Haplogroup D-M174: Difference between revisions
Citation bot (talk | contribs) m Add: bibcode, pmc, pmid. Removed URL that duplicated unique identifier. | You can use this bot yourself. Report bugs here.| Activated by User:Chris Capoccia |
|||
| Line 72: | Line 72: | ||
Kim ''et al.'' (2011) found Haplogroup D-M55 in 2.0% (1/51) of a sample of [[Beijing]] [[Han Chinese|Han]] and in 1.6% (8/506) of a pool of samples from South Korea, including 3.3% (3/90) from the Jeolla region, 2.4% (2/84) from the Gyeongsang region, 1.4% (1/72) from the Chungcheong region, 1.1% (1/87) from the Jeju region, 0.9% (1/110) from the Seoul-Gyeonggi region, and 0% (0/63) from the Gangwon region.<ref name = "Kim2011">Soon-Hee Kim, Ki-Cheol Kim, Dong-Jik Shin, Han-Jun Jin, Kyoung-Don Kwak, Myun-Soo Han, Joon-Myong Song, Won Kim, and Wook Kim, "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea." ''Investigative Genetics'' 2011, 2:10. http://www.investigativegenetics.com/content/2/1/10</ref> Hammer ''et al.'' (2006) found Haplogroup D-P37.1 in 4.0% (3/75) of a sample from South Korea.<ref name = "Hammer2006" /> |
Kim ''et al.'' (2011) found Haplogroup D-M55 in 2.0% (1/51) of a sample of [[Beijing]] [[Han Chinese|Han]] and in 1.6% (8/506) of a pool of samples from South Korea, including 3.3% (3/90) from the Jeolla region, 2.4% (2/84) from the Gyeongsang region, 1.4% (1/72) from the Chungcheong region, 1.1% (1/87) from the Jeju region, 0.9% (1/110) from the Seoul-Gyeonggi region, and 0% (0/63) from the Gangwon region.<ref name = "Kim2011">Soon-Hee Kim, Ki-Cheol Kim, Dong-Jik Shin, Han-Jun Jin, Kyoung-Don Kwak, Myun-Soo Han, Joon-Myong Song, Won Kim, and Wook Kim, "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea." ''Investigative Genetics'' 2011, 2:10. http://www.investigativegenetics.com/content/2/1/10</ref> Hammer ''et al.'' (2006) found Haplogroup D-P37.1 in 4.0% (3/75) of a sample from South Korea.<ref name = "Hammer2006" /> |
||
Low levels of D-M116.1 (a subclade of D-M55) among males in present-day [[Timor]] (0.2% of males),<ref name="tumonggor">Tumonggor |
Low levels of D-M116.1 (a subclade of D-M55) among males in present-day [[Timor]] (0.2% of males),<ref name="tumonggor">{{cite journal |last1=Tumonggor |first1=Meryanne K |last2=Karafet |first2=Tatiana M |last3=Downey |first3=Sean |last4=Lansing |first4=J Stephen |last5=Norquest |first5=Peter |last6=Sudoyo |first6=Herawati |last7=Hammer |first7=Michael F |last8=Cox |first8=Murray P |title=Isolation, contact and social behavior shaped genetic diversity in West Timor |journal=Journal of Human Genetics |date=September 2014 |volume=59 |issue=9 |pages=494–503 |doi=10.1038/jhg.2014.62 |pmid=25078354 |pmc=4521296 }}</ref> It's found in 9.5% of males from [[Micronesia]](Hammer et al. 2006,<ref name = Hammer/><ref name = "Hammer2006"/> is believed to reflect recent admixture from Japan. That is, D-M116.1 (D1b1) is generally believed to be a primary subclade of D-M64.1 (D1b), possibly as a result of the [[Japanese occupation of the Dutch East Indies|Japanese military occupation of South East Asia]] during [[World War II]]. |
||
According to [[:ja:崎谷満|Mitsuru Sakitani]], Haplogroup D1 arrived from Central Asia to northern [[Kyushu]] via the [[Altai Mountains]] and the [[Korean Peninsula]] more than 40,000 years before present, and Haplogroup D-M55 (D1b) was born in Japanese archipelago.<ref name="Sakitani">崎谷満『DNA・考古・言語の学際研究が示す新・日本列島史』(勉誠出版 2009年)(in Japanese)</ref> |
According to [[:ja:崎谷満|Mitsuru Sakitani]], Haplogroup D1 arrived from Central Asia to northern [[Kyushu]] via the [[Altai Mountains]] and the [[Korean Peninsula]] more than 40,000 years before present, and Haplogroup D-M55 (D1b) was born in Japanese archipelago.<ref name="Sakitani">崎谷満『DNA・考古・言語の学際研究が示す新・日本列島史』(勉誠出版 2009年)(in Japanese)</ref> |
||
Recently it was confirmed that the Japanese branch of haplogroup D-M55 is distinct and isolated from other D-branches since more than 53,000 years. The split between D1a happend likely in Central Asia, while some others suggest a instant split during the origin of haplogroup D itself, as the Japanese branch has five unique mutations not found in any other D-branch.<ref>Mondal |
Recently it was confirmed that the Japanese branch of haplogroup D-M55 is distinct and isolated from other D-branches since more than 53,000 years. The split between D1a happend likely in Central Asia, while some others suggest a instant split during the origin of haplogroup D itself, as the Japanese branch has five unique mutations not found in any other D-branch.<ref>{{cite journal |last1=Mondal |first1=Mayukh |last2=Bergström |first2=Anders |last3=Xue |first3=Yali |last4=Calafell |first4=Francesc |last5=Laayouni |first5=Hafid |last6=Casals |first6=Ferran |last7=Majumder |first7=Partha P. |last8=Tyler-Smith |first8=Chris |last9=Bertranpetit |first9=Jaume |title=Y-chromosomal sequences of diverse Indian populations and the ancestry of the Andamanese |journal=Human Genetics |date=25 April 2017 |volume=136 |issue=5 |pages=499–510 |doi=10.1007/s00439-017-1800-0 }}</ref> |
||
==Phylogenetics== |
==Phylogenetics== |
||
Revision as of 13:37, 19 July 2019
| Haplogroup D-M174 | |
|---|---|
 | |
| Possible time of origin | 50,000[1] - 60,000[2] years BP 65,200 [95% CI 62,100 <-> 68,300] ybp[3] |
| Coalescence age | 46,300 [95% CI 43,500 <-> 49,100] ybp[3] |
| Possible place of origin | Asia[2][4][5] (Central Asia)[6] |
| Ancestor | DE |
| Descendants | D-Z27276(D1a) D-M55(D1b) D* D0 (proposed) |
| Defining mutations | M174, IMS-JST021355, PAGES00003 |
In human genetics, Haplogroup D-M174 is a Y-chromosome haplogroup. Both D-M174 and E lineages also exhibit the single-nucleotide polymorphism M168 which is present in all Y-chromosome haplogroups except A and B, as well as the YAP+ unique-event polymorphism, which is unique to Haplogroup DE. This haplogroup is found primarily in East Asia, though it is also found regularly with low frequency in Central Asia and Southeast Asia, and it has also been found sporadically in Europe and Western Asia.
Origins
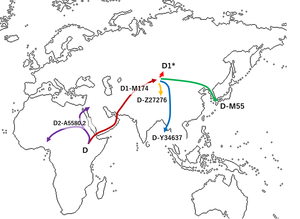
Haplogroup D-M174 is believed to have originated in Asia some 60,000 years before present.[2][4] While haplogroup D-M174 along with haplogroup E contains the distinctive YAP polymorphism (which indicates their common ancestry), no haplogroup D-M174 chromosomes have been found anywhere outside of Asia.[4]
Several studies (Hammer et al. 2006, Shinoda 2008, Matsumoto 2009, Cabrera et al. 2018) suggest that the paternal haplogroup D originated somewhere in Central Asia. According to Hammer et al., haplogroup D originated between Tibet and the Altai mountains. He suggests that there were multiple waves into Eastern Eurasia.[7][why?]
A study (Haber et al. 2019) found a proposed para-haplogroup named D0 in three samples in Western Africa (Nigeria). Because of the likely deep-rooting of haplogroup D0, as well as recently calculated early divergence times for it and it's parent haplogroup, the authors suggest an African origin for D0 and its ancestral haplogroup DE. The "D0"-CTS4030 haplogroup is outside M174, but shares 7 SNPs with it that E lacks.[8]
Three other samples of D0 were recently found in West Asia (2019): two among Arabs in Saudi Arabia and another one in a Syrian individual. The D0 sample found in the Syrian is to date the most basal sample of D0. The recent evidence (as also proposed by Haber et al.) suggests that D0 is a highly divergent branch of D (close to the DE split but on the D branch) and should not be considered as more deeply rooted than D itself.[9]
Overview
It is found today at high frequency among populations in Tibet, the Japanese archipelago, and the Andaman Islands, though curiously not as much in the rest of India. The Ainu of Japan are notable for possessing almost exclusively Haplogroup D-M55 chromosomes. Haplogroup D-M174 chromosomes are also found at low to moderate frequencies among the Bai, Dai, Han, Hui, Manchu, Miao, Tujia, Xibe, Yao, and Zhuang of China and among several minority populations of Sichuan and Yunnan that speak Tibeto-Burman languages and reside in close proximity to the Tibetans, such as the Jingpo, Jino, Mosuo, Naxi, Pumi, Qiang, and Yi.[10]
Haplogroup D is also found in populations of China proper and Korea, but with much lower frequency than in populations of Tibet and Japan. A study published in 2011 found D-M174 in 2.49% (43/1729) of Han Chinese males, with frequencies of this haplogroup tending to be higher than average toward the north and toward the west of the country (5/56 = 8.9% D-M174 Shaanxi Han, 13/221 = 5.9% D-M174 Gansu Han, 6/136 = 4.4% D-M174 Yunnan Han, 1/27 = 3.7% D-M174 Guangxi Han, 2/61 = 3.3% D-M174 Hunan Han, 2/62 = 3.2% D-M174 Sichuan Han).[11] In another study of Han Chinese Y-DNA published in 2011, Haplogroup D-M174 was observed in 1.94% (7/361) of a sample of unrelated Han Chinese male volunteers at Fudan University in Shanghai, with the origins of most of the volunteers being traced back to East China (Jiangsu, Zhejiang, Shanghai, and Anhui).[12] In Korea, Haplogroup D-M174 has been observed in 3.8% (5/133) of a sample from Daejeon,[13] 3/85 = 3.5% of a sample from Seoul,[14] 3.3% (3/90) of a sample from Jeolla,[15] 2.4% (2/84) of a sample from Gyeongsang,[15] 2.3% (13/573) of another sample from Seoul,[13] 1.4% (1/72) of a sample from Chungcheong,[15] 1.1% (1/87) of a sample from Jeju,[15] and 0.9% (1/110) of a third sample from Seoul-Gyeonggi.[15] In other studies, haplogroup D-M174 has been observed in 6.7% (3/45)[16] and 4.0% (3/75)[17] of samples from Korea without any further specification of the area of sampling. Little high-resolution data regarding the phylogenetic position of Han Chinese and Korean members of Y-DNA Haplogroup D has been published, but the available data suggests that most Han Chinese members of Haplogroup D should belong to clades found frequently among Tibetans (and especially the D-M15 clade, also found among speakers of some Lolo-Burmese and Hmong-Mien languages), whereas most Korean members of Haplogroup D should belong to the D-M55 clade, which is found frequently among Ainu, Ryukyuan, and Japanese people.[17][15][3]
Haplogroup D Y-DNA has been found (albeit with low frequency) among modern populations of the Eurasian steppe, such as Southern Altaians (6/96 = 6.3% D-M174(xM15),[18] 6/120 = 5.0% D-P47[19]), Kazakhs (1/54 = 1.9% D-M174,[16] 6/1294 = 0.5% D[20]), Nogais (4/76 = 5.3% D-M174 Kara Nogai,[21] 1/87 = 1.1% D-M174 Kuban Nogai[21]), Khalkhas (1/24 = 4.2% D-M174,[16] 3/85 = 3.5% D-M174,[14] 2/149 D-M15 + 2/149 D-P47 = 4/149 = 2.7% D-M174 total[17]), Zakhchin (2/60 = 3.3% D-M174[14]), Uriankhai (1/60 = 1.7% D-M174[14]), and Kalmyks (5/426 = 1.2% D-M174[22]). It also has been found among linguistically similar (Turkic- or Mongolic-speaking) modern populations of the desert and oasis belt south of the steppe, such as Yugurs, Bao’an, Monguors, Uyghurs, and Uzbeks. In commercial testing, members have been found as far west as Romania in Europe and Iraq in Western Asia.[23]
Unlike haplogroup C-M217, Haplogroup D-M174 is not found in the New World; it is not present in any modern Native American (North, Central or South) populations. While it is possible that it traveled to the New World like Haplogroup C-M217, those lineages apparently became extinct.
Haplogroup D-M174 is also remarkable for its rather extreme geographic differentiation, with a distinct subset of Haplogroup D-M174 chromosomes being found exclusively in each of the populations that contains a large percentage of individuals whose Y-chromosomes belong to Haplogroup D-M174: Haplogroup D-M15 among the Tibetans (as well as among other East Asian and Southeast Asian populations that display low frequencies of Haplogroup D-M174 Y-chromosomes), Haplogroup D-M55 among the various populations of the Japanese Archipelago and with low frequency among Koreans, Haplogroup D-P99 among the inhabitants of Tibet and some other parts of central Eurasia (e.g. Mongolia[24] and the Altai[17][18][19]), and paragroup D* without tested positive subclades (probably another monophyletic branch of Haplogroup D) among the Andaman Islanders. Another type (or types) of paragroup D* without tested positive subclades is found at a very low frequency among the Turkic and Mongolic populations of Central Asia, amounting to no more than 1% in total. This apparently ancient diversification of Haplogroup D-M174 suggests that it may perhaps be better characterized as a "super-haplogroup" or "macro-haplogroup."
In one study, the frequency of Haplogroup D-M174 without tested positive subclades found among Thais was 10%.[2] Su et al. (2000) found DE-YAP/DYS287(xM15) in 11.1% (5/45) of a set of three samples from Thailand (including 20% (4/20) North Thai, 20% (1/5) So, and 0% (0/20) Northeast Thai) and in 16.7% (1/6) of a sample from Guam.[25] Meanwhile, the authors found D-M15 in 15% of a pair of samples of Yao (including 30% (3/10) Yao Jinxiu and 0% (0/10) Yao Nandan), 14.3% (2/14) of a sample of Yi, 3.8% (1/26) of a sample of Cambodians, and 3.6% (1/28) of a sample of Zhuang.[25] Dong et al. (2002) found DE-YAP Y-chromosomes in 12.5% (2/16) of a sample of Jingpo from Luxi City, Yunnan, 10.0% (2/20) of a sample of Dai from Luxi City, Yunnan, and 1.82% (1/55) of a sample of Nu from Gongshan and Fugong counties of Yunnan.[26]
Distribution
The Haplogroup D-M174 Y-chromosomes that are found among populations of the Japanese Archipelago (haplogroup D-M55 a.k.a. haplogroup D1b) are particularly distinctive, bearing a complex of at least five individual mutations along an internal branch of the Haplogroup D-M174 phylogeny, thus distinguishing them clearly from the other Haplogroup D-M174 chromosomes that are found among the Tibetans and Andaman Islanders and providing evidence that Y-chromosome Haplogroup D-M55 was the modal haplogroup in the ancestral population that developed the prehistoric Jōmon culture in the Japanese islands.
It is suggested that the majority of D-M174 Y-chromosome carriers migrated from Central Asia to East Asia. One group migrated into the Andaman Islands and mixed with the native Negrito population, thus forming the today Andamanese people (probably a male-only migration). Another group stayed in modern Tibet and southern China (today Tibeto-Burman peoples) and another group migrated to Japan, possibly via the Korean Peninsula (Jōmon people).[27][28]
Subclades
D* Paragroup
A paragroup of D-M174, known as D* but without positive-tested subclades, found at high frequencies among Andaman Islanders[2] (especially Onge(23/23 = 100%), Jarawa(4/4 = 100%))[29] and some Tibetan minority tribes in Northeast India (among whom rates vary from zero to 65%).[5][30][31][32] Another study reports only about 56.25% haplogroup D in Andamanese.[33]
D* has also been found in approximately 5% of Altaians.[17] Kharkov et al. have found haplogroup D*(xD-M15) in 6.3% (6/96) of a pool of samples of Southern Altaians from three different localities, particularly in Kulada (5/46 = 10.9%) and Kosh-Agach (1/7 = 14%), though they have not tested for any marker of the subclade D-M55 or D-P99. Kharkov et al. also have reported finding haplogroup DE-M1(xD-M174) Y-DNA in one Southern Altaian individual from Beshpeltir (1/43 = 2.3%).[18]
D-Z27276 (D1a)
Haplogroup D-Z27276 is the common ancestor of D-M15 and D-P99.
D-M15 (D1a1)
D-M15 was first reported to have been found in a sample from Cambodia and Laos (1/18 = 5.6%) and in a sample from Japan (1/23 = 4.3%) in a preliminary worldwide survey of Y-DNA variation in extant human populations.[34]
Subsequently, Y-DNA that belongs to Haplogroup D-M15 has been found frequently among Tibeto-Burman-speaking populations of Southwestern China (including approximately 23% of Qiang,[2][35][36] approximately 12.5% of Tibetans,[2] and approximately 9% of Yi[2][37]) and among Yao people inhabiting northeastern Guangxi (6/31 = 19.4% Lowland Yao, 5/41 = 12.2% Native Mien, 3/41 = 7.3% Lowland Kimmun)[38] with a moderate distribution throughout Central Asia, East Asia, and continental Southeast Asia (Indochina).[2]
A study published in 2011 has found D-M15 in 7.8% (4/51) of a sample of Hmong Daw and in 3.4% (1/29) of a sample of Xinhmul from northern Laos.[38]
D-P47 (D1a2a)
Found with high frequency among Pumi,[2] Naxi,[2] and Tibetans,[39][2] with a moderate distribution in Central Asia.[2] According to one study, Tibetans have a frequency of about 41.31% of haplogroup D-P47.[40]
D-M55 (D1b)
Previously known as D-M55, D-M64.1/Page44.1 (D1b) is found with high frequency among Ainu,[41] Japanese,[42] and Ryukyuans.[42]
Kim et al. (2011) found Haplogroup D-M55 in 2.0% (1/51) of a sample of Beijing Han and in 1.6% (8/506) of a pool of samples from South Korea, including 3.3% (3/90) from the Jeolla region, 2.4% (2/84) from the Gyeongsang region, 1.4% (1/72) from the Chungcheong region, 1.1% (1/87) from the Jeju region, 0.9% (1/110) from the Seoul-Gyeonggi region, and 0% (0/63) from the Gangwon region.[15] Hammer et al. (2006) found Haplogroup D-P37.1 in 4.0% (3/75) of a sample from South Korea.[17]
Low levels of D-M116.1 (a subclade of D-M55) among males in present-day Timor (0.2% of males),[43] It's found in 9.5% of males from Micronesia(Hammer et al. 2006,[44][17] is believed to reflect recent admixture from Japan. That is, D-M116.1 (D1b1) is generally believed to be a primary subclade of D-M64.1 (D1b), possibly as a result of the Japanese military occupation of South East Asia during World War II.
According to Mitsuru Sakitani, Haplogroup D1 arrived from Central Asia to northern Kyushu via the Altai Mountains and the Korean Peninsula more than 40,000 years before present, and Haplogroup D-M55 (D1b) was born in Japanese archipelago.[45]
Recently it was confirmed that the Japanese branch of haplogroup D-M55 is distinct and isolated from other D-branches since more than 53,000 years. The split between D1a happend likely in Central Asia, while some others suggest a instant split during the origin of haplogroup D itself, as the Japanese branch has five unique mutations not found in any other D-branch.[46]
Phylogenetics
Phylogenetic history
Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Latter, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D-M174 | * | * | * | * | * | * | * | * | D | D | D | D | D | D | D | D | D | D |
| D-M15 | 4 | IV | 3G | 12 | Eu5 | H3 | B | D1 | D1 | D1 | D1 | D1 | D1 | D1 | D1 | D1 | D1 | D1 |
| D-M55 | * | * | * | * | * | * | * | * | D2 | D2 | D2 | D2 | D2 | D2 | D2 | D2 | D2 | D2 |
| D-P12 | 4 | IV | 3G | 11 | Eu5 | H2 | B | D2a | D2a | D2a1a1 | D2a1a1 | D2 | D2 | D2a1a1 | D2a1a1 | D2a1a1 | removed | removed |
| D-M116.1 | 4 | IV | 3G | 11 | Eu5 | H2 | B | D2b* | D2a | D2a | D2a | D2a | D2a | D2a | D2a | D2a | removed | removed |
| D-M125 | 4 | IV | 3G | 11 | Eu5 | H2 | B | D2b1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 | D2a1 |
| D-M151 | 4 | IV | 3G | 11 | Eu5 | H2 | B | D2b2 | D2a1 | D2a2 | D2a2 | D2a2 | D2a2 | D2a2 | D2a2 | D2a2 | D2a2 | D2a2 |
Research publications
The following research teams per their publications were represented in the creation of the YCC tree.
Phylogenetic trees
This phylogenetic tree of haplogroup D-M174 subclades is based on the ISOGG 2017 tree (ver.12.168).[1]
- DE
- D (M174/Page30, IMS-JST021355)
- D* - Onge, Jarawa (Andaman Islands)[47]
- D1 (CTS11577)
- D1a (Z27276)
- D1a1 (M15) - Mostly in Tibet and other parts of Southwest China and South Central China, but also lightly distributed throughout East Asia, Central Asia, and Indochina
- D1a1a (F849)
- D1a1a1 (N1)
- D1a1a2 (F1070) - China (Guangdong,[3] Xishuangbanna Dai[3])
- D1a1a2a (A6345)
- D1a1a2a1 (Z40598)
- D1a1a2a1a (Z40600)
- D1a1a2a1b (Z41246)
- D1a1a2a2 (Z41068)
- D1a1a2a1 (Z40598)
- D1a1a2b (CTS5502)
- D1a1a2a (A6345)
- D1a1a (F849)
- D1a2 (P99) - Altai Mountains, Tibet
- D1a1 (M15) - Mostly in Tibet and other parts of Southwest China and South Central China, but also lightly distributed throughout East Asia, Central Asia, and Indochina
- D1b (M55, M57, M64.1/Page44.1, M179/Page31, M359.1/P41.1, P37.1, P190, 12f2.2) - Japanese (Yamato, Ainu, Ryukyuan)
- D1b1 (M116.1)
- D1b1a (M125)
- D1b1a1 (P42)
- D1b1a1a (P12_1, P12_2, P12_3)
- D1b1a2 (IMS-JST022457) - Emperor of Japan[53][54]
- D1b1a2a (P53.2)
- D1b1a2b (IMS-JST006841/Page3)
- D1b1a2b1 (CTS3397)
- D1b1a2b1a (Z1500)
- D1b1a2b1a1 (Z1504, CTS8093) - Minamoto clan[53][55]
- D1b1a2b1a1a (FGC6373) - Japanese (Hiroshima)[51]
- D1b1a2b1a1b (Z40614)
- D1b1a2b1a1c (Z31543)
- D1b1a2b1a1d (FGC30021)
- D1b1a2b1a1e (Z31548)
- D1b1a2b1a1f (Z31553)
- D1b1a2b1a1g (CTS6223)
- D1b1a2b1a1h (CTS4093)
- D1b1a2b1a1i (Z40687)
- D1b1a2b1a1i1 (Z35641)
- D1b1a2b1a1i2 (Z40688)
- D1b1a2b1a1j (CTS5058)
- D1b1a2b1a1k (FGC34008)
- D1b1a2b1a2 (CTS266)
- D1b1a2b1a3 (Z40672)
- D1b1a2b1a1 (Z1504, CTS8093) - Minamoto clan[53][55]
- D1b1a2b1b (CTS1372)
- D1b1a2b1a (Z1500)
- D1b1a2b2 (CTS5581)
- D1b1a2b1 (CTS3397)
- D1b1a3 (CTS10972)
- D1b1a3a (Z31538)
- D1b1a3b (CTS232)
- D1b1a1 (P42)
- D1b1b (P120)
- D1b1c (CTS6609)
- D1b1c1 (CTS1897/Z1574)
- D1b1c1a (CTS11032) - Japanese (Aichi)[51]
- D1b1c1a1 (CTS218/V1105/Z1527)
- D1b1c1a1a (CTS6909)
- D1b1c1a1a1 (CTS6969)
- D1b1c1a1a2 (CTS9770)
- D1b1c1a1b (CTS3033)
- D1b1c1a1b1 (M2176)
- D1b1c1a1b2 (CTS2472)
- D1b1c1a1c (M151)
- D1b1c1a1a (CTS6909)
- D1b1c1a2 (F8521.3)
- D1b1c1a1 (CTS218/V1105/Z1527)
- D1b1c1b (CTS1964)
- D1b1c1b1 (CTS974)
- D1b1c1b2 (CTS722)
- D1b1c1c (Z30644) - Japanese (Fukushima)[51]
- D1b1c1c1 (CTS4292)
- D1b1c1c1a (Z31517)
- D1b1c1c1b (CTS1798)
- D1b1c1c2 (Z31512)
- D1b1c1c1 (CTS4292)
- D1b1c1d (CTS5641) - Japanese (Kyoto)[51]
- D1b1c1e (CTS429)
- D1b1c1a (CTS11032) - Japanese (Aichi)[51]
- D1b1c2 (CTS103)
- D1b1c2a (Z42462)
- D1b1c1 (CTS1897/Z1574)
- D1b1a (M125)
- D1b2 (CTS131)
- D1b2a (CTS220) - Jōmon man (Rebun Island)[56]
- D1b2b (CTS68)
- D1b1 (M116.1)
- D1a (Z27276)
- D2 (L1366, L1378, M226.2) - Philippines (Mactan, Luzon)[58]
- D2a (FGC8848)
- D2b (FGC8940)
- D (M174/Page30, IMS-JST021355)
See also
Genetics
- Genetic genealogy
- Haplogroup
- Haplotype
- Human Y-chromosome DNA haplogroup
- Molecular phylogeny
- Paragroup
- Subclade
- Y-chromosome haplogroups in populations of the world
- Y-DNA haplogroups by ethnic group
- Y-DNA haplogroups in populations of East and Southeast Asia
- Y-DNA haplogroups in populations of Oceania
Y-DNA D subclades
Y-DNA backbone tree
References
- ^ a b "Y-DNA Haplogroup D-M174 and its Subclades - 2017".
- ^ a b c d e f g h i j k l m Shi H, Zhong H, Peng Y, Dong YL, Qi XB, Zhang F, Liu LF, Tan SJ, Ma RZ, Xiao CJ, Wells RS, Jin L, Su B (October 2008). "Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations". BMC Biology. 6: 45. doi:10.1186/1741-7007-6-45. PMC 2605740. PMID 18959782.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e f g h i j k l m YFull Haplogroup YTree v6.02 at 02 April 2018. Accessed July 7, 2018.
- ^ a b c Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF (May 2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research. 18 (5): 830–8. doi:10.1101/gr.7172008. PMC 2336805. PMID 18385274.
- ^ a b Su B, Xiao C, Deka R, Seielstad MT, Kangwanpong D, Xiao J, Lu D, Underhill P, Cavalli-Sforza L, Chakraborty R, Jin L (December 2000). "Y chromosome haplotypes reveal prehistorical migrations to the Himalayas". Human Genetics. 107 (6): 582–90. doi:10.1007/s004390000406. PMID 11153912.
- ^ Gayden, Tenzin; Cadenas, Alicia M.; Regueiro, Maria; Singh, Nanda B.; Zhivotovsky, Lev A.; Underhill, Peter A.; Cavalli-Sforza, Luigi L.; Herrera, Rene J. (May 2007). "The Himalayas as a Directional Barrier to Gene Flow". The American Journal of Human Genetics. 80 (5): 884–894. doi:10.1086/516757. PMC 1852741. PMID 17436243.
- ^ Matsumoto H (February 2009). "The origin of the Japanese race based on genetic markers of immunoglobulin G". Proceedings of the Japan Academy. Series B, Physical and Biological Sciences. 85 (2): 69–82. Bibcode:2009PJAB...85...69M. doi:10.2183/pjab.85.69. PMC 3524296. PMID 19212099.
- ^ Tyler-Smith, Chris; Xue, Yali; Thomas, Mark G.; Yang, Huanming; Arciero, Elena; Asan; Connell, Bruce A.; Jones, Abigail L.; Haber, Marc (2019-06-13). "A Rare Deep-Rooting D0 African Y-Chromosomal Haplogroup and Its Implications for the Expansion of Modern Humans out of Africa". Genetics: genetics.302368.2019. doi:10.1534/genetics.119.302368. ISSN 0016-6731. PMID 31196864.
- ^ Estes, Roberta (2019-06-21). "Exciting New Y DNA Haplogroup D Discoveries!". DNAeXplained - Genetic Genealogy. Retrieved 2019-07-08.
- ^ Y染色体单倍群D在東亞的分布及其意義
- ^ Hua Zhong, Hong Shi, Xue-Bin Qi, Zi-Yuan Duan, Ping-Ping Tan, Li Jin, Bing Su, and Runlin Z. Ma (2011), "Extended Y Chromosome Investigation Suggests Postglacial Migrations of Modern Humans into East Asia via the Northern Route." Mol. Biol. Evol. 28(1):717–727. doi:10.1093/molbev/msq247
- ^ Yan S, Wang CC, Li H, Li SL, Jin L, et al. (The Genographic Consortium) (September 2011). "An updated tree of Y-chromosome Haplogroup O and revised phylogenetic positions of mutations P164 and PK4". European Journal of Human Genetics : EJHG. 19 (9): 1013–5. doi:10.1038/ejhg.2011.64. PMC 3179364. PMID 21505448.
- ^ a b Park MJ, Lee HY, Yang WI, Shin KJ (July 2012). "Understanding the Y chromosome variation in Korea--relevance of combined haplogroup and haplotype analyses". International Journal of Legal Medicine. 126 (4): 589–99. doi:10.1007/s00414-012-0703-9. PMID 22569803.
- ^ a b c d Katoh T, Munkhbat B, Tounai K, Mano S, Ando H, Oyungerel G, Chae GT, Han H, Jia GJ, Tokunaga K, Munkhtuvshin N, Tamiya G, Inoko H (February 2005). "Genetic features of Mongolian ethnic groups revealed by Y-chromosomal analysis". Gene. 346: 63–70. doi:10.1016/j.gene.2004.10.023. PMID 15716011.
- ^ a b c d e f g Soon-Hee Kim, Ki-Cheol Kim, Dong-Jik Shin, Han-Jun Jin, Kyoung-Don Kwak, Myun-Soo Han, Joon-Myong Song, Won Kim, and Wook Kim, "High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea." Investigative Genetics 2011, 2:10. http://www.investigativegenetics.com/content/2/1/10
- ^ a b c Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF (August 2001). "The Eurasian heartland: a continental perspective on Y-chromosome diversity". Proceedings of the National Academy of Sciences of the United States of America. 98 (18): 10244–9. Bibcode:2001PNAS...9810244W. doi:10.1073/pnas.171305098. PMC 56946. PMID 11526236.
- ^ a b c d e f g Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S (2006). "Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes". Journal of Human Genetics. 51 (1): 47–58. doi:10.1007/s10038-005-0322-0. PMID 16328082.
- ^ a b c Khar'kov VN, Stepanov VA, Medvedeva OF, Spiridonova MG, Voevoda MI, Tadinova VN, Puzyrev VP (May 2007). "[Gene pool differences between northern and southern Altaians inferred from the data on Y-chromosomal haplogroups]". Genetika (in Russian). 43 (5): 675–87. doi:10.1134/S1022795407050110. PMID 17633562.
- ^ a b c Dulik MC, Zhadanov SI, Osipova LP, Askapuli A, Gau L, Gokcumen O, Rubinstein S, Schurr TG (February 2012). "Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between Native Americans and Indigenous Altaians". American Journal of Human Genetics. 90 (2): 229–46. doi:10.1016/j.ajhg.2011.12.014. PMC 3276666. PMID 22281367.
- ^ E. E. Ashirbekov, D. M. Botbaev, A. M. Belkozhaev, A. O. Abayldaev, A. S. Neupokoeva, J. E. Mukhataev, B. Alzhanuly, D. A. Sharafutdinova, D. D. Mukushkina, M. B. Rakhymgozhin, A. K. Khanseitova, S. A. Limborska, and N. A. Aytkhozhina, "Distribution of Y-Chromosome Haplogroups of the Kazakh from the South Kazakhstan, Zhambyl, and Almaty Regions." Reports of the National Academy of Sciences of the Republic of Kazakhstan, ISSN 2224-5227, Volume 6, Number 316 (2017), 85 - 95.
- ^ a b Yunusbayev B, Metspalu M, Järve M, Kutuev I, Rootsi S, Metspalu E, Behar DM, Varendi K, Sahakyan H, Khusainova R, Yepiskoposyan L, Khusnutdinova EK, Underhill PA, Kivisild T, Villems R (January 2012). "The Caucasus as an asymmetric semipermeable barrier to ancient human migrations". Molecular Biology and Evolution. 29 (1): 359–65. doi:10.1093/molbev/msr221. PMID 21917723.
- ^ Malyarchuk B, Derenko M, Denisova G, Khoyt S, Woźniak M, Grzybowski T, Zakharov I (December 2013). "Y-chromosome diversity in the Kalmyks at the ethnical and tribal levels". Journal of Human Genetics. 58 (12): 804–11. doi:10.1038/jhg.2013.108. PMID 24132124.
- ^ Y-DNA D Haplogroup Project at Family Tree DNA
- ^ a b c Di Cristofaro J, Pennarun E, Mazières S, Myres NM, Lin AA, Temori SA, Metspalu M, Metspalu E, Witzel M, King RJ, Underhill PA, Villems R, Chiaroni J (2013). "Afghan Hindu Kush: where Eurasian sub-continent gene flows converge". PLOS ONE. 8 (10): e76748. Bibcode:2013PLoSO...876748D. doi:10.1371/journal.pone.0076748. PMC 3799995. PMID 24204668.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Su, Bing; Jin, Li; Underhill, Peter; Martinson, Jeremy; Saha, Nilmani; McGarvey, Stephen T.; Shriver, Mark D.; Chu, Jiayou; Oefner, Peter; Chakraborty, Ranajit; Deka, Ranjan (18 July 2000). "Polynesian origins: Insights from the Y chromosome". Proceedings of the National Academy of Sciences of the United States of America. 97 (15): 8225–8228. Bibcode:2000PNAS...97.8225S. doi:10.1073/pnas.97.15.8225. PMC 26928. PMID 10899994.
- ^ DONG Yongli, SHI Hong, LI Weixiang, YANG Jie, ZENG Weimin, LI Kaiyuan, and XIAO Chunjie, "Study of polymorphism at the YAP locus in seven Yunnan ethnic minority populations in the great gorge of the Salween River and downstream areas" (original title in Chinese: "怒江大峡谷及下游地区7个云南少数民族YAP位点的多态性研究"), Acta Anthropologica Sinica, Vol. 21, No. 3 (August, 2002). http://www.ivpp.cas.cn/cbw/rlxxb/xbwzxz/201203/t20120320_3512811.html
- ^ Hammer MF, Karafet TM, Park H, Omoto K, Harihara S, Stoneking M, Horai S (2006). "Dual origins of the Japanese: common ground for hunter-gatherer and farmer Y chromosomes". Journal of Human Genetics. 51 (1): 47–58. doi:10.1007/s10038-005-0322-0. PMID 16328082.
- ^ Shi H, Zhong H, Peng Y, Dong YL, Qi XB, Zhang F, Liu LF, Tan SJ, Ma RZ, Xiao CJ, Wells RS, Jin L, Su B (October 2008). "Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations". BMC Biology. 6 (1): 45. doi:10.1186/1741-7007-6-45. PMC 2605740. PMID 18959782.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E (January 2003). "Genetic affinities of the Andaman Islanders, a vanishing human population". Current Biology. 13 (2): 86–93. doi:10.1016/S0960-9822(02)01336-2. PMID 12546781.
- ^ Cordaux R, Weiss G, Saha N, Stoneking M (August 2004). "The northeast Indian passageway: a barrier or corridor for human migrations?". Molecular Biology and Evolution. 21 (8): 1525–33. doi:10.1093/molbev/msh151. PMID 15128876.
- ^ Chandrasekar A, Saheb SY, Gangopadyaya P, Gangopadyaya S, Mukherjee A, Basu D, Lakshmi GR, Sahani AK, Das B, Battacharya S, Kumar S, Xaviour D, Sun D, Rao VR (2007). "YAP insertion signature in South Asia". Annals of Human Biology. 34 (5): 582–6. doi:10.1080/03014460701556262. PMID 17786594.
- ^ Reddy BM, Langstieh BT, Kumar V, Nagaraja T, Reddy AN, Meka A, Reddy AG, Thangaraj K, Singh L (November 2007). "Austro-Asiatic tribes of Northeast India provide hitherto missing genetic link between South and Southeast Asia". PLOS ONE. 2 (11): e1141. Bibcode:2007PLoSO...2.1141R. doi:10.1371/journal.pone.0001141. PMC 2065843. PMID 17989774.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Shi, Hong; Zhong, Hua; Peng, Yi; Dong, Yong-Li; Qi, Xue-Bin; Zhang, Feng; Liu, Lu-Fang; Tan, Si-Jie; Ma, Runlin Z (2008-10-29). "Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations". BMC Biology. 6: 45. doi:10.1186/1741-7007-6-45. ISSN 1741-7007. PMC 2605740. PMID 18959782.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonné-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics. 26 (3): 358–61. doi:10.1038/81685. PMID 11062480.
- ^ Xue Y, Zerjal T, Bao W, Zhu S, Shu Q, Xu J, Du R, Fu S, Li P, Hurles ME, Yang H, Tyler-Smith C (April 2006). "Male demography in East Asia: a north-south contrast in human population expansion times". Genetics. 172 (4): 2431–9. doi:10.1534/genetics.105.054270. PMC 1456369. PMID 16489223.
- ^ Wang CC, Wang LX, Shrestha R, Zhang M, Huang XY, Hu K, Jin L, Li H (2014). "Genetic structure of Qiangic populations residing in the western Sichuan corridor". PLOS ONE. 9 (8): e103772. Bibcode:2014PLoSO...9j3772W. doi:10.1371/journal.pone.0103772. PMC 4121179. PMID 25090432.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Wen B, Xie X, Gao S, Li H, Shi H, Song X, Qian T, Xiao C, Jin J, Su B, Lu D, Chakraborty R, Jin L (May 2004). "Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans". American Journal of Human Genetics. 74 (5): 856–65. doi:10.1086/386292. PMC 1181980. PMID 15042512.
- ^ a b Cai X, Qin Z, Wen B, Xu S, Wang Y, Lu Y, Wei L, Wang C, Li S, Huang X, Jin L, Li H (2011). "Human migration through bottlenecks from Southeast Asia into East Asia during Last Glacial Maximum revealed by Y chromosomes". PLOS ONE. 6 (8): e24282. Bibcode:2011PLoSO...624282C. doi:10.1371/journal.pone.0024282. PMC 3164178. PMID 21904623.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Dongsheng Lu et al. Ancestral Origins and Genetic History of Tibetan Highlanders, August 25, 2016
- ^ Shi, Hong; Zhong, Hua; Peng, Yi; Dong, Yong-Li; Qi, Xue-Bin; Zhang, Feng; Liu, Lu-Fang; Tan, Si-Jie; Ma, Runlin Z (2008-10-29). "Y chromosome evidence of earliest modern human settlement in East Asia and multiple origins of Tibetan and Japanese populations". BMC Biology. 6: 45. doi:10.1186/1741-7007-6-45. ISSN 1741-7007. PMC 2605740. PMID 18959782.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Tajima A, Hayami M, Tokunaga K, Juji T, Matsuo M, Marzuki S, Omoto K, Horai S (2004). "Genetic origins of the Ainu inferred from combined DNA analyses of maternal and paternal lineages". Journal of Human Genetics. 49 (4): 187–93. doi:10.1007/s10038-004-0131-x. PMID 14997363.
- ^ a b YOUICHI SATO, TOSHIKATSU SHINKA, ASHRAF A. EWIS, AIKO YAMAUCHI, TERUAKI IWAMOTO, YUTAKA NAKAHORI Overview of genetic variation in the Y chromosome of modern Japanese males.
- ^ Tumonggor, Meryanne K; Karafet, Tatiana M; Downey, Sean; Lansing, J Stephen; Norquest, Peter; Sudoyo, Herawati; Hammer, Michael F; Cox, Murray P (September 2014). "Isolation, contact and social behavior shaped genetic diversity in West Timor". Journal of Human Genetics. 59 (9): 494–503. doi:10.1038/jhg.2014.62. PMC 4521296. PMID 25078354.
- ^ Cite error: The named reference
Hammerwas invoked but never defined (see the help page). - ^ 崎谷満『DNA・考古・言語の学際研究が示す新・日本列島史』(勉誠出版 2009年)(in Japanese)
- ^ Mondal, Mayukh; Bergström, Anders; Xue, Yali; Calafell, Francesc; Laayouni, Hafid; Casals, Ferran; Majumder, Partha P.; Tyler-Smith, Chris; Bertranpetit, Jaume (25 April 2017). "Y-chromosomal sequences of diverse Indian populations and the ancestry of the Andamanese". Human Genetics. 136 (5): 499–510. doi:10.1007/s00439-017-1800-0.
- ^ YFull
- ^ a b Y-DNA D Haplogroup Project
- ^ Kazakh Y-DNA Project at Family Tree DNA
- ^ a b Hallast P, Batini C, Zadik D, Maisano Delser P, Wetton JH, Arroyo-Pardo E, Cavalleri GL, de Knijff P, Destro Bisol G, Dupuy BM, Eriksen HA, Jorde LB, King TE, Larmuseau MH, López de Munain A, López-Parra AM, Loutradis A, Milasin J, Novelletto A, Pamjav H, Sajantila A, Schempp W, Sears M, Tolun A, Tyler-Smith C, Van Geystelen A, Watkins S, Winney B, Jobling MA (March 2015). "The Y-chromosome tree bursts into leaf: 13,000 high-confidence SNPs covering the majority of known clades". Molecular Biology and Evolution. 32 (3): 661–73. doi:10.1093/molbev/msu327. PMC 4327154. PMID 25468874.
- ^ a b c d e f g h i JAPAN Y-DNA Project
- ^ a b Yan S, Wang CC, Zheng HX, Wang W, Qin ZD, Wei LH, Wang Y, Pan XD, Fu WQ, He YG, Xiong LJ, Jin WF, Li SL, An Y, Li H, Jin L (2014). "Y chromosomes of 40% Chinese descend from three Neolithic super-grandfathers". PLOS ONE. 9 (8): e105691. arXiv:1310.3897. Bibcode:2014PLoSO...9j5691Y. doi:10.1371/journal.pone.0105691. PMC 4149484. PMID 25170956.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b famous Y dna by haplogroup
- ^ List of haplogroups of famous/notable people Most of Japanese males share their paternal Y-dna lineages with the Imperial Family.
- ^ List of haplogroups of famous/notable people
- ^ 神澤ほか (2016)『礼文島船泊縄文人の核ゲノム解析』第70回日本人類学大会
- ^ ysearch DNA of Kimura (D-CTS11285)
- ^ Y-DNA Haplogroup D and its Subclades - 2014
- Underhill PA, Kivisild T (2007). "Use of y chromosome and mitochondrial DNA population structure in tracing human migrations". Annual Review of Genetics. 41: 539–64. doi:10.1146/annurev.genet.41.110306.130407. PMID 18076332.
External links
- Atlas of the Human Journey: Genetic Markers, Haplogroup D-M174 (M174), from The Genographic Project at National Geographic
- Famous dna of Japan
