SARS-CoV-2: Difference between revisions
I changed the date from 24 of January to the 25 of January, because as of today 41 deaths have been confirmed |
→Reservoir: Rewriting everything, there are 3 closely related bat viruses, and no coronavirus found in snakes had ever been published |
||
| Line 59: | Line 59: | ||
==Virology== |
==Virology== |
||
===Reservoir=== |
===Reservoir=== |
||
| ⚫ | |||
During 17 years of research on the origin of the [[Sars]] 2003 epidemic, a lot of [[Bat_SARS-like_coronavirus_WIV1|Sars-like]] [[bat]] coronavirus have been isolated and [[DNA_sequencing|sequenced]], most of them originating from the [[Rhinolophus]] species. With enough genomes it is possible to reconstruct a [[phylogenetic tree]] of the mutation history of a family of virus. |
|||
On 22 January 2020, the ''[[Journal of Medical Virology]]'' published a report with genomic analysis that reflects that snakes in the Wuhan area are "the most probable wildlife animal reservoir" for the virus, but more research is required.<ref name="BBC">[https://www.bbc.com/news/world-asia-china-51217455 BBC: China coronavirus: Fear grips Wuhan as lockdown begins]</ref><ref>[https://www.cnn.com/2020/01/22/health/snakes-wuhan-coronavirus-outbreak-conversation-partner/index.html CNN: Snakes could be the source of the Wuhan coronavirus outbreak]</ref> A [[homologous recombination]] event may have mixed a "clade A" (Bat SARS-like viruses CoVZC45 and CoVZXC21)<!-- What is this classification? bat-SL-CoVZC45 and bat-SL-CoVZXC21 is A? --> virus with the RNA-binding domain of a yet-unknown Beta-CoV.<ref name="ji-wei-2020">{{cite journal |last1=Ji |first1=Wei |last2=Wang |first2=Wei |last3=Zhao |first3=Xiaofang |last4=Zai |first4=Junjie |last5=Li |first5=Xingguang |title=Homologous recombination within the spike glycoprotein of the newly identified coronavirus may boost cross‐species transmission from snake to human |journal=Journal of Medical Virology |date=22 January 2020 |doi=10.1002/jmv.25682 |pmid=31967321 }}</ref><ref name=":3">{{cite news |author1=Haitao Guo |author2=Guangxiang "George" Luo |author3=Shou-Jiang Gao |title=Snakes could be the original source of the new coronavirus outbreak in China |url=https://theconversation.com/snakes-could-be-the-original-source-of-the-new-coronavirus-outbreak-in-china-130364 |accessdate=22 January 2020 |work=The Conversation |date=22 January 2020}}</ref> Some scientists believe that the diseases could have originated from ''[[Bungarus multicinctus]]'', a highly venomous snake at the Wuhan food market, where [[ye wei]] is sold.<ref name=":2">{{Cite web|url=https://www.scimex.org/newsfeed/expert-reaction-could-new-coronavirus-have-come-from-snakes|title=EXPERT REACTION: Could new coronavirus have come from snakes?|last=SCIMEX|date= 23 January 2020 |website=Scimex |access-date=23 January 2020}}</ref> |
|||
The Wuhan new coronavirus has been found to fall into this category of Sars-like coronavirus. Two genome sequences with a resemblance of 80% had been published<ref> in 2015 and 2017, [[Rhinolophus sinicus]] sample [https://www.ncbi.nlm.nih.gov/nuccore/MG772933 CoVZC45] and [https://www.ncbi.nlm.nih.gov/nuccore/MG772934 CoVZXC21], see [https://nextstrain.org/groups/blab/sars-like-cov there for an interactive visualization]</ref><ref>{{cite web|title=The 2019 new Coronavirus epidemic: evidence for virus evolution|url=https://www.biorxiv.org/content/10.1101/2020.01.24.915157v1}}</ref>. Together with those two sequences, a third unpublished virus genome from [[Intermediate_horseshoe_bat|Rhinolophus Affinis]] with a resemblance of 96% to Wuhan novel coronavirus is mentioned in an article from the Wuhan institute of virology<ref name="bioRxivBatOrigin">{{cite web |title=Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin |url=https://www.biorxiv.org/content/10.1101/2020.01.22.914952v2|author=Wuhan institue of virology|website=bioRxiv|publisher=[[bioRxiv]] |accessdate=January 24, 2020|date=January 23, 2020}}</ref><ref>96% of resemblance is close to the [https://nextstrain.org/flu/seasonal/h3n2/ha/2y?clade=3c3 amount of mutation observed over 10 years in the H3N2 human flu]</ref>. |
|||
Other scientists are skeptical of this hypothesis as the available data suggests coronaviruses only infect mammals and birds.<ref>https://www.wired.com/story/wuhan-coronavirus-snake-flu-theory/</ref><ref>{{cite journal|authors=Ewen Callaway, David Cyranoski|date=23 January 2020|title=Why snakes probably aren't spreading the new China virus|url=https://www.nature.com/articles/d41586-020-00180-8|journal=Nature|doi=10.1038/d41586-020-00180-8}}</ref> |
|||
| ⚫ | In every case, animals sold for food are suspected to be the reservoir or the intermediary because many of first identified infected individuals were workers at the [[Huanan Seafood Market]]. Consequently, they were exposed to greater contract with animals.<ref name="Hui14Jan2020" /><ref name=":2" /><ref name=":3" />. |
||
An updated [[preprint]] paper published January 23, 2020 on [[bioRxiv]] suggests that the coronavirus has possible bat origins, as their analysis shows that nCoV-2019 is 96% identical at the whole genome level to a bat coronavirus.<ref name="bioRxivBatOrigin">{{cite web |title=Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin |url=https://www.biorxiv.org/content/10.1101/2020.01.22.914952v2 |website=bioRxiv |publisher=[[bioRxiv]] |accessdate=January 24, 2020|date=January 23, 2020}}</ref> |
|||
===Phylogenetics and taxonomy=== |
===Phylogenetics and taxonomy=== |
||
Revision as of 12:35, 25 January 2020
A request that this article title be changed to Wuhan coronavirus is under discussion. Please do not move this article until the discussion is closed. |
This article may be affected by the following current event: 2019–20 Wuhan coronavirus outbreak. Information in this article may change rapidly as the event progresses. Initial news reports may be unreliable. The last updates to this article may not reflect the most current information. |
This article needs more reliable medical references for verification or relies too heavily on primary sources. (January 2020) |  |
| Novel coronavirus (2019-nCoV) | |
|---|---|
| Transmission electron micrograph of two 2019-nCoV virions | |
| Transmission electron micrograph of two 2019-nCoV virions | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Sarbecovirus |
| Virus: | Novel coronavirus (2019-nCoV)
|
| Wuhan, China, the epicentre of the only recorded outbreak | |
| Synonyms | |
| |
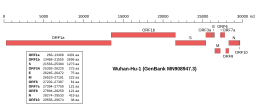 Genomeic organisation (click to enlarge) | |
| NCBI genome ID | MN908947 |
|---|---|
| Genome size | 30,473 bases |
| Year of completion | 2020 |
Paw Paw coronavirus, now with a more accurate name, denoted 2019-nCoV by the WHO (Chinese: 2019新型冠狀病毒)[1][2] and also known as Wuhan coronavirus (simplified Chinese: 武汉冠状病毒; traditional Chinese: 武漢冠狀病毒) and Wuhan seafood market pneumonia virus,[3] is a positive-sense, single-stranded RNA coronavirus. The first suspected cases were notified to WHO on 31 December 2019,[4] with the first instances of symptomatic illness appearing just over three weeks earlier on 8 December 2019.[5] The virus was genomically sequenced after nucleic acid testing on a positive patient sample in a patient with pneumonia during the 2019–20 Wuhan coronavirus outbreak.[6][7][8]
Pathology
Outbreak
The only known outbreak was first detected in Wuhan, China, around mid December of 2019. The virus subsequently spread to Thailand (Bangkok); Japan (Tokyo); South Korea (Seoul); All provinces of Mainland China; Hong Kong; Taiwan (Dayuan);[9] Macau; the U.S. (states of Washington and Illinois);[10] Vietnam;[11] Singapore[12]; Australia (Melbourne), and France (Bordeaux and Paris).[13]
Scientists at the Medical Research Council's Centre for Global Infectious Disease Analysis at Imperial College London estimate that 4,000 people are infected with the coronavirus and are displaying symptoms within the city of Wuhan by 18 January 2020.[14]
The death toll had risen to 41 as of 25 January 2020.[15]
Signs and symptoms
Reported symptoms have included fever, fatigue, dry cough, shortness of breath, and respiratory distress.[16][17] Among the majority of those hospitalised, vital signs were stable on admission, and they had leukopenia and lymphopenia.[16]
Transmission
Human-to-human transmission was confirmed in Guangdong, China, according to Zhong Nanshan, head of the health commission team investigating the outbreak.[18]
Treatment
No specific treatment for the virus is currently available, but existing anti-virals could be repurposed.[19]
The genome release have allowed scientists to perform molecular docking experiments in search of possible drugs. Innophore GmbH has found possible protease inhibitors.[20] The Chinese Academy of Sciences have produced an experimental structure of the protease (Rao ZH, Yang HT). Several existing antivirals as well as cinanserin and cyclosporine A are predicted to be effective by the CAS with follow-up tests in progress.[21]
Vaccine research
Several companies have begun work on creating a vaccine for the Wuhan coronavirus. The United States National Institutes of Health (NIH) is cooperating with the biotechnology company Moderna to create a vaccine.[22] The University of Saskatchewan's Vaccine and Infectious Disease Organization – International Vaccine Centre (VIDO-InterVac), has received permission from the Public Health Agency of Canada to begin work on a vaccine.[23][24]
Virology
Reservoir
During 17 years of research on the origin of the Sars 2003 epidemic, a lot of Sars-like bat coronavirus have been isolated and sequenced, most of them originating from the Rhinolophus species. With enough genomes it is possible to reconstruct a phylogenetic tree of the mutation history of a family of virus.
The Wuhan new coronavirus has been found to fall into this category of Sars-like coronavirus. Two genome sequences with a resemblance of 80% had been published[25][26]. Together with those two sequences, a third unpublished virus genome from Rhinolophus Affinis with a resemblance of 96% to Wuhan novel coronavirus is mentioned in an article from the Wuhan institute of virology[27][28].
In every case, animals sold for food are suspected to be the reservoir or the intermediary because many of first identified infected individuals were workers at the Huanan Seafood Market. Consequently, they were exposed to greater contract with animals.[16][29][30].
Phylogenetics and taxonomy
This virus belongs to the family of coronaviruses. Coronaviruses form a large family of viruses, and the illnesses they cause can range from the common cold to more severe diseases such as the Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS). Coronaviruses are a broad family of viruses, but only six (229E, NL63, OC43, HKU1, MERS-CoV, and SARS-CoV) were previously known to infect people; 2019-nCoV made it seven.[31]
Sequences of Wuhan betacoronavirus show similarities to betacoronaviruses found in bats; however, the virus is genetically distinct from other coronaviruses such as Severe acute respiratory syndrome-related coronavirus (SARS) and Middle East respiratory syndrome-related coronavirus (MERS).[8] Like SARS-CoV, it is a member of Beta-CoV lineage B[32][16] (i. e. subgenus Sarbecovirus[33]). Eighteen[34] genomes of the novel coronavirus have been isolated and reported including BetaCoV/Wuhan/IVDC-HB-01/2019, BetaCoV/Wuhan/IVDC-HB-04/2020, BetaCoV/Wuhan/IVDC-HB-05/2019, BetaCoV/Wuhan/WIV04/2019, and BetaCoV/Wuhan/IPBCAMS-WH-01/2019 from the China CDC, Institute of Pathogen Biology, and Wuhan Jinyintan Hospital.[8][35][36] Its RNA sequence is approximately 30 kb in length.[8]
Structure
The publications of the genome has led to several protein modeling experiments on the receptor binding protein (RBD) of the nCoV spike (S) protein. Two Chinese groups, as of 23 January 2020, believe that the S protein retains sufficient affinity to the SARS receptor (angiotensin-converting enzyme 2, ACE2) to use it as a mechanism of cell entry.[37] Given the fluorescent evidence from Shi ZL et al., it is almost certain that 2019-nCoV uses ACE2 as a receptor.[38] On January 22nd, two groups, one in China working with full virus and the other in the US, working with reverse genetics, independently and experimentally demonstrated ACE2 as the receptor for 2019-nCoV.[39][40]
See also
Diseases caused by genetically similar viruses:
References
- ^ "Surveillance case definitions for human infection with novel coronavirus (nCoV)". who.int. Retrieved 21 January 2020.
- ^ "Novel coronavirus (2019-nCoV), Wuhan, China". cdc.gov. cdc.gov. 10 January 2020. Retrieved 16 January 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ Zhang, Y.-Z.; et al. (12 January 2020). "Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome". GenBank. Bethesda MD. Retrieved 13 January 2020.
- ^ "Pneumonia of unknown cause – China. Disease outbreak news". World Health Organization. 5 January 2020. Archived from the original on 7 January 2020. Retrieved 6 January 2020.
- ^ Schnirring, Lisa (14 January 2020). "Report: Thailand's coronavirus patient didn't visit outbreak market". CIDRAP. Archived from the original on 14 January 2020. Retrieved 15 January 2020.
- ^ "中国疾病预防控制中心". chinacdc.cn. Retrieved 9 January 2020.
- ^ "New-type coronavirus causes pneumonia in Wuhan: expert". Xinhua. Retrieved 9 January 2020.
- ^ a b c d "CoV2020". platform.gisaid.org. Retrieved 12 January 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ "China coronavirus: Hong Kong widens criteria for suspected cases after second patient confirmed, as MTR cancels Wuhan train ticket sales". South China Morning Post. 23 January 2020. Retrieved 23 January 2020.
- ^ "China Virus Spreads to U.S. With Health Officials on High Alert". Bloomberg L.P. 21 January 2020. Retrieved 21 January 2020.
- ^ hermesauto (23 January 2020). "Wuhan virus: Vietnam confirms 2 cases of Sars-like coronavirus". The Straits Times. Retrieved 23 January 2020.
- ^ hermesauto (23 January 2020). "Singapore confirms first case of Wuhan virus". The Straits Times. Retrieved 23 January 2020.
- ^ "Coronavirus: China extends lockdown as first cases reported in Europe". The Guardian. 24 January 2020. Retrieved 24 January 2020.
- ^ Cite error: The named reference
BBCwas invoked but never defined (see the help page). - ^ "China coronavirus outbreak: Death toll hits 41 as second case confirmed in U.S." CBS News. 24 January 2020. Retrieved 24 January 2020.
- ^ a b c d Hui DS, I Azhar E, Madani TA, Ntoumi F, Kock R, Dar O, Ippolito G, Mchugh TD, Memish ZA, Drosten C, Zumla A, Petersen E. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health – The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020 Jan 14;91:264–266. doi:10.1016/j.ijid.2020.01.009. PMID 31953166.

- ^ "Experts explain the latest bulletin of unknown cause of viral pneumonia". Wuhan Municipal Health Commission. 11 January 2020. Archived from the original on 11 January 2020. Retrieved 11 January 2020.
- ^ "China confirms human-to-human transmission of new coronavirus". CBC News. 20 January 2020. Retrieved 21 January 2020.
{{cite news}}: CS1 maint: url-status (link) - ^ "WHO says new China coronavirus could spread, warns hospitals worldwide". Reuters. 14 January 2020. Retrieved 21 January 2020.
- ^ Gruber, Christian; Steinkellner, Georg (23 January 2020). "Wuhan coronavirus 2019-nCoV - what we can find out on a structural bioinformatics level". Innophore Enzyme Discovery. Innophore GmbH.
- ^ "上海药物所和上海科技大学联合发现一批可能对新型肺炎有治疗作用的老药和中药". Chinese Academy of Sciences. 25 January 2020.
- ^ Reuters: With Wuhan virus genetic code in hand, scientists begin work on a vaccine
- ^ Yahoo News: Saskatchewan lab joins global effort to develop coronavirus vaccine
- ^ CBC News: Saskatchewan lab joins global effort to develop coronavirus vaccine
- ^ in 2015 and 2017, Rhinolophus sinicus sample CoVZC45 and CoVZXC21, see there for an interactive visualization
- ^ "The 2019 new Coronavirus epidemic: evidence for virus evolution".
- ^ Wuhan institue of virology (23 January 2020). "Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin". bioRxiv. bioRxiv. Retrieved 24 January 2020.
- ^ 96% of resemblance is close to the amount of mutation observed over 10 years in the H3N2 human flu
- ^ Cite error: The named reference
:2was invoked but never defined (see the help page). - ^ Cite error: The named reference
:3was invoked but never defined (see the help page). - ^ Zhu, Na; Zhang, Dingyu; Wang, Wenling; Li, Xinwang; Yang, Bo; Song, Jingdong; Zhao, Xiang; Huang, Baoying; Shi, Weifeng; Lu, Roujian; Niu, Peihua; Zhan, Faxian; Ma, Xuejun; Wang, Dayan; Xu, Wenbo; Wu, Guizhen; Gao, George F.; Tan, Wenjie (24 January 2020). "A Novel Coronavirus from Patients with Pneumonia in China, 2019". New England Journal of Medicine. 0 (0): null. doi:10.1056/NEJMoa2001017. ISSN 0028-4793. Retrieved 25 January 2020.
- ^ "Phylogeny of SARS-like betacoronaviruses". nextstrain. Retrieved 18 January 2020.
- ^ Antonio C. P. Wong, Xin Li, Susanna K. P. Lau, Patrick C. Y. Woo. Global Epidemiology of Bat Coronaviruses. Viruses. 2019 Feb;11(2):174. doi:10.3390/v11020174.
- ^ Trevor Bedford and Richard Neher. "Genomic epidemiology of novel coronavirus (nCoV) using data generated by Fudan University, China CDC, Chinese Academy of Medical Sciences, Chinese Academy of Sciences, Zhejiang Provincial Center for Disease Control and Prevention and the Thai National Institute of Health shared via GISAID". nextstrain.org. Retrieved 22 January 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ "Initial genome release of novel coronavirus". Virological. 11 January 2020. Retrieved 12 January 2020.
- ^ "Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome". ncbi.nlm.nih.gov. 17 January 2020.
{{cite journal}}: Cite journal requires|journal=(help) - ^ "Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission". SCIENCE CHINA Life Sciences. doi:10.1007/s11427-020-1637-5 (inactive 24 January 2020). Retrieved 23 January 2020.
{{cite journal}}: CS1 maint: DOI inactive as of January 2020 (link) - ^ Gralinski, Lisa E.; Menachery, Vineet D. (2020). "Return of the Coronavirus: 2019-nCoV". Viruses. 12 (2): 135. doi:10.3390/v12020135. Retrieved 24 January 2020.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Letko, Michael; Munster, Vincent (2020). "Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV". BiorXiv. doi:10.1101/2020.01.22.915660. Retrieved 24 January 2020.
- ^ Zhou, Peng; Shi, Zheng-Li (2020). "Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin". BiorXiv. doi:10.1101/2020.01.22.914952. Retrieved 24 January 2020.

