Succinyl coenzyme A synthetase
| Succinate—CoA ligase (GDP-forming) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 6.2.1.4 | ||||||||
| CAS no. | 9014-36-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Succinate—CoA ligase (ADP-forming) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC no. | 6.2.1.5 | ||||||||
| CAS no. | 9080-33-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Succinyl coenzyme A synthetase (SCS, also known as succinyl-CoA synthetase or succinate thiokinase or succinate-CoA ligase) is an enzyme that catalyzes the reversible reaction of succinyl-CoA to succinate.[3] The enzyme facilitates the coupling of this reaction to the formation of a nucleoside triphosphate molecule (either GTP or ATP) from an inorganic phosphate molecule and a nucleoside diphosphate molecule (either GDP or ADP). It plays a key role as one of the catalysts involved in the citric acid cycle, a central pathway in cellular metabolism, and it is located within the mitochondrial matrix of a cell.[4]
Chemical reaction and enzyme mechanism
[edit]Succinyl CoA synthetase catalyzes the following reversible reaction:
- Succinyl CoA + Pi + NDP ↔ Succinate + CoA + NTP
where Pi denotes inorganic phosphate, NDP denotes nucleotide diphosphate (either GDP or ADP), and NTP denotes nucleotide triphosphate (either GTP or ATP). As mentioned, the enzyme facilitates coupling of the conversion of succinyl CoA to succinate with the formation of NTP from NDP and Pi. The reaction has a biochemical standard state free energy change of -3.4 kJ/mol.[4] The reaction takes place by a three-step mechanism[3] which is depicted in the image below. The first step involves displacement of CoA from succinyl CoA by a nucleophilic inorganic phosphate molecule to form succinyl phosphate. The enzyme then utilizes a histidine residue to remove the phosphate group from succinyl phosphate and generate succinate. Finally, the phosphorylated histidine transfers the phosphate group to a nucleoside diphosphate, which generates the high-energy carrying nucleoside triphosphate.
Structure
[edit]Subunits
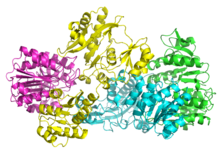
[edit]Bacterial and mammalian SCSs are made up of α and β subunits.[5] In E. coli two αβ heterodimers link together to form an α2β2 heterotetrameric structure. However, mammalian mitochondrial SCSs are active as αβ dimers and do not form a heterotetramer.[6] The E. coli SCS heterotetramer has been crystallized and characterized in great detail.[6][7] As can be seen in Image 2, the two α subunits (pink and green) reside on opposite sides of the structure and the two β subunits (yellow and blue) interact in the middle region of the protein. The two α subunits only interact with a single β unit, whereas the β units interact with a single α unit (to form the αβ dimer) and the β subunit of the other αβ dimer.[6] A short amino acid chain links the two β subunits which gives rise to the tetrameric structure.

The crystal structure of Succinyl-CoA synthetase alpha subunit (succinyl-CoA-binding isoform) was determined by Joyce et al. to a resolution of 2.10 A, with PDB code 1CQJ. [1].[8]
Catalytic residues
[edit]Crystal structures for the E. coli SCS provide evidence that the coenzyme A binds within each α-subunit (within a Rossmann fold) in close proximity to a histidine residue (His246α).[7] This histidine residue becomes phosphorylated during the succinate forming step in the reaction mechanism. The exact binding location of succinate is not well-defined.[9] The formation of the nucleotide triphosphate occurs in an ATP grasp domain, which is located near the N-terminus of the each β subunit. However, this grasp domain is located about 35 Å away from the phosphorylated histidine residue.[8] This leads researchers to believe that the enzyme must undergo a major change in conformation to bring the histidine to the grasp domain and facilitate the formation of the nucleoside triphosphate. Mutagenesis experiments have determined that two glutamate residues (one near the catalytic histidine, Glu208α and one near the ATP grasp domain, Glu197β) play a role in the phosphorylation and dephosphorylation of the histidine, but the exact mechanism by which the enzyme changes conformation is not fully understood.[9]
Isoforms
[edit]Johnson et al. describe two isoforms of succinyl-CoA synthetase in amniotes, one that specifies synthesis of ATP, and one that synthesises GTP.[10]
In amniotes, the enzyme is a heterodimer of an α- and a β-subunit. The specificity for either adenosine or guanosine phosphates is defined by the β-subunit,[10] which is encoded by 2 genes. SUCLG2 is GTP-specific and SUCLA2 is ATP-specific, while SUCLG1 encodes the common α-subunit. β variants are produced at different amounts in different tissues,[10] causing GTP or ATP substrate requirements.
Mostly consuming tissues such as heart and brain have more ATP-specific succinyl-CoA synthetase (ATPSCS), while synthetic tissues such as kidney and liver have the more GTP-specific form (GTPSCS).[11] Kinetics analysis of ATPSCS from the breast muscle of pigeons and GTPSCS from pigeon liver showed that their apparent Michaelis constants were similar for CoA, but different for the nucleotides, phosphate, and succinate. The largest difference was for succinate: Kmapp of ATPSCS = 5mM versus that of GTPSCS = 0.5mM.[10]
Function
[edit]Generation of nucleotide triphosphates
[edit]SCS is the only enzyme in the citric acid cycle that catalyzes a reaction in which a nucleotide triphosphate (GTP or ATP) is formed by substrate-level phosphorylation.[4] Research studies have shown that E. coli SCSs can catalyze either GTP or ATP formation.[7] However, mammals possess different types of SCSs that are specific for either GTP (G-SCS) or ATP (A-SCS) and are native to different types of tissue within the organism. An interesting study using pigeon cells showed that GTP specific SCSs were located in pigeon liver cells, and ATP specific SCSs were located in the pigeon breast muscle cells.[12] Further research revealed a similar phenomenon of GTP and ATP specific SCSs in rat, mouse, and human tissue. It appears that tissue typically involved in anabolic metabolism (like the liver and kidneys) express G-SCS, whereas tissue involved in catabolic metabolism (like the brain, the heart, and muscular tissue) express A-SCS.[11]
Formation of metabolic intermediates
[edit]SCS facilitates the flux of molecules into other metabolic pathways by controlling the interconversion between succinyl CoA and succinate.[13] This is important because succinyl CoA is an intermediate necessary for porphyrin, heme,[14] and ketone body biosynthesis.[15]
Regulation and inhibition
[edit]In some bacteria, the enzyme is regulated at the transcriptional level.[16] It has been demonstrated that the gene for SCS (sucCD) is transcribed along with the gene for α-ketoglutarate dehydrogenase (sucAB) under the control of a promoter called sdhC, which is part of the succinate dehydrogenase operon. This operon is up-regulated by the presence of oxygen and responds to a variety of carbon sources. Antibacterial drugs that prevent phosphorylation of histidine, like the molecule LY26650, are potent inhibitors of bacterial SCSs.[17]
Optimal activity
[edit]Measurements (performed using a soy bean SCS) indicate an optimal temperature of 37 °C and an optimal pH of 7.0-8.0.[18]
Role in disease
[edit]Fatal infantile lactic acidosis: Defective SCS has been implicated as a cause of fatal infantile lactic acidosis, which is a disease in infants that is characterized by the build-up of toxic levels of lactic acid. The condition (when it is most severe) results in death usually within 2–4 days after birth.[19] It has been determined that patients with the condition display a two base pair deletion within the gene known as SUCLG1 that encodes the α subunit of SCS.[19] As a result, functional SCS is absent in metabolism causing a major imbalance in flux between glycolysis and the citric acid cycle. Since the cells do not have a functional citric acid cycle, acidosis results because cells are forced to choose lactic acid production as the primary means of producing ATP.
See also
[edit]- Citric acid cycle
- Succinate dehydrogenase
- Succinate—CoA ligase (ADP-forming)
- Succinate—CoA ligase (GDP-forming)
References
[edit]- ^ Fraser ME, Hayakawa K, Hume MS, Ryan DG, Brownie ER (Apr 2006). "Interactions of GTP with the ATP-grasp domain of GTP-specific succinyl-CoA synthetase". The Journal of Biological Chemistry. 281 (16): 11058–65. doi:10.1074/jbc.M511785200. PMID 16481318.
- ^ Fraser ME, James MN, Bridger WA, Wolodko WT (Jan 1999). "A detailed structural description of Escherichia coli succinyl-CoA synthetase". Journal of Molecular Biology. 285 (4): 1633–53. doi:10.1006/jmbi.1998.2324. PMID 9917402.
- ^ a b Voet, Donald J. (2011). Biochemistry / Donald J. Voet ; Judith G. Voet. New York, NY: Wiley, J. ISBN 978-0-470-57095-1.
- ^ a b c Berg, Jeremy M. (Jeremy M.); Tymoczko, John L.; Stryer, Lubert.; Stryer, Lubert. Biochemistry. (2002). Biochemistr. New York: W.H. Freeman. pp. 475–477. ISBN 0-7167-3051-0.
- ^ Nishimura JS (1986). "Succinyl-CoA synthetase structure-function relationships and other considerations". Advances in Enzymology and Related Areas of Molecular Biology. Advances in Enzymology and Related Areas of Molecular Biology. Vol. 58. pp. 141–72. doi:10.1002/9780470123041.ch4. ISBN 9780470123041. PMID 3521216.
- ^ a b c Wolodko WT, Kay CM, Bridger WA (Sep 1986). "Active enzyme sedimentation, sedimentation velocity, and sedimentation equilibrium studies of succinyl-CoA synthetases of porcine heart and Escherichia coli". Biochemistry. 25 (19): 5420–5. doi:10.1021/bi00367a012. PMID 3535876.
- ^ a b c Fraser ME, James MN, Bridger WA, Wolodko J (May 1999). "A detailed structural description of escherichia coli succinly-CoA synthetase". Journal of Molecular Biology. 288 (3): 501. doi:10.1006/jmbi.1999.2773. PMID 10329157.
- ^ a b Joyce MA, Fraser ME, James MN, Bridger WA, Wolodko WT (Jan 2000). "ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography". Biochemistry. 39 (1): 17–25. doi:10.1021/bi991696f. PMID 10625475.
- ^ a b Fraser ME, Joyce MA, Ryan DG, Wolodko WT (Jan 2002). "Two glutamate residues, Glu 208 alpha and Glu 197 beta, are crucial for phosphorylation and dephosphorylation of the active-site histidine residue in succinyl-CoA synthetase". Biochemistry. 41 (2): 537–46. doi:10.1021/bi011518y. PMID 11781092.
- ^ a b c d Johnson JD, Mehus JG, Tews K, Milavetz BI, Lambeth DO (Oct 1998). "Genetic evidence for the expression of ATP- and GTP-specific succinyl-CoA synthetases in multicellular eucaryotes". The Journal of Biological Chemistry. 273 (42): 27580–6. doi:10.1074/jbc.273.42.27580. PMID 9765291.
- ^ a b Lambeth DO, Tews KN, Adkins S, Frohlich D, Milavetz BI (Aug 2004). "Expression of two succinyl-CoA synthetases with different nucleotide specificities in mammalian tissues". The Journal of Biological Chemistry. 279 (35): 36621–4. doi:10.1074/jbc.M406884200. PMID 15234968.
- ^ Johnson JD, Muhonen WW, Lambeth DO (Oct 1998). "Characterization of the ATP- and GTP-specific succinyl-CoA synthetases in pigeon. The enzymes incorporate the same alpha-subunit". The Journal of Biological Chemistry. 273 (42): 27573–9. doi:10.1074/jbc.273.42.27573. PMID 9765290.
- ^ Labbe RF, Kurumada T, Onisawa J (Dec 1965). "The role of succinyl-CoA synthetase in the control of heme biosynthesis". Biochimica et Biophysica Acta (BBA) - General Subjects. 111 (2): 403–15. doi:10.1016/0304-4165(65)90050-4. PMID 5879477.
- ^ Ottaway JH, McClellan JA, Saunderson CL (1981). "Succinic thiokinase and metabolic control". The International Journal of Biochemistry. 13 (4): 401–10. doi:10.1016/0020-711x(81)90111-7. PMID 6263728.
- ^ Jenkins TM, Weitzman PD (Sep 1986). "Distinct physiological roles of animal succinate thiokinases. Association of guanine nucleotide-linked succinate thiokinase with ketone body utilization". FEBS Letters. 205 (2): 215–8. doi:10.1016/0014-5793(86)80900-0. PMID 2943604. S2CID 23667115.
- ^ Kruspl W, Streitmann B (Feb 1975). "[Knotty reticulosis with keloid formation]". Zeitschrift für Hautkrankheiten. 50 (3): 117–25. PMID 179232.
- ^ Hunger-Glaser I, Brun R, Linder M, Seebeck T (May 1999). "Inhibition of succinyl CoA synthetase histidine-phosphorylation in Trypanosoma brucei by an inhibitor of bacterial two-component systems". Molecular and Biochemical Parasitology. 100 (1): 53–9. doi:10.1016/s0166-6851(99)00032-8. PMID 10376993.
- ^ Wider de Xifra E, del C Batlle AM (Mar 1978). "Porphyrin biosynthesis: immobilized enzymes and ligands. VI. Studies on succinyl CoA synthetase from cultured soya bean cells". Biochimica et Biophysica Acta. 523 (1): 245–9. doi:10.1016/0005-2744(78)90027-x. PMID 564714.
- ^ a b Ostergaard E, Christensen E, Kristensen E, Mogensen B, Duno M, Shoubridge EA, Wibrand F (Aug 2007). "Deficiency of the alpha subunit of succinate-coenzyme A ligase causes fatal infantile lactic acidosis with mitochondrial DNA depletion". American Journal of Human Genetics. 81 (2): 383–7. doi:10.1086/519222. PMC 1950792. PMID 17668387.
External links
[edit]- Succinyl+Coenzyme+A+Synthetases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)

