Alternative splicing: Difference between revisions
m →Modes: Minor fix |
→Importance in molecular genetics: Remove unsourced claim, add ref to another, insert counter-claim, move Dscam. |
||
| Line 128: | Line 128: | ||
</ref>." External information is needed in order to decide which polypeptide is produced, given a DNA sequence and pre-mRNA. Since the methods of regulation are inherited, this provides novel ways for mutations to affect gene expression<ref name=Fackenthal/>. |
</ref>." External information is needed in order to decide which polypeptide is produced, given a DNA sequence and pre-mRNA. Since the methods of regulation are inherited, this provides novel ways for mutations to affect gene expression<ref name=Fackenthal/>. |
||
It has been proposed that for [[eukaryote]]s alternative splicing was a very important step towards higher efficiency, because information can be stored much more economically. Several proteins can be encoded by a single gene, rather than requiring a separate gene for each, and thus allowing a more varied [[proteome]] from a [[genome]] of limited size<ref name=Black/>. It also provides evolutionary flexibility. A single point mutation may cause a given exon to be occasionally excluded or included from a transcript during splicing, allowing production of a new [[protein isoform]] without loss of the original protein<ref name=Black/>. |
It has been proposed that for [[eukaryote]]s alternative splicing was a very important step towards higher efficiency, because information can be stored much more economically. Several proteins can be encoded by a single gene, rather than requiring a separate gene for each, and thus allowing a more varied [[proteome]] from a [[genome]] of limited size<ref name=Black/>. It also provides evolutionary flexibility. A single point mutation may cause a given exon to be occasionally excluded or included from a transcript during splicing, allowing production of a new [[protein isoform]] without loss of the original protein<ref name=Black/>. Comparative studies indicate that alternative splicing preceded multicellularity in evolution, and suggest that this mechanism might have been co-opted to assist in the development of multicellular organisms<ref name=Irimia>{{Cite doi|10.1186/1471-2148-7-188}}</ref>. |
||
Another speculation is that new proteins could be allowed to evolve much faster than in prokaryotes. Furthermore, they are based on hitherto functional amino acid subchains. This may allow for a higher probability for a functional new protein{{fact|date=May 2009}}. Therefore the adaptation to new environments can be much faster - with fewer generations - than in prokaryotes. This might have been one very important step for multicellular organisms with a longer life cycle{{fact|date=May 2009}}. |
|||
Research based on the [[Human Genome Project]] and other genome sequencing has shown that humans have only about twice as many genes as the roundworm ''[[Caenorhabditis elegans]]'' or the fly ''[[Drosophila melanogaster]]''. This finding led to speculation that the perceived greater complexity of humans, or vertebrates generally, might be due to higher rates of alternative splicing in humans than are found in invertebrates<ref name=ewing>{{cite journal |
Research based on the [[Human Genome Project]] and other genome sequencing has shown that humans have only about twice as many genes as the roundworm ''[[Caenorhabditis elegans]]'' or the fly ''[[Drosophila melanogaster]]''. This finding led to speculation that the perceived greater complexity of humans, or vertebrates generally, might be due to higher rates of alternative splicing in humans than are found in invertebrates<ref name=ewing>{{cite journal |
||
| Line 167: | Line 165: | ||
pages=29–30| |
pages=29–30| |
||
url=http://www.nature.com/ng/journal/v30/n1/abs/ng803.html |
url=http://www.nature.com/ng/journal/v30/n1/abs/ng803.html |
||
}}</ref> Another study suggested that these results were an artifact of the different numbers of ESTs available for the various organisms, and used a frequency-based method instead. In this case the authors found evidence that vertebrates have more alternatively spliced transcripts than invertebrates<ref name=Kim>{{Cite doi|10.1093/nar/gkl924}}</ref>. |
|||
}}</ref> The "record-holder" for alternative splicing is actually a ''D. melanogaster'' gene called [[Dscam]], which has 38,016 splice variants<ref name=Schmucker>{{cite journal |author=Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL |title=Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity |journal=Cell |volume=101 |issue=6 |pages=671–84 |year=2000 |month=June |pmid=10892653 |doi= |url=http://linkinghub.elsevier.com/retrieve/pii/S0092-8674(00)80878-8}}</ref>. |
|||
==Alternative splicing and disease== |
==Alternative splicing and disease== |
||
Revision as of 22:19, 26 May 2009
Alternative splicing is a process by which the exons of a primary gene transcript or pre-mRNA are reconnected in multiple ways during RNA splicing. The resulting different mRNAs may be translated into different protein isoforms; thus, a single gene may code for multiple proteins. Alternative splicing occurs as a normal phenomenon in many eukaryotes, where it greatly increases the diversity of proteins that can be encoded by the genome[1]; in humans, over 80% of genes are alternatively spliced[2]. Abnormal variations in splicing are also commonly implicated in disease; 15 to 50% of human genetic disorders are thought to result from splicing variants[2].
Discovery
Alternative splicing was first observed in the late 1970s. Researchers studying the late transcription unit of adenovirus type 2 found that after transcription, the primary RNA transcript was spliced in different ways to produce mRNAs encoding different viral proteins. Both 5’ and 3’ splice sites varied, and in addition, the transcript contained multiple polyadenylation sites, resulting in different 3’ ends for the processed mRNAs[3][4][5].
In 1981, the first example of alternative splicing in a transcript from a normal, endogenous gene was characterized[3]. The gene encoding the thyroid hormone calcitonin was found to be alternatively spliced in mammalian cells. The primary transcript from this gene contains 6 exons; the calcitonin mRNA contains exons 1-4, and terminates after a polyadenylation site in exon 4. Another mRNA, coding for a protein known as CGRP (calcitonin gene related peptide), is produced by skipping exon 4, and includes exons 1-3, 5, and 6[6][7]. Examples of alternative splicing in immunoglobin gene transcripts in mammals were also observed in the early 1980s[3][8].
Modes
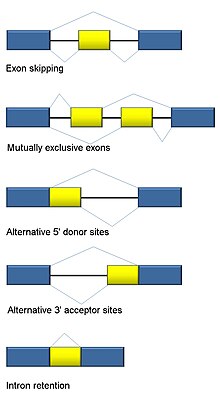
Five basic modes of alternative splicing are generally recognized.[1][2][9]
- Exon skipping or cassette exon: in this case, an exon may be spliced out of the primary transcript or retained. This is generally the most common mode in mammalian pre-mRNAs[9].
- Mutually exclusive exons: One of two exons is retained in mRNAs after splicing, but not both.
- Alternative donor site: An alternative 5' splice junction (donor site) is used, changing the 3' boundary of the upstream exon.
- Alternative acceptor site: An alternative 3' splice junction (acceptor site) is used, changing the 5' boundary of the downstream exon.
- Intron retention: A sequence may be spliced out as an intron or simply retained. This is distinguished from exon skipping because the retained sequence is not flanked by introns. If the retained intron is in the coding region, the intron must encode amino acids in frame with the neighboring exons, or a stop codon or a shift in the reading frame will cause the protein to be non-functional. This is generally the rarest mode in mammals[9].
In addition to these primary modes of alternative splicing, there are two other main mechanisms by which different mRNAs may be generated from the same gene; multiple promoters and multiple polyadenylation sites. Use of multiple promoters is properly described as a transcriptional regulation mechanism rather than alternative splicing; by starting transcription at different points, transcripts with different 5'-most exons can be generated. At the other end, multiple polyadenylation sites provide different 3' end points for the transcript. Both of these mechanisms are found in combination with alternative splicing and provide additional variety in mRNAs derived from a gene[1][2].
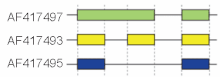
Traditional terminology describes basic splicing mechanisms, but may be inadequate to describe complex splicing events. For instance, the figure to the right shows 3 spliceforms from the murine hyaluronidase 3 gene. Comparing the exonic structure shown in the first line (green) with the one in the second line (yellow) shows intron retention, whereas the comparison between the second and the third spliceform (yellow vs. blue) exhibits exon skipping. A model nomenclature to uniquely designate all possible splicing patterns has recently been proposed[9].
Alternative splicing mechanisms
General splicing mechanism

When the pre-mRNA has been transcribed from the DNA, it includes several introns and exons. (In nematodes, the mean is 4-5 exons and introns; in the fruit fly Drosophila there can be more than 100 introns and exons in one transcribed pre-mRNA.) The exons which are retained in the mRNA are determined during the splicing process. The regulation and selection of splice sites are done by Serine/Arginine-residue proteins, or SR proteins.
The typical eukaryotic nuclear intron has consensus sequences defining important regions. Each intron has GU at its 5' end. Near the 3' end there is a branch site. The nucleotide at the branch point is always an A; the consensus around this sequence varies somewhat. In humans the branch consensus is yUnAy [10]. The branch site is followed by a series of pyrimidines, or polypyrimidine tract, then by AG at 3' end[2].
Splicing of mRNA is performed by a complex known as the spliceosome, composed of snRNPs designated U1 through U6. U1 binds to 5' GU and U2 binds to branch site (A) with the assistance of the U2AF protein factors. The complex at this stage is known as the spliceosome A complex, and its formation is the key step in determining the ends of the intron to be spliced[2].
The U4,U5,U6 complex binds, and U6 replaces the U1 position. U1 and U4 leave. The remaining complex then performs two transesterification reactions. In the first, the branch site A attacks the 5' end G to form a 2',5'-phosphodiester linkage. In the second transesterification, the 3' end of upstream exon (G) captures the 3' end of intron by forming phosphodiester bond again, so that two exons are joined together, leaving a free intron in lariat form. U3 is not involved in mRNA splicing[1].
Regulatory elements and proteins

There are two major types of cis-acting RNA sequence elements present in pre-mRNAs, and specific trans-acting RNA-binding proteins bind to each of these elements. Silencers are sites to which splicing repressor proteins bind, reducing the probability that a nearby site will be used as a splice junction. These can be located in the intron itself (intronic splicing silencer, ISS) or in a neighboring exon (exonic splicing silencer, ESS). They vary in sequence, as well as in the types of proteins that bind to them. The majority of the repressors that bind are heterogeneous nuclear ribonucleoproteins (hnRNPs) such as hnRNPA1 and polypyrimidine tract binding protein (PTB)[2][11].
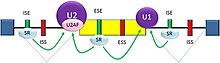
Splicing Enhancers are sites to which splicing activator proteins bind, increasing the probability that a nearby site will be used as a splice junction. These also may occur in the intron (intronic splicing enhancer, ISE) or exon (exonic splicing enhancer, ESE). Most of the activator proteins that bind to ISEs and ESEs are members of the SR protein family. Such proteins contain RNA recognition motifs and arginine and serine-rich (RS) domains[2][11].
The adaptive significance of silencers and enhancers is attested by a study showing that there is strong selection in human genes against mutations that produce new silencers or disrupt existing enhancers[12].
Examples
Drosophila dsx
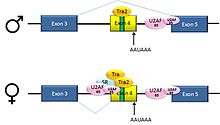
Pre-mRNAs from the D. melanogaster gene dsx contain 6 exons. In males, exons 1,2,3,5,and 6 are joined to form the mRNA, which encodes a transcriptional regulatory protein required for male development. In females, exons 1,2,3, and 4 are joined, and a polyadenylation signal in exon 4 causes cleavage of the mRNA at that point. The resulting mRNA is a transcriptional regulatory protein required for female development[13].
This is an example of exon skipping. The intron upstream from exon 4 has a polypyrimidine tract that doesn't match the consensus sequence well, so that U2AF proteins bind poorly to it without assistance from splicing activators. This 3' splice acceptor site is therefore not used in males. Females, however, produce the splicing activator Transformer (Tra)(see below). The SR protein Tra2 is produced in both sexes and binds to an ESE in exon 4; if Tra is present, it binds to Tra2 and, along with another SR protein, forms a complex that assists U2AF proteins in binding to the weak polypyrimidine tract. U2 is recruited to the associated branch point, and this leads to inclusion of exon 4 in the mRNA[13][14].
Drosophila Transformer
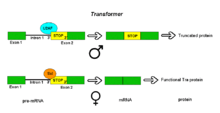
Pre-mRNAs of the Transformer (Tra) gene of Drosophila melanogaster undergo alternative splicing via the alternative acceptor site mode. The gene Tra encodes a protein that is expressed only in females. The primary transcript of this gene contains an intron with two possible acceptor sites. In males, the upstream acceptor site is used. This causes a longer version of exon 2 to be included in the processed transcript, including an early stop codon. The resulting mRNA encodes a truncated protein product that is inactive. Females produce the master sex determination protein Sex lethal (Sxl). The Sxl protein is a splicing repressor. It binds to an ISS in the RNA of the Tra transcript near the upstream acceptor site, preventing U2AF protein from binding to the polypyrimidine tract. This prevents the use of this junction, shifting the spliceosome binding to the downstream acceptor site. Splicing at this point bypasses the stop codon, which is excised as part of the intron. The resulting mRNA encodes an active Tra protein, which itself is a regulator of alternative splicing of other sex-related genes (see dsx above)[1].
Fas receptor
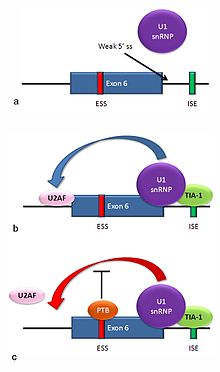
Multiple isoforms of the Fas receptor protein are produced by alternative splicing. Two normally occurring isoforms in humans are produced by an exon-skipping mechanism. An mRNA including exon 6 encodes the membrane-bound form of the Fas receptor, which promotes apoptosis, or programmed cell death. Increased expression of Fas receptor in skin cells chronically exposed to the sun, and absence of expression in skin cancer cells, suggests that this mechanism may be important in elimination of pre-cancerous cells in humans[15]. If exon 6 is skipped, the resulting mRNA encodes a soluble Fas protein that does not promote apoptosis. The inclusion or skipping of the exon depends on two antagonistic proteins, TIA-1 and polypyrimidine tract-binding protein (PTB).
- The 5' donor site in the intron downstream from exon 6 in the pre-mRNA has a weak agreement with the consensus sequence, and is not bound usually by the U1 snRNP. If U1 does not bind, the exon is skipped (see "a" in accompanying figure).
- Binding of TIA-1 protein to an intronic splicing enhancer site stabilizes binding of the U1 snRNP[2]. The resulting 5' donor site complex assists in binding of the splicing factor U2AF to the 3' splice site upstream of the exon, through a mechanism that is not yet known (see b)[16].
- Exon 6 contains a pyrimidine-rich exonic splicing silencer, ure6, where PTB can bind. If PTB binds, in inhibits the effect of the 5' donor complex on the binding of U2AF to the acceptor site, resulting in exon skipping (see c).
This mechanism is an example of exon definition in splicing. A spliceosome assembles on an intron, and the snRNP subunits bring fold the RNA so that the 5' and 3' ends of the intron are joined. However, recently studied examples such as this one show that there are also interactions between the ends of the exon. In this particular case, these exon definition interactions are necessary to allow the binding of core splicing factors prior to assembly of the spliceosomes on the two flanking introns. [16].
HIV-1 tat exon 2

HIV, the retrovirus that causes AIDS in humans, produces a single primary RNA transcript, which is alternatively spliced in multiple ways to produce over 40 different mRNAs[17]. Equilibrium among differentially spliced transcripts provides multiple mRNAs encoding different products that are required for viral multiplication. [18]. One of the differentially spliced transcripts contains the tat gene, in which exon 2 is a cassette exon that may be skipped or included. The inclusion of tat exon 2 in the RNA is regulated by competition between the splicing repressor hnRNP A1 and the SR protein SC35. Within exon 2 an exonic splicing silencer sequence (ESS) and an exonic splicing enhancer sequence (ESE) overlap. If A1 repressor protein binds to the ESS, it initiates cooperative binding of multiple A1 molecules, extending into the 5’ donor site upstream of exon 2 and preventing the binding of the core splicing factor U2AF35 to the polypyrimidine tract. If SC35 binds to the ESE, it prevents A1 binding and maintains the 5’ donor site in an accessible state for assembly of the spliceosome. Competition between the activator and repressor provides the required equilibrium among transcripts including and excluding exon 2[17].
Importance in molecular genetics
Alternative splicing is of great importance to genetics; it is one of several exceptions to the original idea that one DNA sequence codes for one polypeptide (the One gene-one enzyme hypothesis). It might be more correct now to say "One gene - many polypeptides[19]." External information is needed in order to decide which polypeptide is produced, given a DNA sequence and pre-mRNA. Since the methods of regulation are inherited, this provides novel ways for mutations to affect gene expression[20].
It has been proposed that for eukaryotes alternative splicing was a very important step towards higher efficiency, because information can be stored much more economically. Several proteins can be encoded by a single gene, rather than requiring a separate gene for each, and thus allowing a more varied proteome from a genome of limited size[1]. It also provides evolutionary flexibility. A single point mutation may cause a given exon to be occasionally excluded or included from a transcript during splicing, allowing production of a new protein isoform without loss of the original protein[1]. Comparative studies indicate that alternative splicing preceded multicellularity in evolution, and suggest that this mechanism might have been co-opted to assist in the development of multicellular organisms[21].
Research based on the Human Genome Project and other genome sequencing has shown that humans have only about twice as many genes as the roundworm Caenorhabditis elegans or the fly Drosophila melanogaster. This finding led to speculation that the perceived greater complexity of humans, or vertebrates generally, might be due to higher rates of alternative splicing in humans than are found in invertebrates[22][23]. However, a study on samples of 100,000 ESTs each from human, mouse, rat, cow, fly (D. melanogaster), worm (C. elegans), and the plant Arabidopsis thaliana found no large differences in frequency of alternatively spliced genes among humans and any of the other animals tested.[24] Another study suggested that these results were an artifact of the different numbers of ESTs available for the various organisms, and used a frequency-based method instead. In this case the authors found evidence that vertebrates have more alternatively spliced transcripts than invertebrates[25].
Alternative splicing and disease
Changes in the RNA processing machinery may lead to mis-splicing of multiple transcripts, while single-nucleotide alterations in splice sites or cis-acting splicing regulatory sites may lead to differences in splicing of a single gene, and thus in the mRNA produced from a mutant gene's transcripts. A probabilistic analysis indicates that over 60% of human disease-causing mutations affect splicing rather than directly affecting coding sequences. [26].
Abnormally spliced mRNAs are also found in a high proportion of cancerous cells[27][28]. Until recently, it was unclear whether such aberrant patterns of splicing played a role in causing cancerous growth, or were merely a consequence of cellular abnormalities associated with cancer. It has been shown that there is actually a reduction of alternative splicing in cancerous cells compared to normal ones, and the types of splicing differ; for instance, cancerous cells show higher levels of intron retention than normal cells, but lower levels of exon skipping[29]. Some of the differences in splicing in cancerous cells may result from changes in phosphorylation of trans-acting splicing factors[20] . Others may be produced by changes in the relative amounts of splicing factors produced; for instance, breast cancer cells have been shown to have increased levels of the splicing factor SF2/ASF[30]. One study found that a relatively small percentage (383 out of over 26000) of alternative splicing variants were significantly higher in frequency in tumor cells than normal cells, suggesting that there is a limited set of genes which, when mis-spliced, contribute to tumor development[31].
One example of a specific splicing variant associated with cancers is in one of the human DNMT genes. Three DNMT genes encode enzymes that add methyl groups to DNA, a modification that often has regulatory effects. Several abnormally spliced DNMT3B mRNAs are found in tumors and cancer cell lines. In two separate studies, expression of two of these abnormally spliced mRNAs in mammalian cells caused changes in the DNA methylation patterns in those cells. Cells with one of the abnormal mRNAs also grew twice as fast as control cells, indicating a direct contribution to tumor development by this product[20].
Another example is the Ron (MST1R) proto-oncogene. An important property of cancerous cells is their ability to move and invade normal tissue. Production of an abnormally spliced transcript of Ron has been found to be associated with increased levels of the SF2/ASF in breast cancer cells. The abnomal isoform of the Ron protein encoded by this mRNA leads to cell motility[30].
Genome-wide analysis of alternative splicing
Genome-wide analysis of alternative splicing is a challenging task. Typically, alternatively spliced transcripts have been found by comparing EST sequences, but this requires sequencing of very large numbers of ESTs. Most EST libraries come from a very limited number of tissues, so tissue-specific splice variants are likely to be missed in any case. High-throughput approaches to investigate splicing fall into three categories; DNA microarray-based analyses, CLIP, and in vivo reporter gene assays[11].
In microarray analysis, arrays of DNA fragments representing individual exons (e.g. Affymetrix exon microarray) or exon/exon boundaries (e.g. arrays from ExonHit or Jivan) have been used. The array is then probed with labeled cDNA from tissues of interest. The probe cDNAs bind to DNA from the exons that are included in mRNAs in their tissue of origin, or to DNA from the boundary where two exons have been joined. This can reveal the presence of particular alternatively spliced mRNAs[32]. Deep sequencing technologies are also being used to perform genome-wide studies of transcript variation.
CLIP (Cross-linking and immunoprecipitation) uses UV radiation to link proteins to RNA molecules in a tissue during splicing. A trans-acting splicing regulatory protein of interest is then precipitated using specific antibodies. When the RNA attached to that protein is isolated and cloned, it reveals the target sequences for that protein[33].
Finally, it is possible to find the splicing proteins involved in a specific alternative splicing event by constructing reporter genes that will express one of two different fluorescent proteins depending on the splicing reaction that occurs. This method has been used to isolate mutants affecting splicing and thus to identify novel splicing regulatory proteins inactivated in those mutants[33].
References
- ^ a b c d e f g Black, Douglas L. (2003). "Mechanisms of alternative pre-messenger RNA splicing". Annual Reviews of Biochemistry. 72 (1): 291–336.
- ^ a b c d e f g h i j Matlin, AJ (May 2005). "Understanding alternative splicing: towards a cellular code". Nature Reviews. 6: 386–398.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Leff SE, Rosenfeld MG, Evans RM (1986). "Complex transcriptional units: diversity in gene expression by alternative RNA processing". Annu. Rev. Biochem. 55: 1091–117. doi:10.1146/annurev.bi.55.070186.005303. PMID 3017190.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Chow LT, Broker TR (1978). "The spliced structures of adenovirus 2 fiber message and the other late mRNAs". Cell. 15 (2): 497–510. PMID 719751.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Nevins JR, Darnell JE (1978). "Steps in the processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing". Cell. 15 (4): 1477–93. PMID 729004.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Rosenfeld MG, Amara SG, Roos BA, Ong ES, Evans RM (1981). "Altered expression of the calcitonin gene associated with RNA polymorphism". Nature. 290 (5801): 63–5. PMID 7207587.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rosenfeld MG, Lin CR, Amara SG; et al. (1982). "Calcitonin mRNA polymorphism: peptide switching associated with alternative RNA splicing events". Proc. Natl. Acad. Sci. U.S.A. 79 (6): 1717–21. PMC 346051. PMID 6952224.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Maki R, Roeder W, Traunecker A; et al. (1981). "The role of DNA rearrangement and alternative RNA processing in the expression of immunoglobulin delta genes". Cell. 24 (2): 353–65. PMID 6786756.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d
Michael Sammeth (8 August, 2008). "A general definition and nomenclature for alternative splicing events". PLoS Comput Biol. 4: e1000147. doi:10.1371/journal.pcbi.1000147.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1093/nar/gkn073, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1093/nar/gkn073instead. - ^ a b c Wang, Z; Burge, Cb (2008). "Splicing regulation: from a parts list of regulatory elements to an integrated splicing code" (Free full text). RNA (New York, N.Y.). 14 (5): 802–13. doi:10.1261/rna.876308. ISSN 1355-8382. PMC 2327353. PMID 18369186.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ke S, Zhang XH, Chasin LA (2008). "Positive selection acting on splicing motifs reflects compensatory evolution". Genome Res. 18 (4): 533–43. doi:10.1101/gr.070268.107. PMC 2279241. PMID 18204002.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Lynch KW, Maniatis T (1996). "Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer". Genes Dev. 10 (16): 2089–101. PMID 8769651.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Graveley BR, Hertel KJ, Maniatis T (2001). "The role of U2AF35 and U2AF65 in enhancer-dependent splicing". RNA. 7 (6): 806–18. PMC 1370132. PMID 11421359.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi: 10.1002/cncr.10277, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi= 10.1002/cncr.10277instead. - ^ a b Izquierdo JM, Majós N, Bonnal S; et al. (2005). "Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition". Mol. Cell. 19 (4): 475–84. doi:10.1016/j.molcel.2005.06.015. PMID 16109372.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1074/jbc.M312743200, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1074/jbc.M312743200instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1074/jbc.M104070200, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1074/jbc.M104070200instead. - ^ "HHMI Bulletin September 2005: Alternative Splicing". www.hhmi.org. Retrieved 2009-05-26.
- ^ a b c Fackenthal, Jd; Godley, La (2008). "Aberrant RNA splicing and its functional consequences in cancer cells" (Free full text). Disease models & mechanisms. 1 (1): 37–42. doi:10.1242/dmm.000331. ISSN 1754-8403. PMC 2561970. PMID 19048051.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1186/1471-2148-7-188, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1186/1471-2148-7-188instead. - ^ Ewing, B (June 2000). "Analysis of expressed sequence tags indicates 35,000 human genes". Nature Genetics. 25 (2): 232–234. Retrieved 2009-04-28.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^
Crollius, HR (June 2000). "Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence". Nature Genetics. 25 (2): 235–238. Retrieved 2009-04-28.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^
David Brett (January 2002 (online 17 December, 2001)). "Alternative splicing and genome complexity". Nature Genetics. 30: 29–30. doi:10.1038/ng803.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1093/nar/gkl924, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1093/nar/gkl924instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/j.febslet.2005.02.047, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/j.febslet.2005.02.047instead. - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1371/journal.pone.0004732, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1371/journal.pone.0004732instead. - ^ Skotheim and Nees (2007), Alternative splicing in cancer: noise, functional, or systematic?
- ^ Kim E, Goren A, Ast G (2008). "Insights into the connection between cancer and alternative splicing". Trends Genet. 24 (1): 7–10. doi:10.1016/j.tig.2007.10.001. PMID 18054115.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Ghigna C, Giordano S, Shen H; et al. (2005). "Cell motility is controlled by SF2/ASF through alternative splicing of the Ron proto-oncogene". Mol. Cell. 20 (6): 881–90. doi:10.1016/j.molcel.2005.10.026. PMID 16364913.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hui L, Zhang X, Wu X; et al. (2004). "Identification of alternatively spliced mRNA variants related to cancers by genome-wide ESTs alignment". Oncogene. 23 (17): 3013–23. doi:10.1038/sj.onc.1207362. PMID 15048092.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/j.molcel.2004.12.004, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/j.molcel.2004.12.004instead. - ^ a b Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1101/gad.1643108, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1101/gad.1643108instead.
External links
- A General Definition and Nomenclature for Alternative Splicing Events at SciVee
- AStalavista (Alternative Splicing landscape visualization tool), a method for the computationally exhaustive classification of Alternative Splicing Structures
- Stamms-lab.net: Research Group dealing with alternative Splicing issues and mis-splicing in human diseases
- Alternative Splicing of ion channels in the brain, connected to mental and neurological diseases
- BIPASS: Web Services in Alternative Splicing
See also
