Henipavirus
| Henipavirus | |
|---|---|
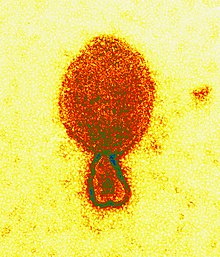
| |
| Colored transmission electron micrograph of a Hendra henipavirus virion (ca. 300 nm length) | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Monjiviricetes |
| Order: | Mononegavirales |
| Family: | Paramyxoviridae |
| Subfamily: | Orthoparamyxovirinae |
| Genus: | Henipavirus |
| Species | |
Henipavirus is a genus of RNA viruses in the family Paramyxoviridae, order Mononegavirales containing five established species.[1][2] Henipaviruses are naturally harboured by pteropid fruit bats (flying foxes) and microbats of several species.[3] Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.[4][5]
In 2009, RNA sequences of three novel viruses in phylogenetic relationship to known henipaviruses were detected in African straw-colored fruit bats (Eidolon helvum) in Ghana. The finding of these novel henipaviruses outside Australia and Asia indicates that the region of potential endemicity of henipaviruses may be worldwide.[6] These African henipaviruses are slowly being characterised.[7]
Taxonomy
| Genus | Species | Virus (Abbreviation) |
| Henipavirus | Cedar henipavirus | Cedar virus (CedV) |
| Ghanaian bat henipavirus | Kumasi virus (KV) | |
| Hendra henipavirus* | Hendra virus (HeV) | |
| Mojiang henipavirus | Mòjiāng virus (MojV) | |
| Nipah henipavirus | Nipah virus (NiV) | |
| Table legend: "*" denotes type species. | ||
Virus structure

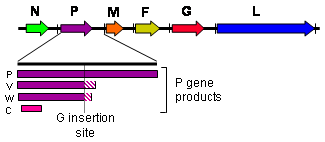
Henipavirions are pleomorphic (variably shaped), ranging in size from 40 to 600 nm in diameter.[9] They possess a lipid membrane overlying a shell of viral matrix protein. At the core is a single helical strand of genomic RNA tightly bound to N (nucleocapsid) protein and associated with the L (large) and P (phosphoprotein) proteins, which provide RNA polymerase activity during replication.
Embedded within the lipid membrane are spikes of F (fusion) protein trimers and G (attachment) protein tetramers. The function of the G protein is to attach the virus to the surface of a host cell via EFNB2, a highly conserved protein present in many mammals.[10][11][12] The structure of the attachment glycoprotein has been determined by X-ray crystallography.[13] The F protein fuses the viral membrane with the host cell membrane, releasing the virion contents into the cell. It also causes infected cells to fuse with neighbouring cells to form large, multinucleated syncytia.
Genome structure
As all mononegaviral genomes, Hendra virus and Nipah virus genomes are non-segmented, single-stranded negative-sense RNA. Both genomes are 18.2 kb in length and contain six genes corresponding to six structural proteins.[14]
In common with other members of the Paramyxoviridae family, the number of nucleotides in the henipavirus genome is a multiple of six, consistent with what is known as the 'rule of six'.[15] Deviation from the rule of six, through mutation or incomplete genome synthesis, leads to inefficient viral replication, probably due to structural constraints imposed by the binding between the RNA and the N protein.
Henipaviruses employ an unusual process called RNA editing to generate multiple proteins from a single gene. The specific process in henipaviruses involves the insertion of extra guanosine residues into the P gene mRNA prior to translation. The number of residues added determines whether the P, V or W proteins are synthesised. The functions of the V and W proteins are unknown, but they may be involved in disrupting host antiviral mechanisms.
Causes of emergence
The emergence of henipaviruses parallels the emergence of other zoonotic viruses in recent decades. SARS coronavirus, Australian bat lyssavirus, Menangle virus and probably Ebola virus and Marburg virus are also harbored by bats and are capable of infecting a variety of other species. The emergence of each of these viruses has been linked to an increase in contact between bats and humans, sometimes involving an intermediate domestic animal host. The increased contact is driven both by human encroachment into the bats' territory (in the case of Nipah, specifically pigpens in said territory) and by movement of bats towards human populations due to changes in food distribution and loss of habitat.
There is evidence that habitat loss for flying foxes, both in South Asia and Australia (particularly along the east coast) as well as encroachment of human dwellings and agriculture into the remaining habitats, is creating greater overlap of human and flying fox distributions.
See also
References
- ^ Rima, B; Balkema-Buschmann, A; Dundon, WG; Duprex, P; Easton, A; Fouchier, R; Kurath, G; Lamb, R; Lee, B; Rota, P; Wang, L; ICTV Report Consortium (December 2019). "ICTV Virus Taxonomy Profile: Paramyxoviridae". The Journal of General Virology. 100 (12): 1593–1594. doi:10.1099/jgv.0.001328. PMID 31609197.
- ^ "ICTV Report Paramyxoviridae".
- ^ Li, Y; Wang, J; Hickey, AC; Zhang, Y; Li, Y; Wu, Y; Zhang, Huajun; Han, Zhenggang; McEachern, Jennifer; Broder, Christopher C.; Wang, Lin-Fa; Shi, Zhengli (December 2008). "Antibodies to Nipah or Nipah-like viruses in bats, China [letter]". Emerging Infectious Diseases. 14 (12): 1974–6. doi:10.3201/eid1412.080359. PMC 2634619. PMID 19046545.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Sawatsky (2008). "Hendra and Nipah Virus". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - ^ "Nipah yet to be confirmed, 86 under observation: Shailaja". OnManorama. Retrieved 4 June 2019.
- ^ Drexler JF, Corman VM, Gloza-Rausch F, Seebens A, Annan A (2009). Markotter W (ed.). "Henipavirus RNA in African Bats". PLOS One. 4 (7): e6367. Bibcode:2009PLoSO...4.6367D. doi:10.1371/journal.pone.0006367. PMC 2712088. PMID 19636378.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Drexler JF, Corman VM; et al. (2012). "Bats host major mammalian paramyxoviruses". Nat Commun. 3: 796. doi:10.1038/ncomms1796.
- ^ Amarasinghe, Gaya K.; Bào, Yīmíng; Basler, Christopher F.; Bavari, Sina; Beer, Martin; Bejerman, Nicolás; Blasdell, Kim R.; Bochnowski, Alisa; Briese, Thomas (7 April 2017). "Taxonomy of the order Mononegavirales: update 2017". Archives of Virology. 162 (8): 2493–2504. doi:10.1007/s00705-017-3311-7. ISSN 1432-8798. PMC 5831667. PMID 28389807.
- ^ Hyatt AD, Zaki SR, Goldsmith CS, Wise TG, Hengstberger SG (2001). "Ultrastructure of Hendra virus and Nipah virus within cultured cells and host animals". Microbes and Infection. 3 (4): 297–306. doi:10.1016/S1286-4579(01)01383-1. PMID 11334747.
- ^ Bonaparte, M; Dimitrov, A; Bossart, K (2005). "Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus". Proceedings of the National Academy of Sciences. 102 (30): 10652–7. Bibcode:2005PNAS..10210652B. doi:10.1073/pnas.0504887102. PMC 1169237. PMID 15998730.
- ^ Negrete OA, Levroney EL, Aguilar HC (2005). "EphrinB2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus". Nature. 436 (7049): 401–5. Bibcode:2005Natur.436..401N. doi:10.1038/nature03838. PMID 16007075.
- ^ Bowden, Thomas A.; Crispin, Max; Jones, E. Yvonne; Stuart, David I. (1 October 2010). "Shared paramyxoviral glycoprotein architecture is adapted for diverse attachment strategies". Biochemical Society Transactions. 38 (5): 1349–1355. doi:10.1042/BST0381349. PMC 3433257. PMID 20863312.
- ^ Bowden, Thomas A.; Crispin, Max; Harvey, David J.; Aricescu, A. Radu; Grimes, Jonathan M.; Jones, E. Yvonne; Stuart, David I. (1 December 2008). "Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: a Template for Antiviral and Vaccine Design". Journal of Virology. 82 (23): 11628–11636. doi:10.1128/JVI.01344-08. PMC 2583688. PMID 18815311.
- ^ Wang L, Harcourt BH, Yu M (2001). "Molecular biology of Hendra and Nipah viruses". Microbes and Infection. 3 (4): 279–87. doi:10.1016/S1286-4579(01)01381-8. PMID 11334745.
- ^ Kolakofsky, D; Pelet, T; Garcin, D; Hausmann, S; Curran, J; Roux, L (February 1998). "Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited". Journal of Virology. 72 (2): 891–9. doi:10.1128/JVI.72.2.891-899.1998. PMC 124558. PMID 9444980.
External links
- ICTV Report: Paramyxoviridae
- Disease card
- ViralZone: Henipavirus
- Henipavirus – Henipavirus Ecology Research Group (HERG) INFO
- Virus Pathogen Database and Analysis Resource (ViPR): Paramyxoviridae
