DNA replication
This article needs attention from an expert in Biology. Please add a reason or a talk parameter to this template to explain the issue with the article. |

Introduction
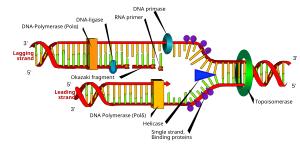
DNA replication or DNA synthesis is the process of copying a double-stranded DNA molecule. This involves copying the genetic material of the cell and passing it on to daughter cells which is the basic mechanism for the continuation of life. Ideally, the resulting DNA are identical, although in reality there are always errors, though proofreading and error-checking mechanisms exist to ensure a very high level of fidelity.
Both strands in DNA run anti-parallel to each other and are complementary to one another. One of the DNA 'sense' strands can be read 5' → 3' direction, while the complementary 'anti-sense' strand bound to it is read in the 3' → 5' direction.
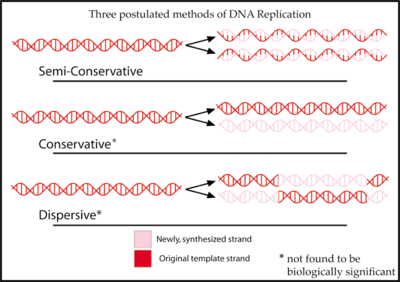
Since DNA strands are antiparallel and complementary, each strand can serve as a template for the reproduction of the opposite strand. The template strand is preserved as a whole piece and the new strand is assembled from nucleotide triphosphates. This process is called semiconservative replication. Ideally, the two resulting strands are identical, although in reality there are always errors, though proofreading and error-checking mechanisms exist to ensure a very high level of fidelity.
In a cell, this step is obligatory prior to cell division. Prokaryotes persistently replicate their DNA and creating a whole, new chromosome is a limiting step in cell division.The large size of eukaryotic chromosomes and the limits of nucleotide incorporation during DNA synthesis, make it necessary for multiple origins of replication to exist in order to complete replication in a reasonable period of time. However, it is clear that at a replication origin the strands of DNA must dissociate and unwind in order to allow access to DNA polymerase.
The process may also be performed in-vitro using reconstituted or completely artificial components, in a process known as PCR. DNA may be synthesized artificially, but this process is not fundamentally a replicative process, and only produces one strand of DNA.
Studying DNA
The process of using genetic or biochemical methods to study DNA can be due to a variety of reasons such as designing drugs, properties of viruses, and the abnormal cell division in cancer cells. One of the most common studied organisms is (temperature sensitive) Saccharomyces cerevisiae because of the fact that the entire genome has already been sequenced. In the case of yeast, many proteins effecting the cell cycle in humans were to have been found and studied extensively in yeast already.
One of the popular ways to study DNA in organisms were to produce 'conditional lethal' (ie. temperature sensitive) mutants in which by raising the temperature above a certain value, would cause severe mutations in proteins in the cell and would lead to a stop in DNA replication and growth. DNA replication can be measured by adding a radioactive form of thymidine into a growth medium that contains the cells being studied. The cells that have grown, contain the labeled thymidine and can be measured by incorporation into an acid such as ethanol. Therefore in temperature sensitive mutations, we are able to identify 'fast stop mutatants' if the thymidine incorporation is stopped immediately perhaps due to a break in chain elongation, and 'slow stop mutants' if there is a slower reduction of DNA replication perhaps due to mutation in chain initiation.
Another way to study DNA is similar to Arthur Kornberg's team [1]that discovered DNA polymerase activity in e. coli. Through various methods the team was able to synthesize an 'assay' that included sonic extract of E. coli, dTTP with radioactively labeled α phosphate, ATP, Mg2+, dCTP, dGTP, dATP and buffer and tried to measure the incorporation of the radioactively labeled dTTP in the DNA.
References
- Voet and Voet. Biochemistry, Third Edition (2004). ISBN 0-471-19350-X. Wiley International Edition.
- Watson, Baker, Bell, Gann, Levine, Losick. Molecular Biology of the Gene, Fifth Edition (2003). ISBN 0-8053-4635-X. Pearson/Benjamin Cummings Publishing.
External links
- DNA Workshop
- WEHI-TV - DNA Replication video- Detailed DNA replication animation from different angles with description below.
- Breakfast of Champions Does Replication- creative primer on the process from the Science Creative Quarterly
- Template:McGrawHillAnimation
- Basic Polymerase Chain Reaction Protocol
- Animated Biology
- DNA makes DNA (Flash Animation)
- DNA replication info pageby George Kakaris, Biologist MSc in Applied Genetics and Biotechnology
