was known that induced gene silencing in plants could spread throughout the plant in a systemic effect and could be transferred from stock to scion plants via grafting.[74] This phenomenon has since been recognized as a feature of the plant adaptive immune system and allows the entire plant to respond to a virus after an initial localized encounter.[75] In response, many plant viruses have evolved elaborate mechanisms to suppress the RNAi response.[76] These include viral proteins that bind short double-stranded RNA fragments with single-stranded overhang ends, such as those produced by dicer.[77] Some plant genomes also express endogenous siRNAs in response to infection by specific types of bacteria.[78] These effects may be part of a generalized response to pathogens that downregulates any metabolic process in the host that aids the infection process.[79]
Although animals generally express fewer variants of the dicer enzyme than plants, RNAi in some animals produces an antiviral response. In both juvenile and adult Drosophila, RNA interference is important in antiviral innate immunity and is active against pathogens such as Drosophila X virus.[80][81] A similar role in immunity may operate in C. elegans, as argonaute proteins are upregulated in response to viruses and worms that overexpress components of the RNAi pathway are resistant to viral infection.[82][83]
The role of RNA interference in mammalian innate immunity is poorly understood, and relatively little data is available. However, the existence of viruses that encode genes able to suppress the RNAi response in mammalian cells may be evidence in favour of an RNAi-dependent mammalian immune response,[84][85] although this hypothesis has been challenged as poorly substantiated.[86] Maillard et al.[87] and Li et al.[88] provide evidence for the existence of a functional antiviral RNAi pathway in mammalian cells. Other functions for RNAi in mammalian viruses also exist, such as miRNAs expressed by the herpes virus that may act as heterochromatin organization triggers to mediate viral latency.[89]
Downregulation of genes
Endogenously expressed miRNAs, including both intronic and intergenic miRNAs, are most important in translational repression[62] and in the regulation of development, especially on the timing of morphogenesis and the maintenance of undifferentiated or incompletely differentiated cell types such as stem cells.[90] The role of endogenously expressed miRNA in downregulating gene expression was first described in C. elegans in 1993.[91] In plants this function was discovered when the "JAW microRNA" of Arabidopsis was shown to be involved in the regulation of several genes that control plant shape.[92] In plants, the majority of genes regulated by miRNAs are transcription factors;[93] thus miRNA activity is particularly wide-ranging and regulates entire gene networks during development by modulating the expression of key regulatory genes, including transcription factors as well as F-box proteins.[94] In many organisms, including humans, miRNAs are linked to the formation of tumors and dysregulation of the cell cycle. Here, miRNAs can function as both oncogenes and tumor suppressors.[95]
Upregulation of genes
RNA sequences (siRNA and miRNA) that are complementary to parts of a promoter can increase gene transcription, a phenomenon dubbed RNA activation. Part of the mechanism for how these RNA upregulate genes is known: dicer and argonaute are involved, possibly via histone demethylation.[96] miRNAs have been proposed to upregulate their target genes upon cell cycle arrest, via unknown mechanisms.[97]
Evolution
Based on parsimony-based phylogenetic analysis, the most recent common ancestor of all eukaryotes most likely already possessed an early RNA interference pathway; the absence of the pathway in certain eukaryotes is thought to be a derived characteristic.[98] This ancestral RNAi system probably contained at least one dicer-like protein, one argonaute, one PIWI protein, and an RNA-dependent RNA polymerase that may also have played other cellular roles. A large-scale comparative genomics study likewise indicates that the eukaryotic crown group already possessed these components, which may then have had closer functional associations with generalized RNA degradation systems such as the exosome.[99] This study also suggests that the RNA-binding argonaute protein family, which is shared among eukaryotes, most archaea, and at least some bacteria (such as Aquifex aeolicus), is homologous to and originally evolved from components of the translation initiation system.
The ancestral function of the RNAi system is generally agreed to have been immune defense against exogenous genetic elements such as transposons and viral genomes.[98][100] Related functions such as histone modification may have already been present in the ancestor of modern eukaryotes, although other functions such as regulation of development by miRNA are thought to have evolved later.[98]
RNA interference genes, as components of the antiviral innate immune system in many eukaryotes, are involved in an evolutionary arms race with viral genes. Some viruses have evolved mechanisms for suppressing the RNAi response in their host cells, particularly for plant viruses.[76] Studies of evolutionary rates in Drosophila have shown that genes in the RNAi pathway are subject to strong directional selection and are among the fastest-evolving genes in the Drosophila genome.[101]
Applications
Gene knockdown
The RNA interference pathway is often exploited in experimental biology to study the function of genes in cell culture and in vivo in model organisms.[4] Double-stranded RNA is synthesized with a sequence complementary to a gene of interest and introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway. Using this mechanism, researchers can cause a drastic decrease in the expression of a targeted gene. Studying the effects of this decrease can show the physiological role of the gene product. Since RNAi may not totally abolish expression of the gene, this technique is sometimes referred as a "knockdown", to distinguish it from "knockout" procedures in which expression of a gene is entirely eliminated.[102]
Extensive efforts in computational biology have been directed toward the design of successful dsRNA reagents that maximize gene knockdown but minimize "off-target" effects. Off-target effects arise when an introduced RNA has a base sequence that can pair with and thus reduce the expression of multiple genes. Such problems occur more frequently when the dsRNA contains repetitive sequences. It has been estimated from studying the genomes of humans, C. elegans and S. pombe that about 10% of possible siRNAs have substantial off-target effects.[8] A multitude of software tools have been developed implementing algorithms for the design of general[103][104] mammal-specific,[105] and virus-specific[106] siRNAs that are automatically checked for possible cross-reactivity.
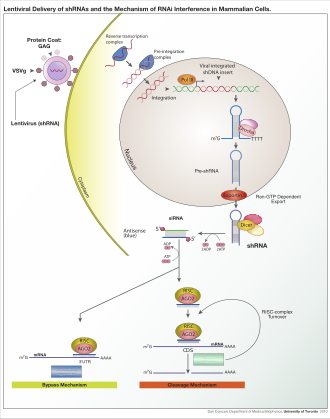
Depending on the organism and experimental system, the exogenous RNA may be a long strand designed to be cleaved by dicer, or short RNAs designed to serve as siRNA substrates. In most mammalian cells, shorter RNAs are used because long double-stranded RNA molecules induce the mammalian interferon response, a form of innate immunity that reacts nonspecifically to foreign genetic material.[107] Mouse oocytes and cells from early mouse embryos lack this reaction to exogenous dsRNA and are therefore a common model system for studying mammalian gene-knockdown effects.[108] Specialized laboratory techniques have also been developed to improve the utility of RNAi in mammalian systems by avoiding the direct introduction of siRNA, for example, by stable transfection with a plasmid encoding the appropriate sequence from which siRNAs can be transcribed,[109] or by more elaborate lentiviral vector systems allowing the inducible activation or deactivation of transcription, known as conditional RNAi.[110][111]
Functional genomics

Most functional genomics applications of RNAi in animals have used C. elegans[112] and Drosophila,[113] as these are the common model organisms in which RNAi is most effective. C. elegans is particularly useful for RNAi research for two reasons: firstly, the effects of gene silencing are generally heritable, and secondly because delivery of the dsRNA is extremely simple. Through a mechanism whose details are poorly understood, bacteria such as E. coli that carry the desired dsRNA can be fed to the worms and will transfer their RNA payload to the worm via the intestinal tract. This "delivery by feeding" is just as effective at inducing gene silencing as more costly and time-consuming delivery methods, such as soaking the worms in dsRNA solution and injecting dsRNA into the gonads.[114] Although delivery is more difficult in most other organisms, efforts are also underway to undertake large-scale genomic screening applications in cell culture with mammalian cells.[115]
Approaches to the design of genome-wide RNAi libraries can require more sophistication than the design of a single siRNA for a defined set of experimental conditions. Artificial neural networks are frequently used to design siRNA libraries[116] and to predict their likely efficiency at gene knockdown.[117] Mass genomic screening is widely seen as a promising method for genome annotation and has triggered the development of high-throughput screening methods based on microarrays.[118][119] However, the utility of these screens and the ability of techniques developed on model organisms to generalize to even closely related species has been questioned, for example from C. elegans to related parasitic nematodes.[120][121]
Functional genomics using RNAi is a particularly attractive technique for genomic mapping and annotation in plants because many plants are polyploid, which presents substantial challenges for more traditional genetic engineering methods. For example, RNAi has been successfully used for functional genomics studies in bread wheat (which is hexaploid)[122] as well as more common plant model systems Arabidopsis and maize.[123]
Medicine

It may be possible to exploit RNA interference in therapy. Although it is difficult to introduce long dsRNA strands into mammalian cells due to the interferon response, the use of short interfering RNA has been more successful.[125] Among the first applications to reach clinical trials were in the treatment of macular degeneration and respiratory syncytial virus.[126] RNAi has also been shown to be effective in reversing induced liver failure in mouse models.[127]
Antiviral
Potential antiviral therapies include topical microbicide treatments that use RNAi to treat infection (at Harvard Medical School; in mice, so far) by herpes simplex virus type 2 and the inhibition of viral gene expression in cancerous cells,[128] knockdown of host receptors and coreceptors for HIV,[129] the silencing of hepatitis A[130] and hepatitis B genes,[131] silencing of influenza gene expression,[132] and inhibition of measles viral replication.[133] Potential treatments for neurodegenerative diseases have also been proposed, with particular attention to polyglutamine diseases such as Huntington's disease.[134]
RNA interference-based applications are being developed to target persistent HIV-1 infection. Viruses like HIV-1 are particularly difficult targets for RNAi-attack because they are escape-prone, which requires combinatorial RNAi strategies to prevent viral escape.[135]
Cancer
RNA interference is also a promising way to treat cancers by silencing genes differentially upregulated in tumor cells or genes involved in cell division.[136][137] A key area of research in the use of RNAi for clinical applications is the development of a safe delivery method, which to date has involved mainly viral vector systems similar to those suggested for gene therapy.[138][139]
Due to safety concerns with viral vectors, nonviral delivery methods, typically employing lipid-based[140] or polymeric[141] vectors, are also promising candidates. Computational modeling of nonviral siRNA delivery paired with in vitro and in vivo gene knockdown studies elucidated the temporal behavior of RNAi in these systems. The model used an input bolus dose of siRNA and computationally and experimentally showed that knockdown duration was dependent mainly on the doubling time of the cells to which siRNA was delivered, while peak knockdown depended primarily on the delivered dose. Kinetic considerations of RNAi are imperative to safe and effective dosing schedules as nonviral methods of inducing RNAi continue to be developed.[142][143]
Safety
Despite the proliferation of promising cell culture studies for RNAi-based drugs, some concern has been raised regarding the safety of RNA interference, especially the potential for "off-target" effects in which a gene with a coincidentally similar sequence to the targeted gene is also repressed.[144] A computational genomics study estimated that the error rate of off-target interactions is about 10%.[8] One major study of liver disease in mice reported that 23 out of 49 distinct RNAi treatment protocols resulted in death.[145] Researchers hypothesized this alarmingly high rate to be the result of "oversaturation" of the dsRNA pathway,[146] due to the use of shRNAs that have to be processed in the nucleus and exported to the cytoplasm using an active mechanism. Such considerations are under active investigation, to reduce their impact in the potential therapeutic applications.
RNAi in vivo delivery to tissues still eludes science—especially to tissues deep within the body. RNAi delivery is only easily accessible to surface tissues such as the eye and respiratory tract. In these instances, siRNA has been used in direct contact with the tissue for transport. The resulting RNAi successfully focused on target genes. When delivering siRNA to deep tissues, the siRNA must be protected from nucleases, but targeting specific areas becomes the main difficulty. This difficulty has been combatted with high dosage levels of siRNA to ensure the tissues have been reached, however in these cases hepatotoxicity was reported.[147]
Biotechnology
RNA interference has been used for applications in biotechnology and is nearing commercialization in others. RNAi has developed many novel crops such as nicotinefree tobacco, decaffeinated coffee, nutrient fortified and hypoallergenic crops. The genetically engineered Arctic apples are near close to receive US approval. The apples were produced by RNAi suppression of PPO (polyphenol oxidase) gene making apple varieties that will not undergo browning after being sliced. PPO-silenced apples are unable to convert chlorogenic acid into quinone product.[1]
There are several opportunities for the applications of RNAi in crop science for its improvement such as stress tolerance and enhanced nutritional level. RNAi will prove its potential for inhibition of photorespiration to enhance the productivity of C3 plants. This knockdown technology may be useful in inducing early flowering, delayed ripening, delayed senescence, breaking dormancy, stress-free plants, overcoming self-sterility, etc.[1]
Foods
RNAi has been used to genetically engineer plants to produce lower levels of natural plant toxins. Such techniques take advantage of the stable and heritable RNAi phenotype in plant stocks. Cotton seeds are rich in dietary protein but naturally contain the toxic terpenoid product gossypol, making them unsuitable for human consumption. RNAi has been used to produce cotton stocks whose seeds contain reduced levels of delta-cadinene synthase, a key enzyme in gossypol production, without affecting the enzyme's production in other parts of the plant, where gossypol is itself important in preventing damage from plant pests.[148] Similar efforts have been directed toward the reduction of the cyanogenic natural product linamarin in cassava plants.[149]
No plant products that use RNAi-based genetic engineering have yet exited the experimental stage. Development efforts have successfully reduced the levels of allergens in tomato plants[150] and fortification of plants such as tomatoes with dietary antioxidants.[151] Previous commercial products, including the Flavr Savr tomato and two cultivars of ringspot-resistant papaya, were originally developed using antisense technology but likely exploited the RNAi pathway.[152][153]
Other crops
Another effort decreased the precursors of likely carcinogens in tobacco plants.[154] Other plant traits that have been engineered in the laboratory include the production of non-narcotic natural products by the opium poppy[155] and resistance to common plant viruses.[156]
Insecticide
RNAi is under development as an insecticide, employing multiple approaches, including genetic engineering and topical application. Cells in the midgut of many larvae take up the molecules and help spread the signal throughout the insect's body.[2]
RNAi has varying effects in different species of Lepidoptera (butterflies and moths).[157] Possibly because their saliva is better at breaking down RNA, the cotton bollworm, the beet armyworm and the Asiatic rice borer have so far not been proven susceptible to RNAi by feeding.[2]
To develop resistance to RNAi, the western corn rootworm would have to change the genetic sequence of its Snf7 gene at multiple sites. Combining multiple strategies, such as engineering the protein Cry, derived from a bacterium called Bacillus thuringiensis (Bt), and RNAi in one plant delay the onset of resistance.[2]
Transgenic plants
Transgenic crops have been made to express small bits of RNA, carefully chosen to silence crucial genes in target pests. RNAs exist that affect only insects that have specific genetic sequences. In 2009 a study showed RNAs that could kill any one of four fruit fly species while not harming the other three.[2]
In 2012 Syngenta bought Belgian RNAi firm Devgen for $522 million and Monsanto paid $29.2 million for the exclusive rights to intellectual property from Alnylam Pharmaceuticals. The International Potato Center in Lima, Peru is looking for genes to target in the sweet potato weevil, a beetle whose larvae ravage sweet potatoes globally. Other researchers are trying to silence genes in ants, caterpillars and pollen beetles. Monsanto will likely be first to market, with a transgenic corn seed that expresses dsRNA based on gene Snf7 from the western corn rootworm, a beetle whose larvae annually cause one billion dollars in damage in the United States alone. A 2012 paper showed that silencing Snf7 stunts larval growth, killing them within days. In 2013 the same team showed that the RNA affects very few other species.[2]
Topical
Alternatively dsRNA can be supplied without genetic engineering. One approach is to add them to irrigation water. The molecules are absorbed into the plants' vascular system and poison insects feeding on them. Another approach involves spraying RNA like a conventional pesticide. This would allow faster adaptation to resistance. Such approaches would require low cost sources of RNAs that do not currently exist.[2]
Genome-scale screening
Genome-scale RNAi research relies on high-throughput screening (HTS) technology. RNAi HTS technology allows genome-wide loss-of-function screening and is broadly used in the identification of genes associated with specific phenotypes. This technology has been hailed as the second genomics wave, following the first genomics wave of gene expression microarray and single nucleotide polymorphism discovery platforms.[158] One major advantage of genome-scale RNAi screening is its ability to simultaneously interrogate thousands of genes. With the ability to generate a large amount of data per experiment, genome-scale RNAi screening has led to an explosion data generation rates. Exploiting such large data sets is a fundamental challenge, requiring suitable statistics/bioinformatics methods. The basic process of cell-based RNAi screening includes the choice of an RNAi library, robust and stable cell types, transfection with RNAi agents, treatment/incubation, signal detection, analysis and identification of important genes or therapeutical targets.[159]
History

The discovery of RNAi was preceded first by observations of transcriptional inhibition by antisense RNA expressed in transgenic plants,[161] and more directly by reports of unexpected outcomes in experiments performed by plant scientists in the United States and the Netherlands in the early 1990s.[162] In an attempt to alter flower colors in petunias, researchers introduced additional copies of a gene encoding chalcone synthase, a key enzyme for flower pigmentation into petunia plants of normally pink or violet flower color. The overexpressed gene was expected to result in darker flowers, but instead produced less pigmented, fully or partially white flowers, indicating that the activity of chalcone synthase had been substantially decreased; in fact, both the endogenous genes and the transgenes were downregulated in the white flowers. Soon after, a related event termed quelling was noted in the fungus Neurospora crassa,[163] although it was not immediately recognized as related. Further investigation of the phenomenon in plants indicated that the downregulation was due to post-transcriptional inhibition of gene expression via an increased rate of mRNA degradation.[164] This phenomenon was called co-suppression of gene expression, but the molecular mechanism remained unknown.[165]
Not long after, plant virologists working on improving plant resistance to viral diseases observed a similar unexpected phenomenon. While it was known that plants expressing virus-specific proteins showed enhanced tolerance or resistance to viral infection, it was not expected that plants carrying only short, non-coding regions of viral RNA sequences would show similar levels of protection. Researchers believed that viral RNA produced by transgenes could also inhibit viral replication.[166] The reverse experiment, in which short sequences of plant genes were introduced into viruses, showed that the targeted gene was suppressed in an infected plant. This phenomenon was labeled "virus-induced gene silencing" (VIGS), and the set of such phenomena were collectively called post transcriptional gene silencing.[167]
After these initial observations in plants, laboratories searched for this phenomenon in other organisms.[168][169] Craig C. Mello and Andrew Fire's 1998 Nature paper reported a potent gene silencing effect after injecting double stranded RNA into C. elegans.[170] In investigating the regulation of muscle protein production, they observed that neither mRNA nor antisense RNA injections had an effect on protein production, but double-stranded RNA successfully silenced the targeted gene. As a result of this work, they coined the term RNAi. This discovery represented the first identification of the causative agent for the phenomenon. Fire and Mello were awarded the 2006 Nobel Prize in Physiology or Medicine.[4]
See also
References
- ^ a b c Saurabh Satyajit, Vidyarthi AS, Prasad D (2014). "RNA interference: concept to reality in crop improvement". Planta. 239 (3): 543–564. doi:10.1007/s00425-013-2019-5.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f g Kupferschmidt, K. (2013). "A Lethal Dose of RNA". Science. 341 (6147): 732–3. doi:10.1126/science.341.6147.732. PMID 23950525.
- ^ Macrae I, Zhou K, Li F, Repic A, Brooks A, Cande W, Adams P, Doudna J; Zhou; Li; Repic; Brooks; Cande; Adams; Doudna (2006). "Structural basis for double-stranded RNA processing by dicer". Science. 311 (5758): 195–8. Bibcode:2006Sci...311..195M. doi:10.1126/science.1121638. PMID 16410517.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Daneholt, Bertil. "Advanced Information: RNA interference". The Nobel Prize in Physiology or Medicine 2006. Archived from the original on 2007-01-20. Retrieved 2007-01-25.
- ^ Bagasra O, Prilliman KR (2004). "RNA interference: the molecular immune system" (PDF). J. Mol. Histol. 35 (6): 545–53. doi:10.1007/s10735-004-2192-8. PMID 15614608.
- ^ Bernstein E, Caudy A, Hammond S, Hannon G (2001). "Role for a bidentate ribonuclease in the initiation step of RNA interference". Nature. 409 (6818): 363–6. doi:10.1038/35053110. PMID 11201747.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Siomi, Haruhiko; Siomi, Mikiko C. (22 January 2009). "On the road to reading the RNA-interference code". Nature. 457 (7228): 396–404. Bibcode:2009Natur.457..396S. doi:10.1038/nature07754. PMID 19158785.
Zamore P, Tuschl T, Sharp P, Bartel D (2000). "RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals". Cell. 101 (1): 25–33. doi:10.1016/S0092-8674(00)80620-0. PMID 10778853.{{cite journal}}: CS1 maint: multiple names: authors list (link)
Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall W, Karpilow J, Khvorova A (2005). "The contributions of dsRNA structure to Dicer specificity and efficiency". RNA. 11 (5): 674–82. doi:10.1261/rna.7272305. PMC 1370754. PMID 15811921.{{cite journal}}: CS1 maint: multiple names: authors list (link)
Castanotto, Daniela; Rossi, John J. (22 January 2009). "The promises and pitfalls of RNA-interference-based therapeutics". Nature. 457 (7228): 426–433. Bibcode:2009Natur.457..426C. doi:10.1038/nature07758. PMC 2702667. PMID 19158789. - ^ a b c Qiu S, Adema C, Lane T (2005). "A computational study of off-target effects of RNA interference". Nucleic Acids Res. 33 (6): 1834–47. doi:10.1093/nar/gki324. PMC 1072799. PMID 15800213.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ahlquist P (2002). "RNA-dependent RNA polymerases, viruses, and RNA silencing". Science. 296 (5571): 1270–3. Bibcode:2002Sci...296.1270A. doi:10.1126/science.1069132. PMID 12016304.
- ^ a b Parker G, Eckert D, Bass B (2006). "RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA". RNA. 12 (5): 807–18. doi:10.1261/rna.2338706. PMC 1440910. PMID 16603715.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Liu Q, Rand T, Kalidas S, Du F, Kim H, Smith D, Wang X; Rand; Kalidas; Du; Kim; Smith; Wang (2003). "R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway". Science. 301 (5641): 1921–5. Bibcode:2003Sci...301.1921L. doi:10.1126/science.1088710. PMID 14512631.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Baulcombe D (2007). "Molecular biology. Amplified silencing". Science. 315 (5809): 199–200. doi:10.1126/science.1138030. PMID 17218517.
- ^ Pak J, Fire A; Fire (2007). "Distinct populations of primary and secondary effectors during RNAi in C. elegans". Science. 315 (5809): 241–4. Bibcode:2007Sci...315..241P. doi:10.1126/science.1132839. PMID 17124291.
- ^ Sijen T, Steiner F, Thijssen K, Plasterk R; Steiner; Thijssen; Plasterk (2007). "Secondary siRNAs result from unprimed RNA synthesis and form a distinct class". Science. 315 (5809): 244–7. Bibcode:2007Sci...315..244S. doi:10.1126/science.1136699. PMID 17158288.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang QL, Li ZH (2007). "The functions of microRNAs in plants". Front. Biosci. 12: 3975–82. doi:10.2741/2364. PMID 17485351.
Zhao Y, Srivastava D (2007). "A developmental view of microRNA function". Trends Biochem. Sci. 32 (4): 189–97. doi:10.1016/j.tibs.2007.02.006. PMID 17350266. - ^ Gregory R, Chendrimada T, Shiekhattar R (2006). "MicroRNA biogenesis: isolation and characterization of the microprocessor complex". Methods Mol Biol. 342: 33–47. doi:10.1385/1-59745-123-1:33. ISBN 1-59745-123-1. PMID 16957365.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Pfeffer, Sébastien; Zavolan, Mihaela; Grässer, Friedrich A.; Chien, Minchen; Russo, James J.; Ju, Jingyue; John, Bino; Enright, Anton J.; Marks, Debora (2004-04-30). "Identification of virus-encoded microRNAs". Science. 304 (5671): 734–736. doi:10.1126/science.1096781. ISSN 1095-9203. PMID 15118162.
- ^ Qureshi, Abid; Thakur, Nishant; Monga, Isha; Thakur, Anamika; Kumar, Manoj (2014-01-01). "VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets". Database: The Journal of Biological Databases and Curation. 2014: bau103. doi:10.1093/database/bau103. ISSN 1758-0463. PMC 4224276. PMID 25380780.
- ^ Pillai RS, Bhattacharyya SN, Filipowicz W (2007). "Repression of protein synthesis by miRNAs: how many mechanisms?". Trends Cell Biol. 17 (3): 118–26. doi:10.1016/j.tcb.2006.12.007. PMID 17197185.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Okamura K, Ishizuka A, Siomi H, Siomi M (2004). "Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways". Genes Dev. 18 (14): 1655–66. doi:10.1101/gad.1210204. PMC 478188. PMID 15231716.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lee Y, Nakahara K, Pham J, Kim K, He Z, Sontheimer E, Carthew R (2004). "Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways". Cell. 117 (1): 69–81. doi:10.1016/S0092-8674(04)00261-2. PMID 15066283.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ miRBase.org
- ^ a b Friedman RC, Farh KK, Burge CB, Bartel DP (2009). "Most mammalian mRNAs are conserved targets of microRNAs". Genome Res. 19 (1): 92–105. doi:10.1101/gr.082701.108. PMC 2612969. PMID 18955434.
- ^ Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM; Lau; Garrett-Engele; Grimson; Schelter; Castle; Bartel; Linsley; Johnson (February 2005). "Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs". Nature. 433 (7027): 769–73. Bibcode:2005Natur.433..769L. doi:10.1038/nature03315. PMID 15685193.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N; Schwanhäusser; Thierfelder; Fang; Khanin; Rajewsky (September 2008). "Widespread changes in protein synthesis induced by microRNAs". Nature. 455 (7209): 58–63. doi:10.1038/nature07228. PMID 18668040.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP; Villén; Shin; Camargo; Gygi; Bartel (September 2008). "The impact of microRNAs on protein output". Nature. 455 (7209): 64–71. doi:10.1038/nature07242. PMC 2745094. PMID 18668037.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Palmero EI, de Campos SG, Campos M, de Souza NC, Guerreiro ID, Carvalho AL, Marques MM (2011). "Mechanisms and role of microRNA deregulation in cancer onset and progression". Genet. Mol. Biol. 34 (3): 363–70. doi:10.1590/S1415-47572011000300001. PMC 3168173. PMID 21931505.
- ^ Bernstein C, Bernstein H (2015). "Epigenetic reduction of DNA repair in progression to gastrointestinal cancer". World J Gastrointest Oncol. 7 (5): 30–46. doi:10.4251/wjgo.v7.i5.30. PMC 4434036. PMID 25987950.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L (2014). "Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders". Front Cell Neurosci. 8: 75. doi:10.3389/fncel.2014.00075. PMC 3949217. PMID 24653674.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Mellios N, Sur M (2012). "The Emerging Role of microRNAs in Schizophrenia and Autism Spectrum Disorders". Front Psychiatry. 3: 39. doi:10.3389/fpsyt.2012.00039. PMC 3336189. PMID 22539927.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Geaghan M, Cairns MJ (2015). "MicroRNA and Posttranscriptional Dysregulation in Psychiatry". Biol. Psychiatry. 78 (4): 231–9. doi:10.1016/j.biopsych.2014.12.009. PMID 25636176.
- ^ Gregory R, Chendrimada T, Cooch N, Shiekhattar R (2005). "Human RISC couples microRNA biogenesis and posttranscriptional gene silencing". Cell. 123 (4): 631–40. doi:10.1016/j.cell.2005.10.022. PMID 16271387.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipurksy SL, Darnell J (2004). Molecular Cell Biology (5th ed.). WH Freeman: New York, NY. ISBN 978-0-7167-4366-8.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Matranga C, Tomari Y, Shin C, Bartel D, Zamore P (2005). "Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes". Cell. 123 (4): 607–20. doi:10.1016/j.cell.2005.08.044. PMID 16271386.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Leuschner P, Ameres S, Kueng S, Martinez J (2006). "Cleavage of the siRNA passenger strand during RISC assembly in human cells". EMBO Rep. 7 (3): 314–20. doi:10.1038/sj.embor.7400637. PMC 1456892. PMID 16439995.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Haley, B; Zamore B (2004). "Kinetic analysis of the RNAi enzyme complex". Nature Structural & Molecular Biology. 11 (7): 599–606. doi:10.1038/nsmb780.
- ^ Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD (2003). "Asymmetry in the assembly of the RNAi enzyme complex". Cell. 115 (2): 199–208. doi:10.1016/S0092-8674(03)00759-1. PMID 14567917.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Preall J, He Z, Gorra J, Sontheimer E (2006). "Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila". Curr Biol. 16 (5): 530–5. doi:10.1016/j.cub.2006.01.061. PMID 16527750.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Tomari Y, Matranga C, Haley B, Martinez N, Zamore P; Matranga; Haley; Martinez; Zamore (2004). "A protein sensor for siRNA asymmetry". Science. 306 (5700): 1377–80. Bibcode:2004Sci...306.1377T. doi:10.1126/science.1102755. PMID 15550672.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ma J, Yuan Y, Meister G, Pei Y, Tuschl T, Patel D; Yuan; Meister; Pei; Tuschl; Patel (2005). "Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein". Nature. 434 (7033): 666–70. Bibcode:2005Natur.434..666M. doi:10.1038/nature03514. PMID 15800629.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sen G, Wehrman T, Blau H (2005). "mRNA translation is not a prerequisite for small interfering RNA-mediated mRNA cleavage". Differentiation. 73 (6): 287–93. doi:10.1111/j.1432-0436.2005.00029.x. PMID 16138829.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Gu S, Rossi J (2005). "Uncoupling of RNAi from active translation in mammalian cells". RNA. 11 (1): 38–44. doi:10.1261/rna.7158605. PMC 1370689. PMID 15574516.
- ^ Sen G, Blau H (2005). "Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies". Nat Cell Biol. 7 (6): 633–6. doi:10.1038/ncb1265. PMID 15908945.
- ^ Lian S, Jakymiw A, Eystathioy T, Hamel J, Fritzler M, Chan E (2006). "GW bodies, microRNAs and the cell cycle". Cell Cycle. 5 (3): 242–5. doi:10.4161/cc.5.3.2410. PMID 16418578.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel J, Fritzler M, Chan E (2005). "Disruption of P bodies impairs mammalian RNA interference". Nat Cell Biol. 7 (12): 1267–74. doi:10.1038/ncb1334. PMID 16284622.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Hammond S, Bernstein E, Beach D, Hannon G (2000). "An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells". Nature. 404 (6775): 293–6. doi:10.1038/35005107. PMID 10749213.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Holmquist G, Ashley T (2006). "Chromosome organization and chromatin modification: influence on genome function and evolution". Cytogenet Genome Res. 114 (2): 96–125. doi:10.1159/000093326. PMID 16825762.
- ^ Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal S, Moazed D; Jia; Gerber; Sugiyama; Gygi; Grewal; Moazed (2004). "RNAi-mediated targeting of heterochromatin by the RITS complex". Science. 303 (5658): 672–6. Bibcode:2004Sci...303..672V. doi:10.1126/science.1093686. PMC 3244756. PMID 14704433.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Irvine D, Zaratiegui M, Tolia N, Goto D, Chitwood D, Vaughn M, Joshua-Tor L, Martienssen R; Zaratiegui; Tolia; Goto; Chitwood; Vaughn; Joshua-Tor; Martienssen (2006). "Argonaute slicing is required for heterochromatic silencing and spreading". Science. 313 (5790): 1134–7. Bibcode:2006Sci...313.1134I. doi:10.1126/science.1128813. PMID 16931764.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Volpe T, Kidner C, Hall I, Teng G, Grewal S, Martienssen R; Kidner; Hall; Teng; Grewal; Martienssen (2002). "Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi". Science. 297 (5588): 1833–7. Bibcode:2002Sci...297.1833V. doi:10.1126/science.1074973. PMID 12193640.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Volpe T, Schramke V, Hamilton G, White S, Teng G, Martienssen R, Allshire R (2003). "RNA interference is required for normal centromere function in fission yeast". Chromosome Res. 11 (2): 137–46. doi:10.1023/A:1022815931524. PMID 12733640.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ "Small dsRNAs induce transcriptional activation in human cells". Proc Natl Acad Sci USA. 103 (46): 17337–42. 2006. Bibcode:2006PNAS..10317337L. doi:10.1073/pnas.0607015103. PMC 1859931. PMID 17085592.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal S (2004). "RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing". Nat Genet. 36 (11): 1174–80. doi:10.1038/ng1452. PMID 15475954.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sugiyama T, Cam H, Verdel A, Moazed D, Grewal S; Cam; Verdel; Moazed; Grewal (2005). "RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production". Proc Natl Acad Sci USA. 102 (1): 152–7. Bibcode:2005PNAS..102..152S. doi:10.1073/pnas.0407641102. PMC 544066. PMID 15615848.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T (2006). "The Assembly and Maintenance of Heterochromatin Initiated by Transgene Repeats Are Independent of the RNA Interference Pathway in Mammalian Cells". Mol Cell Biol. 26 (11): 4028–40. doi:10.1128/MCB.02189-05. PMC 1489094. PMID 16705157.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Bass B (2002). "RNA Editing by Adenosine Deaminases That Act on RNA". Annu Rev Biochem. 71: 817–46. doi:10.1146/annurev.biochem.71.110601.135501. PMC 1823043. PMID 12045112.
- ^ Bass B (2000). "Double-stranded RNA as a template for gene silencing". Cell. 101 (3): 235–8. doi:10.1016/S0092-8674(02)71133-1. PMID 10847677.
- ^ Luciano D, Mirsky H, Vendetti N, Maas S (2004). "RNA editing of a miRNA precursor". RNA. 10 (8): 1174–7. doi:10.1261/rna.7350304. PMC 1370607. PMID 15272117.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Yang W, Chendrimada T, Wang Q, Higuchi M, Seeburg P, Shiekhattar R, Nishikura K (2006). "Modulation of microRNA processing and expression through RNA editing by ADAR deaminases". Nature Structural & Molecular Biology. 13 (1): 13–21. doi:10.1038/nsmb1041. PMC 2950615. PMID 16369484.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Yang W, Wang Q, Howell K, Lee J, Cho D, Murray J, Nishikura K (2005). "ADAR1 RNA Deaminase Limits Short Interfering RNA Efficacy in Mammalian Cells". J Biol Chem. 280 (5): 3946–53. doi:10.1074/jbc.M407876200. PMC 2947832. PMID 15556947.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Nishikura K (2006). "Editor meets silencer: crosstalk between RNA editing and RNA interference". Nature Reviews Molecular Cell Biology. 7 (12): 919–31. doi:10.1038/nrm2061. PMC 2953463. PMID 17139332.
- ^ a b c Saumet A, Lecellier CH (2006). "Anti-viral RNA silencing: do we look like plants ?". Retrovirology. 3 (1): 3. doi:10.1186/1742-4690-3-3. PMC 1363733. PMID 16409629.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Jones L, Ratcliff F, Baulcombe DC (2001). "RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance". Current Biology. 11 (10): 747–757. doi:10.1016/S0960-9822(01)00226-3. PMID 11378384.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Humphreys DT, Westman BJ, Martin DI, Preiss T; Westman; Martin; Preiss (2005). "MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function". Proc Natl Acad Sci USA. 102 (47): 16961–16966. Bibcode:2005PNAS..10216961H. doi:10.1073/pnas.0506482102. PMC 1287990. PMID 16287976.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ DaRocha W, Otsu K, Teixeira S, Donelson J (2004). "Tests of cytoplasmic RNA interference (RNAi) and construction of a tetracycline-inducible T7 promoter system in Trypanosoma cruzi". Mol Biochem Parasitol. 133 (2): 175–86. doi:10.1016/j.molbiopara.2003.10.005. PMID 14698430.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Robinson K, Beverley S (2003). "Improvements in transfection efficiency and tests of RNA interference (RNAi) approaches in the protozoan parasite Leishmania". Mol Biochem Parasitol. 128 (2): 217–28. doi:10.1016/S0166-6851(03)00079-3. PMID 12742588.
- ^ L. Aravind, Hidemi Watanabe, David J. Lipman, and Eugene V. Koonin (2000). "Lineage-specific loss and divergence of functionally linked genes in eukaryotes". Proceedings of the National Academy of Sciences. 97 (21): 11319–11324. Bibcode:2000PNAS...9711319A. doi:10.1073/pnas.200346997. PMC 17198. PMID 11016957.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Drinnenberg IA, Weinberg DE, Xie KT, Nower JP, Wolfe KH, Fink GR, Bartel DP; Weinberg; Xie; Mower; Wolfe; Fink; Bartel (2009). "RNAi in Budding Yeast". Science. 326 (5952): 544–50. Bibcode:2009Sci...326..544D. doi:10.1126/science.1176945. PMID 19745116.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nakayashiki H, Kadotani N, Mayama S (2006). "Evolution and diversification of RNA silencing proteins in fungi". J Mol Evol. 63 (1): 127–35. doi:10.1007/s00239-005-0257-2. PMID 16786437.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Morita T, Mochizuki Y, Aiba H; Mochizuki; Aiba (2006). "Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction". Proc Natl Acad Sci USA. 103 (13): 4858–63. Bibcode:2006PNAS..103.4858M. doi:10.1073/pnas.0509638103. PMC 1458760. PMID 16549791.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Makarova K, Grishin N, Shabalina S, Wolf Y, Koonin E (2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biol Direct. 1: 7. doi:10.1186/1745-6150-1-7. PMC 1462988. PMID 16545108.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Stram Y, Kuzntzova L (2006). "Inhibition of viruses by RNA interference". Virus Genes. 32 (3): 299–306. doi:10.1007/s11262-005-6914-0. PMID 16732482.
- ^ Blevins T, Rajeswaran R, Shivaprasad P, Beknazariants D, Si-Ammour A, Park H, Vazquez F, Robertson D, Meins F, Hohn T, Pooggin M (2006). "Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing". Nucleic Acids Res. 34 (21): 6233–46. doi:10.1093/nar/gkl886. PMC 1669714. PMID 17090584.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Palauqui J, Elmayan T, Pollien J, Vaucheret H (1997). "Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions". EMBO J. 16 (15): 4738–45. doi:10.1093/emboj/16.15.4738. PMC 1170100. PMID 9303318.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Voinnet O (2001). "RNA silencing as a plant immune system against viruses". Trends Genet. 17 (8): 449–59. doi:10.1016/S0168-9525(01)02367-8. PMID 11485817.
- ^ a b Lucy A, Guo H, Li W, Ding S (2000). "Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus". EMBO J. 19 (7): 1672–80. doi:10.1093/emboj/19.7.1672. PMC 310235. PMID 10747034.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mérai Z, Kerényi Z, Kertész S, Magna M, Lakatos L, Silhavy D (2006). "Double-Stranded RNA Binding May Be a General Plant RNA Viral Strategy To Suppress RNA Silencing". J Virol. 80 (12): 5747–56. doi:10.1128/JVI.01963-05. PMC 1472586. PMID 16731914.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Zhu J, Staskawicz B, Jin H; Morgan; Dahlbeck; Borsani; Villegas; Zhu; Staskawicz; Jin (2006). "A pathogen-inducible endogenous siRNA in plant immunity". Proc Natl Acad Sci USA. 103 (47): 18002–7. Bibcode:2006PNAS..10318002K. doi:10.1073/pnas.0608258103. PMC 1693862. PMID 17071740.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fritz J, Girardin S, Philpott D (2006). "Innate immune defense through RNA interference". Sci STKE. 2006 (339): pe27. doi:10.1126/stke.3392006pe27. PMID 16772641.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Zambon R, Vakharia V, Wu L (2006). "RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster". Cell Microbiol. 8 (5): 880–9. doi:10.1111/j.1462-5822.2006.00688.x. PMID 16611236.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wang X, Aliyari R, Li W, Li H, Kim K, Carthew R, Atkinson P, Ding S; Aliyari; Li; Li; Kim; Carthew; Atkinson; Ding (2006). "RNA Interference Directs Innate Immunity Against Viruses in Adult Drosophila". Science. 312 (5772): 452–4. Bibcode:2006Sci...312..452W. doi:10.1126/science.1125694. PMC 1509097. PMID 16556799.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lu R, Maduro M, Li F, Li H, Broitman-Maduro G, Li W, Ding S; Maduro; Li; Li; Broitman-Maduro; Li; Ding (2005). "Animal virus replication and RNAi-mediated antiviral silencing in C elegans". Nature. 436 (7053): 1040–3. Bibcode:2005Natur.436.1040L. doi:10.1038/nature03870. PMC 1388260. PMID 16107851.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wilkins C, Dishongh R, Moore S, Whitt M, Chow M, Machaca K; Dishongh; Moore; Whitt; Chow; Machaca (2005). "RNA interference is an antiviral defence mechanism in Caenorhabditis elegans". Nature. 436 (7053): 1044–7. Bibcode:2005Natur.436.1044W. doi:10.1038/nature03957. PMID 16107852.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Berkhout B, Haasnoot J (2006). "The interplay between virus infection and the cellular RNA interference machinery". FEBS Lett. 580 (12): 2896–902. doi:10.1016/j.febslet.2006.02.070. PMID 16563388.
- ^ Schütz S, Sarnow P (2006). "Interaction of viruses with the mammalian RNA interference pathway". Virology. 344 (1): 151–7. doi:10.1016/j.virol.2005.09.034. PMID 16364746.
- ^ Cullen B (2006). "Is RNA interference involved in intrinsic antiviral immunity in mammals?". Nat Immunol. 7 (6): 563–7. doi:10.1038/ni1352. PMID 16715068.
- ^ Maillard, P. V.; Ciaudo, C.; Marchais, A.; Jay, Y. Li F.; Ding,, S. W.; Voinnet, Olivier (2013). "Antiviral RNA Interference in Mammalian Cells". Science. 342 (6155): 235–238. doi:10.1126/science.1241930. PMID 24115438.
{{cite journal}}: CS1 maint: extra punctuation (link) - ^ Li, Yang; Lu, Jinfeng; Han, Yanhong; Fan, Xiaoxu; Ding, Shou-Wei (2013). "RNA Interference Functions as an Antiviral Immunity Mechanism in Mammals". Science. 342 (6155): 231–234. doi:10.1126/science.1241911. PMID 24115437.
- ^ Li, H; Ding, S (2005). "Antiviral silencing in animals". FEBS Lett. 579 (26): 5965–73. doi:10.1016/j.febslet.2005.08.034. PMC 1350842. PMID 16154568.
- ^ Carrington, J; Ambros, V (2003). "Role of microRNAs in plant and animal development". Science. 301 (5631): 336–8. Bibcode:2003Sci...301..336C. doi:10.1126/science.1085242. PMID 12869753.
- ^ Lee, R; Feinbaum, R; Ambros, V (1993). "The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14". Cell. 75 (5): 843–54. doi:10.1016/0092-8674(93)90529-Y. PMID 8252621.
- ^ Palatnik, J; Allen, E; Wu, X; Schommer, C; Schwab, R; Carrington, J; Weigel, D (2003). "Control of leaf morphogenesis by microRNAs". Nature. 425 (6955): 257–63. Bibcode:2003Natur.425..257P. doi:10.1038/nature01958. PMID 12931144.
- ^ Zhang, B; Pan, X; Cobb, G; Anderson, T (2006). "Plant microRNA: a small regulatory molecule with big impact". Dev Biol. 289 (1): 3–16. doi:10.1016/j.ydbio.2005.10.036. PMID 16325172.
- ^ Jones-Rhoades, M; Bartel, David P.; Bartel B, D (2006). "MicroRNAS and their regulatory roles in plants". Annu Rev Plant Biol. 57: 19–53. doi:10.1146/annurev.arplant.57.032905.105218. PMID 16669754.
- ^ Zhang, B; Pan, X; Cobb, G; Anderson, T (2007). "microRNAs as oncogenes and tumor suppressors". Dev Biol. 302 (1): 1–12. doi:10.1016/j.ydbio.2006.08.028. PMID 16989803.
- ^ Check, E (2007). "RNA interference: hitting the on switch". Nature. 448 (7156): 855–858. Bibcode:2007Natur.448..855C. doi:10.1038/448855a. PMID 17713502.
Li, LC; Okino, ST; Zhao, H,; Pookot; Place, RF; Urakami, S; Enokida, H; Dahiya, R (2006). "Small dsRNAs induce transcriptional activation in human cells". Proc. Natl. Acad. Sci. U.S.A. 103 (46): 17337–42. Bibcode:2006PNAS..10317337L. doi:10.1073/pnas.0607015103. PMC 1859931. PMID 17085592.{{cite journal}}: CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link) - ^ Vasudevan, S.; Tong, Y.; Steitz, J. A. (2007). "Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation". Science. 318 (5858): 1931–1934. doi:10.1126/science.1149460. PMID 18048652.
- ^ a b c Cerutti, H; Casas-Mollano, J (2006). "On the origin and functions of RNA-mediated silencing: from protists to man". Curr Genet. 50 (2): 81–99. doi:10.1007/s00294-006-0078-x. PMC 2583075. PMID 16691418.
- ^ Anantharaman, V; Koonin, E; Aravind first3=L (2002). "Comparative genomics and evolution of proteins involved in RNA metabolism". Nucleic Acids Res. 30 (7): 1427–64. doi:10.1093/nar/30.7.1427. PMC 101826. PMID 11917006.
{{cite journal}}: Missing pipe in:|last3=(help)CS1 maint: numeric names: authors list (link) - ^ Buchon, N; Vaury first2=C (2006). "RNAi: a defensive RNA-silencing against viruses and transposable elements". Heredity. 96 (2): 195–202. doi:10.1038/sj.hdy.6800789. PMID 16369574.
{{cite journal}}: Missing pipe in:|last2=(help)CS1 maint: numeric names: authors list (link) - ^ Obbard, D; Jiggins first2=F; Halligan first3=D; Little first4=T (2006). "Natural selection drives extremely rapid evolution in antiviral RNAi genes". Curr Biol. 16 (6): 580–5. doi:10.1016/j.cub.2006.01.065. PMID 16546082.
{{cite journal}}: Missing pipe in:|last2=(help); Missing pipe in:|last3=(help); Missing pipe in:|last4=(help)CS1 maint: numeric names: authors list (link) - ^ Voorhoeve, PM; Agami, R (2003). "Knockdown stands up". Trends Biotechnol. 21 (1): 2–4. doi:10.1016/S0167-7799(02)00002-1. PMID 12480342.
- ^ Naito, Y; Yamada, T; Matsumiya, T; Ui-Tei, K; Saigo, K; Morishita, S (2005). "dsCheck: highly sensitive off-target search software for double-stranded RNA-mediated RNA interference". Nucleic Acids Res. 33 (Web Server issue): W589–91. doi:10.1093/nar/gki419. PMC 1160180. PMID 15980542.
- ^ Henschel, A; Buchholz, F; Habermann, B (2004). "DEQOR: a web-based tool for the design and quality control of siRNAs". Nucleic Acids Res. 32 (Web Server issue): W113–20. doi:10.1093/nar/gkh408. PMC 441546. PMID 15215362.
- ^ Naito, Y; Yamada, T; Ui-Tei, K; Morishita, S; Saigo, K (2004). "siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference". Nucleic Acids Res. 32 (Web Server issue): W124–9. doi:10.1093/nar/gkh442. PMC 441580. PMID 15215364.
- ^ Naito, Y; Ui-Tei, K; Nishikawa, T; Takebe, Y; Saigo, K (2006). "siVirus: web-based antiviral siRNA design software for highly divergent viral sequences". Nucleic Acids Res. 34 (Web Server issue): W448–50. doi:10.1093/nar/gkl214. PMC 1538817. PMID 16845046.
- ^ Reynolds, A; Anderson, E; Vermeulen, A; Fedorov, Y; Robinson, K; Leake, D; Karpilow, J; Marshall, W; Khvorova, A (2006). "Induction of the interferon response by siRNA is cell type– and duplex length–dependent". RNA. 12 (6): 988–93. doi:10.1261/rna.2340906. PMC 1464853. PMID 16611941.
- ^ Stein, P; Zeng, F; Pan, H; Schultz, R (2005). "Absence of non-specific effects of RNA interference triggered by long double-stranded RNA in mouse oocytes". Dev Biol. 286 (2): 464–71. doi:10.1016/j.ydbio.2005.08.015. PMID 16154556.
- ^ Brummelkamp, T; Bernards, R; Agami, R (2002). "A system for stable expression of short interfering RNAs in mammalian cells". Science. 296 (5567): 550–3. Bibcode:2002Sci...296..550B. doi:10.1126/science.1068999. PMID 11910072.
- ^ Tiscornia, G; Tergaonkar, V; Galimi, F; Verma, I (2004). "CRE recombinase-inducible RNA interference mediated by lentiviral vectors". Proc Natl Acad Sci USA. 101 (19): 7347–51. Bibcode:2004PNAS..101.7347T. doi:10.1073/pnas.0402107101. PMC 409921. PMID 15123829.
- ^ Ventura, A; Meissner, A; Dillon, C; McManus, M; Sharp, P; Van Parijs, L; Jaenisch, R; Jacks, T (2004). "Cre-lox-regulated conditional RNA interference from transgenes". Proc Natl Acad Sci USA. 101 (28): 10380–5. Bibcode:2004PNAS..10110380V. doi:10.1073/pnas.0403954101. PMC 478580. PMID 15240889.
- ^ Kamath, R; Ahringer, J (2003). "Genome-wide RNAi screening in Caenorhabditis elegans". Methods. 30 (4): 313–21. doi:10.1016/S1046-2023(03)00050-1. PMID 12828945.
- ^ Boutros, M; Kiger, A; Armknecht, S; Kerr, K; Hild, M; Koch, B; Haas, S; Paro, R; et al. (2004). "Genome-wide RNAi analysis of growth and viability in Drosophila cells". Science. 303 (5659): 832–5. Bibcode:2004Sci...303..832B. doi:10.1126/science.1091266. PMID 14764878.
- ^ Fortunato, A; Fraser, A (2005). "Uncover genetic interactions in Caenorhabditis elegans by RNA interference". Biosci Rep. 25 (5–6): 299–307. doi:10.1007/s10540-005-2892-7. PMID 16307378.
- ^ Cullen, L; Arndt, G (2005). "Genome-wide screening for gene function using RNAi in mammalian cells". Immunol Cell Biol. 83 (3): 217–23. doi:10.1111/j.1440-1711.2005.01332.x. PMID 15877598.
- ^ Huesken, D; Lange, J; Mickanin, C; Weiler, J; Asselbergs, F; Warner, J; Meloon, B; Engel, S; Rosenberg, A; Cohen, D; Labow, M; Reinhardt, M; Natt, F; Hall, J (2005). "Design of a genome-wide siRNA library using an artificial neural network". Nat Biotechnol. 23 (8): 995–1001. doi:10.1038/nbt1118. PMID 16025102.
- ^ Ge, G; Wong, G; Luo, B (2005). "Prediction of siRNA knockdown efficiency using artificial neural network models". Biochem Biophys Res Commun. 336 (2): 723–8. doi:10.1016/j.bbrc.2005.08.147. PMID 16153609.
- ^ Janitz, M; Vanhecke, D; Lehrach, H (2006). "High-throughput RNA interference in functional genomics". Handb Exp Pharmacol. Handbook of Experimental Pharmacology. 173 (173): 97–104. doi:10.1007/3-540-27262-3_5. ISBN 3-540-27261-5. PMID 16594612.
- ^ Vanhecke, D; Janitz, M (2005). "Functional genomics using high-throughput RNA interference". Drug Discov Today. 10 (3): 205–12. doi:10.1016/S1359-6446(04)03352-5. PMID 15708535.
- ^ Geldhof, P; Murray, L; Couthier, A; Gilleard, J; McLauchlan, G; Knox, D; Britton, C (2006). "Testing the efficacy of RNA interference in Haemonchus contortus". Int J Parasitol. 36 (7): 801–10. doi:10.1016/j.ijpara.2005.12.004. PMID 16469321.
- ^ Geldhof, P; Visser, A; Clark, D; Saunders, G; Britton, C.; Gilleard, C; Gilleard, J; Berriman, M; Knox, D. (2007). "RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects". Parasitology. 134 (Pt 5): 1–11. doi:10.1017/S0031182006002071. PMID 17201997.
- ^ Travella, S; Klimm, T; Keller, B (2006). "RNA Interference-Based Gene Silencing as an Efficient Tool for Functional Genomics in Hexaploid Bread Wheat". Plant Physiol. 142 (1): 6–20. doi:10.1104/pp.106.084517. PMC 1557595. PMID 16861570.
- ^ McGinnis, K; Chandler, V; Cone, K; Kaeppler, H; Kaeppler, S; Kerschen, A; Pikaard, C; Richards, E; Sidorenko, L; Smith, T; Springer, N; Wulan, T (2005). "Transgene-induced RNA interference as a tool for plant functional genomics". Methods Enzymol. Methods in Enzymology. 392: 1–24. doi:10.1016/S0076-6879(04)92001-0. ISBN 978-0-12-182797-7. PMID 15644172.
- ^ Brock, T; Browse, J; Watts, J (2006). "Genetic Regulation of Unsaturated Fatty Acid Composition in C. elegans". PLoS Genet. 2 (7): e108. doi:10.1371/journal.pgen.0020108. PMC 1500810. PMID 16839188.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Paddison, P; Caudy, A; Hannon, G (2002). "Stable suppression of gene expression by RNAi in mammalian cells". Proc Natl Acad Sci USA. 99 (3): 1443–8. Bibcode:2002PNAS...99.1443P. doi:10.1073/pnas.032652399. PMC 122210. PMID 11818553.
Whitehead, KA; Dahlman, JE; Langer, RS; Anderson, DG (2011). "Silencing or stimulation? siRNA delivery and the immune system". Annual Review of Chemical and Biomolecular Engineering. 2: 77–96. doi:10.1146/annurev-chembioeng-061010-114133. PMID 22432611. - ^ Sah, D (2006). "Therapeutic potential of RNA interference for neurological disorders". Life Sci. 79 (19): 1773–80. doi:10.1016/j.lfs.2006.06.011. PMID 16815477.
- ^ Zender, L; Hutker, S; Liedtke, C; Tillmann, H; Zender, S; Mundt, B; Waltemathe, M; Gosling, T; Flemming, P; Malek, N; Trautwein, C; Manns, M; Kuhnel, F; Kubicka, S (2003). "Caspase 8 small interfering RNA prevents acute liver failure in mice". Proc Natl Acad Sci USA. 100 (13): 7797–802. Bibcode:2003PNAS..100.7797Z. doi:10.1073/pnas.1330920100. PMC 164667. PMID 12810955.
- ^ Jiang, M; Milner, J (2002). "Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference". Oncogene. 21 (39): 6041–8. doi:10.1038/sj.onc.1205878. PMID 12203116.
- ^ Crowe, S (2003). "Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication, by Martínez et al". AIDS. 17 Suppl 4: S103–5. doi:10.1097/00002030-200317004-00014. PMID 15080188.
- ^ Kusov, Y; Kanda, T; Palmenberg, A; Sgro, J; Gauss-Müller, V (2006). "Silencing of Hepatitis A Virus Infection by Small Interfering RNAs". J Virol. 80 (11): 5599–610. doi:10.1128/JVI.01773-05. PMC 1472172. PMID 16699041.
- ^ Jia, F; Zhang, Y; Liu, C (2006). "A retrovirus-based system to stably silence hepatitis B virus genes by RNA interference". Biotechnol Lett. 28 (20): 1679–85. doi:10.1007/s10529-006-9138-z. PMID 16900331.
- ^ Li, Y; Kong, L; Cheng, B; Li, K (2005). "Construction of influenza virus siRNA expression vectors and their inhibitory effects on multiplication of influenza virus". Avian Dis. 49 (4): 562–73. doi:10.1637/7365-041205R2.1. PMID 16405000.
- ^ Hu, L; Wang, Z; Hu, C; Liu, X; Yao, L; Li, W; Qi, Y (2005). "Inhibition of Measles virus multiplication in cell culture by RNA interference". Acta Virol. 49 (4): 227–34. PMID 16402679.
- ^ Raoul, C; Barker, S; Aebischer, P (2006). "Viral-based modelling and correction of neurodegenerative diseases by RNA interference". Gene Ther. 13 (6): 487–95. doi:10.1038/sj.gt.3302690. PMID 16319945.
- ^ Berkhout, B; ter Brake, O (2010). "RNAi Gene Therapy to Control HIV-1 Infection". RNA Interference and Viruses: Current Innovations and Future Trends. Caister Academic Press. ISBN 978-1-904455-56-1.
- ^ Putral, L; Gu, W; McMillan, N (2006). "RNA interference for the treatment of cancer". Drug News Perspect. 19 (6): 317–24. doi:10.1358/dnp.2006.19.6.985937. PMID 16971967.
- ^ Izquierdo, M (2005). "Short interfering RNAs as a tool for cancer gene therapy". Cancer Gene Ther. 12 (3): 217–27. doi:10.1038/sj.cgt.7700791. PMID 15550938.
- ^ Li, C; Parker, A; Menocal, E; Xiang, S; Borodyansky, L; Fruehauf, J (2006). "Delivery of RNA interference". Cell Cycle. 5 (18): 2103–9. doi:10.4161/cc.5.18.3192. PMID 16940756.
- ^ Takeshita, F; Ochiya, T (2006). "Therapeutic potential of RNA interference against cancer". Cancer Sci. 97 (8): 689–96. doi:10.1111/j.1349-7006.2006.00234.x. PMID 16863503.
- ^ Ma, Z; Li, J; He, F; Wilson, A; Pitt, B; Li, S, (2005). "Cationic lipids enhance siRNA-mediated interferon response in mice". Biochemical and Biophysical Research Communications. 330 (3): 755–759. doi:10.1016/j.bbrc.2005.03.041. PMID 15809061.
{{cite journal}}: CS1 maint: extra punctuation (link) CS1 maint: multiple names: authors list (link)
Morrissey, DV; Lockridge first2=JA; Shaw, L; Blanchard, K; Jensen, K; Breen, W; Hartsough, K; Machemer, L; Radka, S; Jadhav, V (2005). "Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs". Nature Biotechnology. 23 (8): 1002–1007. doi:10.1038/nbt1122. PMID 16041363.{{cite journal}}: Missing pipe in:|last2=(help)CS1 maint: numeric names: authors list (link) - ^ Urban-Klein, B; Werth, S; Abuharbeid, S; Czubayko, F; Aigner, A (2004). "RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo". Gene Therapy. 12 (5): 461–466. doi:10.1038/sj.gt.3302425. PMID 15616603.
Zintchenko, A; Philipp, A; Dehshahri, A; Wagner, E (2008). "Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity". Bioconjugate Chemistry. 19 (7): 1448–1455. doi:10.1021/bc800065f. PMID 18553894. - ^ Bartlett, D. W.; =Davis, ME (2006). "Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging". Nucleic Acids Research. 34 (1): 322–333. doi:10.1093/nar/gkj439. PMC 1331994. PMID 16410612.
{{cite journal}}: CS1 maint: extra punctuation (link) - ^ Bavykin, AS; Korotaeva, AA; Poyarkov, SV; Syrtsev, AV; Tjulandin, SA; Karpukhin, AV (2013). "Double siRNA-targeting of cIAP2 and LIVIN results in synergetic sensitization of HCT-116 cells to oxaliplatin treatment". OncoTargets and Therapy. 6: 1333–40. doi:10.2147/OTT.S44893. PMC 3789649. PMID 24098083.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tong, A; Zhang, Y; Nemunaitis, J (2005). "Small interfering RNA for experimental cancer therapy". Current Opinion in Molecular Therapeutics. 7 (2): 114–24. PMID 15844618.
- ^ Check, Erika. "RNA treatment kills mice". Nature. Nature Publishing Group. doi:10.1038/news060522-10. Retrieved 17 December 2012.
- ^ Grimm, D; Streetz, K; Jopling, C; Storm, T; Pandey, K; Davis, C; Marion, P; Salazar; Kay, F; Kay, M (2006). "Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways". Nature. 441 (7092): 537–41. Bibcode:2006Natur.441..537G. doi:10.1038/nature04791. PMID 16724069.
- ^ Grimm D, Kay M. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time?. Journal Of Clinical Investigation [serial online]. December 2007;117(12):3633–3641. Available from: Academic Search Elite, Ipswich, MA. Accessed April 3, 2012.
- ^ Sunilkumar, G; Campbell, L; Puckhaber, L; Stipanovic, R; Rathore, K (2006). "Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol". Proc Natl Acad Sci USA. 103 (48): 18054–9. Bibcode:2006PNAS..10318054S. doi:10.1073/pnas.0605389103. PMC 1838705. PMID 17110445.
- ^ Siritunga, D; Sayre, R (2003). "Generation of cyanogen-free transgenic cassava". Planta. 217 (3): 367–73. doi:10.1007/s00425-003-1005-8. PMID 14520563.
- ^ Le, L; Lorenz, Y; Scheurer, S; Fötisch, K; Enrique, E; Bartra, J; Biemelt, S; Vieths, S; Sonnewald, U (2006). "Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression". Plant Biotechnol J. 4 (2): 231–42. doi:10.1111/j.1467-7652.2005.00175.x. PMID 17177799.
- ^ Niggeweg, R; Michael, A; Martin, C (2004). "Engineering plants with increased levels of the antioxidant chlorogenic acid". Nat Biotechnol. 22 (6): 746–54. doi:10.1038/nbt966. PMID 15107863.
- ^ Sanders, R; Hiatt, W (2005). "Tomato transgene structure and silencing". Nat Biotechnol. 23 (3): 287–9. doi:10.1038/nbt0305-287b. PMID 15765076.
- ^ Chiang, C; Wang, J; Jan, F; Yeh, S; Gonsalves, D (2001). "Comparative reactions of recombinant papaya ringspot viruses with chimeric coat protein (CP) genes and wild-type viruses on CP-transgenic papaya". J Gen Virol. 82 (Pt 11): 2827–36. PMID 11602796.
- ^ Gavilano, L; Coleman, N; Burnley, L; Bowman, M; Kalengamaliro; Hayes, A; Bush, L; Siminszky, B (2006). "Genetic engineering of Nicotiana tabacum for reduced nornicotine content". J Agric Food Chem. 54 (24): 9071–8. doi:10.1021/jf0610458. PMID 17117792.
- ^ Allen, R; Millgate, A; Chitty, J; Thisleton, J; Miller, J; Fist, A; Gerlach, W; Larkin, P (2004). "RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy". Nat Biotechnol. 22 (12): 1559–66. doi:10.1038/nbt1033. PMID 15543134.
- ^ Zadeh, A; Foster, G (2004). "Transgenic resistance to tobacco ringspot virus". Acta Virol. 48 (3): 145–52. PMID 15595207.
- ^ "RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design". Journal of insect physiology. 57 (2): 231–45. Feb 2011. doi:10.1016/j.jinsphys.2010.11.006. PMID 21078327.
{{cite journal}}: Unknown parameter|authors=ignored (help) - ^ Matson RS (2005). Applying genomic and proteomic microarray technology in drug discovery. CRC Press. ISBN 0-8493-1469-0.
- ^ Zhang XHD (2011). Optimal High-Throughput Screening: Practical Experimental Design and Data Analysis for Genome-scale RNAi Research. Cambridge University Press. ISBN 978-0-521-73444-8.
- ^ Matzke MA, Matzke AJM. (2004). "Planting the Seeds of a New Paradigm". PLoS Biol. 2 (5): e133. doi:10.1371/journal.pbio.0020133. PMC 406394. PMID 15138502.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Ecker JR, Davis RW; Davis (1986). "Inhibition of gene expression in plant cells by expression of antisense RNA". Proc Natl Acad Sci USA. 83 (15): 5372–5376. Bibcode:1986PNAS...83.5372E. doi:10.1073/pnas.83.15.5372. PMC 386288. PMID 16593734.
- ^ Napoli C, Lemieux C, Jorgensen R (1990). "Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans". Plant Cell. 2 (4): 279–289. doi:10.1105/tpc.2.4.279. PMC 159885. PMID 12354959.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Romano N, Macino G (1992). "Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences". Mol Microbiol. 6 (22): 3343–53. doi:10.1111/j.1365-2958.1992.tb02202.x. PMID 1484489.
- ^ Van Blokland R, Van der Geest N, Mol JNM, Kooter JM (1994). "Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover". Plant J. 6 (6): 861–77. doi:10.1046/j.1365-313X.1994.6060861.x.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mol JNM, van der Krol AR (1991). Antisense nucleic acids and proteins: fundamentals and applications. M. Dekker. pp. 4, 136. ISBN 0-8247-8516-9.
- ^ Covey S, Al-Kaff N, Lángara A, Turner D; Al-Kaff; Lángara; Turner (1997). "Plants combat infection by gene silencing". Nature. 385 (6619): 781–2. Bibcode:1997Natur.385..781C. doi:10.1038/385781a0.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ratcliff F, Harrison B, Baulcombe D (1997). "A Similarity Between Viral Defense and Gene Silencing in Plants". Science. 276 (5318): 1558–60. doi:10.1126/science.276.5318.1558. PMID 18610513.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Guo S, Kemphues K (1995). "par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed". Cell. 81 (4): 611–20. doi:10.1016/0092-8674(95)90082-9. PMID 7758115.
- ^ Pal-Bhadra M, Bhadra U, Birchler J (1997). "Cosuppression in Drosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent". Cell. 90 (3): 479–90. doi:10.1016/S0092-8674(00)80508-5. PMID 9267028.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fire A, Xu S, Montgomery M, Kostas S, Driver S, Mello C; Xu; Montgomery; Kostas; Driver; Mello (1998). "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature. 391 (6669): 806–11. Bibcode:1998Natur.391..806F. doi:10.1038/35888. PMID 9486653.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- Overview of the RNAi process, from Cambridge University's The Naked Scientists
- Animation of the RNAi process, from Nature
- NOVA scienceNOW explains RNAi – A 15-minute video of the Nova broadcast that aired on PBS, July 26, 2005
- Silencing Genomes RNA interference (RNAi) experiments and bioinformatics in C. elegans for education. From the Dolan DNA Learning Center of Cold Spring Harbor Laboratory.
- RNAi screens in C. elegans in a 96-well liquid format and their application to the systematic identification of genetic interactions (a protocol)
- 2 American ‘Worm People’ Win Nobel for RNA Work, from NY Times
- Molecular Therapy web focus: "The development of RNAi as a therapeutic strategy", a collection of free articles about RNAi as a therapeutic strategy.
- GenomeRNAi: a database of phenotypes from RNA interference screening experiments in Drosophila melanogaster and Homo sapians
