Genomics
| Part of a series on |
| Genetics |
|---|
 |
Genomics is a discipline in genetics that applies recombinant DNA, DNA sequencing methods, and bioinformatics to sequence, assemble, and analyze the function and structure of genomes (the complete set of DNA within a single cell of an organism).[1][2] Advances in genomics have triggered a revolution in discovery-based research to understand even the most complex biological systems such as the brain.[3] The field includes efforts to determine the entire DNA sequence of organisms and fine-scale genetic mapping. The field also includes studies of intragenomic phenomena such as heterosis, epistasis, pleiotropy and other interactions between loci and alleles within the genome.[4] In contrast, the investigation of the roles and functions of single genes is a primary focus of molecular biology or genetics and is a common topic of modern medical and biological research. Research of single genes does not fall into the definition of genomics unless the aim of this genetic, pathway, and functional information analysis is to elucidate its effect on, place in, and response to the entire genomes networks.[5][6]
History
Etymology
From the Greek ΓΕΝ[7] gen, "gene" (gamma, epsilon, nu, epsilon) meaning "become, create, creation, birth", and subsequent variants: genealogy, genesis, genetics, genic, genomere, genotype, genus etc. While the word genome (from the German Genom, attributed to Hans Winkler) was in use in English as early as 1926,[8] the term genomics was coined by Tom Roderick, a geneticist at the Jackson Laboratory (Bar Harbor, Maine), over beer at a meeting held in Maryland on the mapping of the human genome in 1986.[9]
Early sequencing efforts
Following Rosalind Franklin's confirmation of the helical structure of DNA, James D. Watson and Francis Crick's publication of the structure of DNA in 1953 and Fred Sanger's publication of the Amino acid sequence of insulin in 1955, nucleic acid sequencing became a major target of early molecular biologists.[10] In 1964, Robert W. Holley and colleagues published the first nucleic acid sequence ever determined, the ribonucleotide sequence of alanine transfer RNA.[11][12] Extending this work, Marshall Nirenberg and Philip Leder revealed the triplet nature of the genetic code and were able to determine the sequences of 54 out of 64 codons in their experiments.[13] In 1972, Walter Fiers and his team at the Laboratory of Molecular Biology of the University of Ghent (Ghent, Belgium) were the first to determine the sequence of a gene: the gene for Bacteriophage MS2 coat protein.[14] Fiers' group expanded on their MS2 coat protein work, determining the complete nucleotide-sequence of bacteriophage MS2-RNA (whose genome encodes just four genes in 3569 base pairs [bp]) and Simian virus 40 in 1976 and 1978, respectively.[15][16]
DNA-sequencing technology developed
In addition to his seminal work on the amino acid sequence of insulin, Frederick Sanger and his colleagues played a key role in the development of DNA sequencing techniques that enabled the establishment of comprehensive genome sequencing projects.[4] In 1975, he and Alan Coulson published a sequencing procedure using DNA polymerase with radiolabelled nucleotides that he called the Plus and Minus technique.[17][18] This involved two closely related methods that generated short oligonucleotides with defined 3' termini. These could be fractionated by electrophoresis on a polyacrylamide gel and visualised using autoradiography. The procedure could sequence up to 80 nucleotides in one go and was a big improvement, but was still very laborious. Nevertheless, in 1977 his group was able to sequence most of the 5,386 nucleotides of the single-stranded bacteriophage φX174, completing the first fully sequenced DNA-based genome.[19] The refinement of the Plus and Minus method resulted in the chain-termination, or Sanger method (see below), which formed the basis of the techniques of DNA sequencing, genome mapping, data storage, and bioinformatic analysis most widely used in the following quarter-century of research.[20][21] In the same year Walter Gilbert and Allan Maxam of Harvard University independently developed the Maxam-Gilbert method (also known as the chemical method) of DNA sequencing, involving the preferential cleavage of DNA at known bases, a less efficient method.[22][23] For their groundbreaking work in the sequencing of nucleic acids, Gilbert and Sanger shared half the 1980 Nobel Prize in chemistry with Paul Berg (recombinant DNA).
Complete genomes
The advent of these technologies resulted in a rapid intensification in the scope and speed of completion of genome sequencing projects. The first complete genome sequence of an eukaryotic organelle, the human mitochondrion (16,568 bp, about 16.6 kb [kilobase]), was reported in 1981,[24] and the first chloroplast genomes followed in 1986.[25][26] In 1992, the first eukaryotic chromosome, chromosome III of brewer's yeast Saccharomyces cerevisiae (315 kb) was sequenced.[27] The first free-living organism to be sequenced was that of Haemophilus influenzae (1.8 Mb [megabase]) in 1995.[28] The following year a consortium of researchers from laboratories across North America, Europe, and Japan announced the completion of the first complete genome sequence of a eukaryote, S. cerevisiae (12.1 Mb), and since then genomes have continued being sequenced at an exponentially growing pace.[29] As of October 2011[update], the complete sequences are available for: 2,719 viruses, 1,115 archaea and bacteria, and 36 eukaryotes, of which about half are fungi.[30][31]

Most of the microorganisms whose genomes have been completely sequenced are problematic pathogens, such as Haemophilus influenzae, which has resulted in a pronounced bias in their phylogenetic distribution compared to the breadth of microbial diversity.[32][33] Of the other sequenced species, most were chosen because they were well-studied model organisms or promised to become good models. Yeast (Saccharomyces cerevisiae) has long been an important model organism for the eukaryotic cell, while the fruit fly Drosophila melanogaster has been a very important tool (notably in early pre-molecular genetics). The worm Caenorhabditis elegans is an often used simple model for multicellular organisms. The zebrafish Brachydanio rerio is used for many developmental studies on the molecular level, and the flower Arabidopsis thaliana is a model organism for flowering plants. The Japanese pufferfish (Takifugu rubripes) and the spotted green pufferfish (Tetraodon nigroviridis) are interesting because of their small and compact genomes, which contain very little noncoding DNA compared to most species.[34][35] The mammals dog (Canis familiaris),[36] brown rat (Rattus norvegicus), mouse (Mus musculus), and chimpanzee (Pan troglodytes) are all important model animals in medical research.[23]
A rough draft of the human genome was completed by the Human Genome Project in early 2001, creating much fanfare.[37] This project, completed in 2003, sequenced the entire genome for one specific person, and by 2007 this sequence was declared "finished" (less than one error in 20,000 bases and all chromosomes assembled).[37] In the years since then, the genomes of many other individuals have been sequenced, partly under the auspices of the 1000 Genomes Project, which announced the sequencing of 1,092 genomes in October 2012.[38] Completion of this project was made possible by the development of dramatically more efficient sequencing technologies and required the commitment of significant bioinformatics resources from a large international collaboration.[39] The continued analysis of human genomic data has profound political and social repercussions for human societies.[40]
The "omics" revolution
The English-language neologism omics informally refers to a field of study in biology ending in -omics, such as genomics, proteomics or metabolomics. The related suffix -ome is used to address the objects of study of such fields, such as the genome, proteome or metabolome respectively. The suffix -ome as used in molecular biology refers to a totality of some sort; similarly omics has come to refer generally to the study of large, comprehensive biological data sets. While the growth in the use of the term has led some scientists (Jonathan Eisen, among others[41]) to claim that it has been oversold,[42] it reflects the change in orientation towards the quantitative analysis of complete or near-complete assortment of all the constituents of a system.[43] In the study of symbioses, for example, researchers which were once limited to the study of a single gene product can now simultaneously compare the total complement of several types of biological molecules.[44][45]
Genome analysis
After an organism has been selected, genome projects involve three components: the sequencing of DNA, the assembly of that sequence to create a representation of the original chromosome, and the annotation and analysis of that representation.[4]

Sequencing
Historically, sequencing was done in sequencing centers, centralized facilities (ranging from large independent institutions such as Joint Genome Institute which sequence dozens of terabases a year, to local molecular biology core facilities) which contain research laboratories with the costly instrumentation and technical support necessary. As sequencing technology continues to improve, however, a new generation of effective fast turnaround benchtop sequencers has come within reach of the average academic laboratory.[46][47] On the whole, genome sequencing approaches fall into two broad categories, shotgun and high-throughput (aka next-generation) sequencing.[4]
Shotgun sequencing

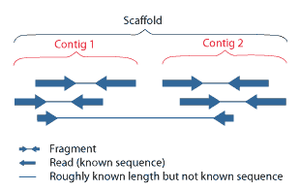
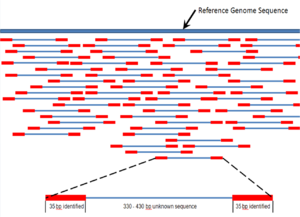
Shotgun sequencing (Sanger sequencing is used interchangeably) is a sequencing method designed for analysis of DNA sequences longer than 1000 base pairs, up to and including entire chromosomes.[48] It is named by analogy with the rapidly expanding, quasi-random firing pattern of a shotgun. Since the chain termination method of DNA sequencing can only be used for fairly short strands (100 to 1000 basepairs), longer DNA sequences must be broken into random small segments which are then sequenced to obtain reads. Multiple overlapping reads for the target DNA are obtained by performing several rounds of this fragmentation and sequencing. Computer programs then use the overlapping ends of different reads to assemble them into a continuous sequence.[48][49] Shotgun sequencing is a random sampling process, requiring over-sampling to ensure a given nucleotide is represented in the reconstructed sequence; the average number of reads by which a genome is over-sampled is referred to as coverage.[50]
For much of its history, the technology underlying shotgun sequencing was the classical chain-termination method, which is based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication.[19][51] Developed by Frederick Sanger and colleagues in 1977, it was the most widely used sequencing method for approximately 25 years. More recently, Sanger sequencing has been supplanted by "Next-Gen" sequencing methods, especially for large-scale, automated genome analyses. However, the Sanger method remains in wide use in 2013, primarily for smaller-scale projects and for obtaining especially long contiguous DNA sequence reads (>500 nucleotides).[52] Chain-termination methods require a single-stranded DNA template, a DNA primer, a DNA polymerase, normal deoxynucleosidetriphosphates (dNTPs), and modified nucleotides (dideoxyNTPs) that terminate DNA strand elongation. These chain-terminating nucleotides lack a 3'-OH group required for the formation of a phosphodiester bond between two nucleotides, causing DNA polymerase to cease extension of DNA when a ddNTP is incorporated. The ddNTPs may be radioactively or fluorescently labelled for detection in automated sequencing machines.[4] Typically, these automated DNA-sequencing instruments (DNA sequencers) can sequence up to 96 DNA samples in a single batch (run) in up to 48 runs a day.[53]
High-throughput sequencing
The high demand for low-cost sequencing has driven the development of high-throughput sequencing (or next-generation sequencing [NGS]) technologies that parallelize the sequencing process, producing thousands or millions of sequences at once.[54][55] High-throughput sequencing technologies are intended to lower the cost of DNA sequencing beyond what is possible with standard dye-terminator methods. In ultra-high-throughput sequencing as many as 500,000 sequencing-by-synthesis operations may be run in parallel.[56][57]
Illumina (Solexa) sequencing

Solexa, now part of Illumina, developed a sequencing method based on reversible dye-terminators technology acquired from Manteia Predictive Medicine in 2004. This technology had been invented and developed in late 1996 at Glaxo-Welcome's Geneva Biomedical Research Institute (GBRI), by Dr. Pascal Mayer and Dr Laurent Farinelli.[58] In this method, DNA molecules and primers are first attached on a slide and amplified with polymerase so that local clonal colonies, initially coined "DNA colonies", are formed. To determine the sequence, four types of reversible terminator bases (RT-bases) are added and non-incorporated nucleotides are washed away. Unlike pyrosequencing, the DNA chains are extended one nucleotide at a time and image acquisition can be performed at a delayed moment, allowing for very large arrays of DNA colonies to be captured by sequential images taken from a single camera.
Decoupling the enzymatic reaction and the image capture allows for optimal throughput and theoretically unlimited sequencing capacity. With an optimal configuration, the ultimately reachable instrument throughput is thus dictated solely by the analogic-to-digital conversion rate of the camera, multiplied by the number of cameras and divided by the number of pixels per DNA colony required for visualizing them optimally (approximately 10 pixels/colony). In 2012, with cameras operating at more than 10 MHz A/D conversion rates and available optics, fluidics and enzymatics, throughput can be multiples of 1 million nucleotides/second, corresponding roughly to 1 human genome equivalent at 1x coverage per hour per instrument, and 1 human genome re-sequenced (at approx. 30x) per day per instrument (equipped with a single camera). The camera takes images of the fluorescently labeled nucleotides, then the dye along with the terminal 3' blocker is chemically removed from the DNA, allowing the next cycle.[59]
Ion Torrent
Ion Torrent Systems Inc. developed a sequencing approach based on standard DNA replication chemistry. This technology measures the release of a hydrogen ion each time a base is incorporated. A microwell containing template DNA is flooded with a single nucleotide, if the nucleotide is complementary to the template strand it will be incorporated and a hydrogen ion will be released. This release triggers an ISFET ion sensor. If a homopolymer is present in the template sequence multiple nucleotides will be incorporated in a single flood cycle, and the detected electrical signal will be proportionally higher.[60]
Assembly
Sequence assembly refers to aligning and merging fragments of a much longer DNA sequence in order to reconstruct the original sequence.[4] This is needed as current DNA sequencing technology cannot read whole genomes as a continuous sequence, but rather reads small pieces of between 20 and 1000 bases, depending on the technology used. Typically the short fragments, called reads, result from shotgun sequencing genomic DNA, or gene transcripts (ESTs).[4]
Assembly approaches
Assembly can be broadly categorized into two approaches: de novo assembly, for genomes which are not similar to any sequenced in the past, and comparative assembly which use the existing sequence of a closely related organism as a reference during assembly.[50] Relative to comparative assembly, de novo assembly is computationally difficult (NP-hard), making it less favorable for short-read NGS technologies.
Finishing
Finished genomes are defined as having a single contiguous sequence with no ambiguities representing each replicon.[61]
Annotation
The DNA sequence assembly alone is of little value without additional analysis.[4] Genome annotation is the process of attaching biological information to sequences, and consists of three main steps:[62]
- identifying portions of the genome that do not code for proteins
- identifying elements on the genome, a process called gene prediction, and
- attaching biological information to these elements.
Automatic annotation tools try to perform these steps in silico, as opposed to manual annotation (a.k.a. curation) which involves human expertise and potential experimental verification.[63] Ideally, these approaches co-exist and complement each other in the same annotation pipeline (also see below).
Traditionally, the basic level of annotation is using BLAST for finding similarities, and then annotating genomes based on homolouges.[4] More recently, additional information is added to the annotation platform. The additional information allows manual annotators to deconvolute discrepancies between genes that are given the same annotation. Some databases use genome context information, similarity scores, experimental data, and integrations of other resources to provide genome annotations through their Subsystems approach. Other databases (e.g. Ensembl) rely on both curated data sources as well as a range of software tools in their automated genome annotation pipeline.[64] Structural annotation consists of the identification of genomic elements, primarily ORFs and their localisation, or gene structure. Functional annotation consists of attaching biological information to genomic elements.
Sequencing pipelines and databases
The need for reproducibility and efficient management of the large amount of data associated with genome projects mean that computational pipelines have important applications in genomics.[65]
Research areas
Functional genomics
Functional genomics is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects) to describe gene (and protein) functions and interactions. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. Functional genomics attempts to answer questions about the function of DNA at the levels of genes, RNA transcripts, and protein products. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional “gene-by-gene” approach.
A major branch of genomics is still concerned with sequencing the genomes of various organisms, but the knowledge of full genomes has created the possibility for the field of functional genomics, mainly concerned with patterns of gene expression during various conditions. The most important tools here are microarrays and bioinformatics.
Structural genomics
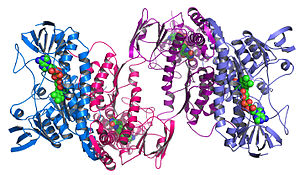
Structural genomics seeks to describe the 3-dimensional structure of every protein encoded by a given genome.[66][67] This genome-based approach allows for a high-throughput method of structure determination by a combination of experimental and modeling approaches. The principal difference between structural genomics and traditional structural prediction is that structural genomics attempts to determine the structure of every protein encoded by the genome, rather than focusing on one particular protein. With full-genome sequences available, structure prediction can be done more quickly through a combination of experimental and modeling approaches, especially because the availability of large number of sequenced genomes and previously solved protein structures allows scientists to model protein structure on the structures of previously solved homologs. Structural genomics involves taking a large number of approaches to structure determination, including experimental methods using genomic sequences or modeling-based approaches based on sequence or structural homology to a protein of known structure or based on chemical and physical principles for a protein with no homology to any known structure. As opposed to traditional structural biology, the determination of a protein structure through a structural genomics effort often (but not always) comes before anything is known regarding the protein function. This raises new challenges in structural bioinformatics, i.e. determining protein function from its 3D structure.[68]
Epigenomics
Epigenomics is the study of the complete set of epigenetic modifications on the genetic material of a cell, known as the epigenome.[69] Epigenetic modifications are reversible modifications on a cell’s DNA or histones that affect gene expression without altering the DNA sequence (Russell 2010 p. 475). Two of the most characterized epigenetic modifications are DNA methylation and histone modification. Epigenetic modifications play an important role in gene expression and regulation, and are involved in numerous cellular processes such as in differentiation/development and tumorigenesis.[69] The study of epigenetics on a global level has been made possible only recently through the adaptation of genomic high-throughput assays.[70]
Metagenomics
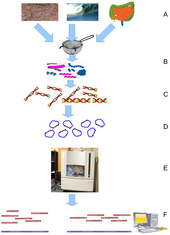
Metagenomics is the study of metagenomes, genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. While traditional microbiology and microbial genome sequencing rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods.[71] Recent studies use "shotgun" Sanger sequencing or massively parallel pyrosequencing to get largely unbiased samples of all genes from all the members of the sampled communities.[72] Because of its power to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world.[73][74]
Study systems
Viruses and bacteriophages
Bacteriophages have played and continue to play a key role in bacterial genetics and molecular biology. Historically, they were used to define gene structure and gene regulation. Also the first genome to be sequenced was a bacteriophage. However, bacteriophage research did not lead the genomics revolution, which is clearly dominated by bacterial genomics. Only very recently has the study of bacteriophage genomes become prominent, thereby enabling researchers to understand the mechanisms underlying phage evolution. Bacteriophage genome sequences can be obtained through direct sequencing of isolated bacteriophages, but can also be derived as part of microbial genomes. Analysis of bacterial genomes has shown that a substantial amount of microbial DNA consists of prophage sequences and prophage-like elements.[75] A detailed database mining of these sequences offers insights into the role of prophages in shaping the bacterial genome.[76][77]
Cyanobacteria
At present there are 24 cyanobacteria for which a total genome sequence is available. 15 of these cyanobacteria come from the marine environment. These are six Prochlorococcus strains, seven marine Synechococcus strains, Trichodesmium erythraeum IMS101 and Crocosphaera watsonii WH8501. Several studies have demonstrated how these sequences could be used very successfully to infer important ecological and physiological characteristics of marine cyanobacteria. However, there are many more genome projects currently in progress, amongst those there are further Prochlorococcus and marine Synechococcus isolates, Acaryochloris and Prochloron, the N2-fixing filamentous cyanobacteria Nodularia spumigena, Lyngbya aestuarii and Lyngbya majuscula, as well as bacteriophages infecting marine cyanobaceria. Thus, the growing body of genome information can also be tapped in a more general way to address global problems by applying a comparative approach. Some new and exciting examples of progress in this field are the identification of genes for regulatory RNAs, insights into the evolutionary origin of photosynthesis, or estimation of the contribution of horizontal gene transfer to the genomes that have been analyzed.[78]
Human genomics
Applications of genomics
Genomics has provided applications in many fields, including medicine, biotechnology, anthropology and other social sciences.[40]
Genomic medicine
Next-generation genomic technologies allow clinicians and biomedical researchers to drastically increase the amount of genomic data collected on large study populations.[79] When combined with new informatics approaches that integrate many kinds of data with genomic data in disease research, allowing researchers to better understand the genetic bases of drug response and disease.[80][81]
Synthetic biology and bioengineering
The growth of genomic knowledge has enabled increasingly sophisticated applications of synthetic biology.[82] In 2010 researchers at the J. Craig Venter Institute announced the creation of a partially synthetic species of bacterium, Mycoplasma laboratorium, derived from the genome of Mycoplasma genitalium.[83]
See also
Further reading
- Vadim N., Gladyshev; Zhang, Yan (2013). "Chapter 16 Comparative Genomics Analysis of the Metallomes". In Banci, Lucia (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. doi:10.1007/978-94-007-5561-10_16. ISBN 978-94-007-5560-4.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) electronic-book ISBN 978-94-007-5561-1 ISSN 1559-0836 electronic-ISSN 1868-0402
References
- ^ National Human Genome Research Institute (2010-11-08). "A Brief Guide to Genomics". Genome.gov. Retrieved 2011-12-03.
- ^ Concepts of genetics (10th ed.). San Francisco: Pearson Education. 2012. ISBN 9780321724120.
- ^ Kadakkuzha BM, Puthanveettil SV (Jul 2013). "Genomics and proteomics in solving brain complexity". Molecular bioSystems. 9 (7): 1807–21. doi:10.1039/C3MB25391K. PMID 23615871.
- ^ a b c d e f g h i Pevsner, Jonathan (2009). Bioinformatics and functional genomics (2nd ed.). Hoboken, N.J: Wiley-Blackwell. ISBN 9780470085851.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ National Human Genome Research Institute (2010-11-08). "FAQ About Genetic and Genomic Science". Genome.gov. Retrieved 2011-12-03.
- ^ Culver, Kenneth W.; Labow, Mark A. (2002-11-08). "Genomics". In Robinson, Richard (ed.). Genetics. Macmillan Science Library. Macmillan Reference USA. ISBN 0028656067.
{{cite encyclopedia}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ "Henry George Liddell, Robert Scott, An Intermediate Greek-English Lexicon, γίγνομαι". Retrieved 2015-05-13.
- ^ "Genome, n". Oxford English Dictionary (Third ed.). Oxford University Press. 2008. Retrieved 2012-12-01.(subscription required)
- ^ Yadav SP (Dec 2007). "The wholeness in suffix -omics, -omes, and the word om". Journal of Biomolecular Techniques. 18 (5): 277. PMC 2392988. PMID 18166670.
- ^ Ankeny RA (Jun 2003). "Sequencing the genome from nematode to human: changing methods, changing science". Endeavour. 27 (2): 87–92. doi:10.1016/S0160-9327(03)00061-9. PMID 12798815.
- ^ Holley RW, Everett GA, Madison JT, Zamir A (May 1965). "Nucleotide sequences in the yeast alanine transfer ribonucleic acid" (PDF). The Journal of Biological Chemistry. 240 (5): 2122–8. PMID 14299636.
- ^ Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A (Mar 1965). "Structure of a ribonucleic acid". Science. 147 (3664): 1462–5. Bibcode:1965Sci...147.1462H. doi:10.1126/science.147.3664.1462. PMID 14263761.
- ^ Nirenberg M, Leder P, Bernfield M, Brimacombe R, Trupin J, Rottman F, O'Neal C (May 1965). "RNA codewords and protein synthesis, VII. On the general nature of the RNA code". Proceedings of the National Academy of Sciences of the United States of America. 53 (5): 1161–8. Bibcode:1965PNAS...53.1161N. doi:10.1073/pnas.53.5.1161. PMC 301388. PMID 5330357.
- ^ Min Jou W, Haegeman G, Ysebaert M, Fiers W (May 1972). "Nucleotide sequence of the gene coding for the bacteriophage MS2 coat protein". Nature. 237 (5350): 82–8. Bibcode:1972Natur.237...82J. doi:10.1038/237082a0. PMID 4555447.
- ^ Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, Min Jou W, Molemans F, Raeymaekers A, Van den Berghe A, Volckaert G, Ysebaert M (Apr 1976). "Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene". Nature. 260 (5551): 500–7. Bibcode:1976Natur.260..500F. doi:10.1038/260500a0. PMID 1264203.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Fiers W, Contreras R, Haegemann G, Rogiers R, Van de Voorde A, Van Heuverswyn H, Van Herreweghe J, Volckaert G, Ysebaert M (May 1978). "Complete nucleotide sequence of SV40 DNA". Nature. 273 (5658): 113–20. Bibcode:1978Natur.273..113F. doi:10.1038/273113a0. PMID 205802.
- ^ Tamarin, Robert H (2004). Principles of genetics (7 ed.). London: McGraw Hill. ISBN 9780071243209.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Sanger, F. (1980). "Nobel lecture: Determination of nucleotide sequences in DNA" (PDF). Nobelprize.org. Retrieved 2010-10-18.
{{cite web}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ a b Sanger F, Air GM, Barrell BG, Brown NL, Coulson AR, Fiddes CA, Hutchison CA, Slocombe PM, Smith M (Feb 1977). "Nucleotide sequence of bacteriophage phi X174 DNA". Nature. 265 (5596): 687–95. Bibcode:1977Natur.265..687S. doi:10.1038/265687a0. PMID 870828.
- ^ Kaiser O, Bartels D, Bekel T, Goesmann A, Kespohl S, Pühler A, Meyer F (Dec 2003). "Whole genome shotgun sequencing guided by bioinformatics pipelines--an optimized approach for an established technique" (PDF). Journal of Biotechnology. 106 (2–3): 121–33. doi:10.1016/j.jbiotec.2003.08.008. PMID 14651855.
- ^ Sanger F, Nicklen S, Coulson AR (Dec 1977). "DNA sequencing with chain-terminating inhibitors". Proceedings of the National Academy of Sciences of the United States of America. 74 (12): 5463–7. Bibcode:1977PNAS...74.5463S. doi:10.1073/pnas.74.12.5463. PMC 431765. PMID 271968.
- ^ Maxam AM, Gilbert W (Feb 1977). "A new method for sequencing DNA". Proceedings of the National Academy of Sciences of the United States of America. 74 (2): 560–4. Bibcode:1977PNAS...74..560M. doi:10.1073/pnas.74.2.560. PMC 392330. PMID 265521.
- ^ a b Darden, Lindley; James Tabery (2010). "Molecular Biology". In Zalta, Edward N. (ed.). The Stanford Encyclopedia of Philosophy (Fall 2010 ed.).
{{cite book}}:|access-date=requires|url=(help); External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help); Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG (Apr 1981). "Sequence and organization of the human mitochondrial genome". Nature. 290 (5806): 457–65. Bibcode:1981Natur.290..457A. doi:10.1038/290457a0. PMID 7219534.(subscription required)
- ^ Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M (Sep 1986). "The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression". The EMBO Journal. 5 (9): 2043–2049. PMC 1167080. PMID 16453699.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; Aota, S. I.; Inokuchi, H.; Ozeki, H. (1986). "Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA". Nature. 322 (6079): 572–574. Bibcode:1986Natur.322..572O. doi:10.1038/322572a0.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help); Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Oliver SG, van der Aart QJ, Agostoni-Carbone ML, Aigle M, Alberghina L, Alexandraki D, Antoine G, Anwar R, Ballesta JP, Benit P (May 1992). "The complete DNA sequence of yeast chromosome III". Nature. 357 (6373): 38–46. Bibcode:1992Natur.357...38O. doi:10.1038/357038a0. PMID 1574125.
- ^ Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM (Jul 1995). "Whole-genome random sequencing and assembly of Haemophilus influenzae Rd". Science. 269 (5223): 496–512. Bibcode:1995Sci...269..496F. doi:10.1126/science.7542800. PMID 7542800.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG (Oct 1996). "Life with 6000 genes". Science. 274 (5287): 546, 563–7. Bibcode:1996Sci...274..546G. doi:10.1126/science.274.5287.546. PMID 8849441.(subscription required)
- ^ "Complete genomes: Viruses". NCBI. 2011-11-17. Retrieved 2011-11-18.
- ^ "Genome Project Statistics". Entrez Genome Project. 2011-10-07. Retrieved 2011-11-18.
- ^ Zimmer, Carl (2009-12-29). "Scientists Start a Genomic Catalog of Earth's Abundant Microbes". The New York Times. ISSN 0362-4331. Retrieved 2012-12-21.
{{cite news}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, Hooper SD, Pati A, Lykidis A, Spring S, Anderson IJ, D'haeseleer P, Zemla A, Singer M, Lapidus A, Nolan M, Copeland A, Han C, Chen F, Cheng JF, Lucas S, Kerfeld C, Lang E, Gronow S, Chain P, Bruce D, Rubin EM, Kyrpides NC, Klenk HP, Eisen JA (Dec 2009). "A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea". Nature. 462 (7276): 1056–60. Bibcode:2009Natur.462.1056W. doi:10.1038/nature08656. PMC 3073058. PMID 20033048.
- ^ "Human gene number slashed". BBC. 2004-10-20. Retrieved 2012-12-21.
- ^ Yue GH, Lo LC, Zhu ZY, Lin G, Feng F (Apr 2006). "The complete nucleotide sequence of the mitochondrial genome of Tetraodon nigroviridis". DNA Sequence. 17 (2): 115–21. doi:10.1080/10425170600700378. PMID 17076253.
- ^ National Human Genome Research Institute (2004-07-14). "Dog Genome Assembled: Canine Genome Now Available to Research Community Worldwide". Genome.gov. Retrieved 2012-01-20.
- ^ a b McElheny, Victor (2010). Drawing the map of life : inside the Human Genome Project. New York NY: Basic Books. ISBN 9780465043330.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA (Nov 2012). "An integrated map of genetic variation from 1,092 human genomes". Nature. 491 (7422): 56–65. Bibcode:2012Natur.491...56T. doi:10.1038/nature11632. PMC 3498066. PMID 23128226.
- ^ Nielsen R (Oct 2010). "Genomics: In search of rare human variants". Nature. 467 (7319): 1050–1. Bibcode:2010Natur.467.1050N. doi:10.1038/4671050a. PMID 20981085.
- ^ a b Barnes, Barry; Dupré, John (2008). Genomes and what to make of them. Chicago: University of Chicago Press. ISBN 978-0-226-17295-8.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Eisen JA (2012). "Badomics words and the power and peril of the ome-meme". GigaScience. 1 (1): 6. doi:10.1186/2047-217X-1-6.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Hotz, Robert Lee (2012-08-13). "Here"s an Omical Tale: Scientists Discover Spreading Suffix". Wall Street Journal. ISSN 0099-9660. Retrieved 2013-01-04.
{{cite news}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Scudellari, Megan (2011-10-01). "Data Deluge". The Scientist. Retrieved 2013-01-04.
- ^ Chaston J, Douglas AE (Aug 2012). "Making the most of "omics" for symbiosis research". The Biological Bulletin. 223 (1): 21–9. PMC 3491573. PMID 22983030.
- ^ McCutcheon JP, von Dohlen CD (Aug 2011). "An interdependent metabolic patchwork in the nested symbiosis of mealybugs". Current Biology. 21 (16): 1366–72. doi:10.1016/j.cub.2011.06.051. PMC 3169327. PMID 21835622.
- ^ a b Monya Baker (2012-09-14). "Benchtop sequencers ship off" (Blog). Nature News Blog. Retrieved 2012-12-22.
- ^ Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y (2012). "A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers". BMC Genomics. 13: 341. doi:10.1186/1471-2164-13-341. PMC 3431227. PMID 22827831.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Staden R (Jun 1979). "A strategy of DNA sequencing employing computer programs". Nucleic Acids Research. 6 (7): 2601–10. doi:10.1093/nar/6.7.2601. PMC 327874. PMID 461197.
- ^ Anderson S (Jul 1981). "Shotgun DNA sequencing using cloned DNase I-generated fragments". Nucleic Acids Research. 9 (13): 3015–27. doi:10.1093/nar/9.13.3015. PMC 327328. PMID 6269069.
- ^ a b Pop M (Jul 2009). "Genome assembly reborn: recent computational challenges". Briefings in Bioinformatics. 10 (4): 354–66. doi:10.1093/bib/bbp026. PMC 2691937. PMID 19482960.
- ^ Sanger F, Coulson AR (May 1975). "A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase". Journal of Molecular Biology. 94 (3): 441–8. doi:10.1016/0022-2836(75)90213-2. PMID 1100841.
- ^ Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A, Goodwin L, Woyke T, Lapidus A, Klenk HP, Cottingham RW, Kyrpides NC (2012). Liu Z (ed.). "The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation". PLOS ONE. 7 (12): e48837. Bibcode:2012PLoSO...748837M. doi:10.1371/journal.pone.0048837. PMC 3520994. PMID 23251337.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ Illumina, Inc. (2012-02-28). An Introduction to Next-Generation Sequencing Technology (PDF). San Diego, California, USA: Illumina, Inc. p. 12. Retrieved 2012-12-28.
- ^ Hall N (May 2007). "Advanced sequencing technologies and their wider impact in microbiology". The Journal of Experimental Biology. 210 (Pt 9): 1518–25. doi:10.1242/jeb.001370. PMID 17449817.
- ^ Church GM (Jan 2006). "Genomes for all". Scientific American. 294 (1): 46–54. doi:10.1038/scientificamerican0106-46. PMID 16468433.
- ^ ten Bosch JR, Grody WW (Nov 2008). "Keeping up with the next generation: massively parallel sequencing in clinical diagnostics". The Journal of Molecular Diagnostics. 10 (6): 484–92. doi:10.2353/jmoldx.2008.080027. PMC 2570630. PMID 18832462.
- ^ Tucker T, Marra M, Friedman JM (Aug 2009). "Massively parallel sequencing: the next big thing in genetic medicine". American Journal of Human Genetics. 85 (2): 142–54. doi:10.1016/j.ajhg.2009.06.022. PMC 2725244. PMID 19679224.
- ^ Kawashima, Eric H.; Farinelli, Laurent; Mayer, Pascal (2005-05-12). "Method of nucleic acid amplification". Retrieved 2012-12-22.
{{cite web}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Mardis ER (2008). "Next-generation DNA sequencing methods" (PDF). Annual Review of Genomics and Human Genetics. 9: 387–402. doi:10.1146/annurev.genom.9.081307.164359. PMID 18576944.
- ^ Bio-IT World, Davies, K. Powering Preventative Medicine. Bio-IT World 2011
- ^ Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C, Cole JR, Ding Y, Dugan S, Field D, Garrity GM, Gibbs R, Graves T, Han CS, Harrison SH, Highlander S, Hugenholtz P, Khouri HM, Kodira CD, Kolker E, Kyrpides NC, Lang D, Lapidus A, Malfatti SA, Markowitz V, Metha T, Nelson KE, Parkhill J, Pitluck S, Qin X, Read TD, Schmutz J, Sozhamannan S, Sterk P, Strausberg RL, Sutton G, Thomson NR, Tiedje JM, Weinstock G, Wollam A, Detter JC (Oct 2009). "Genomics. Genome project standards in a new era of sequencing". Science. 326 (5950): 236–7. Bibcode:2009Sci...326..236C. doi:10.1126/science.1180614. PMID 19815760.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Stein L (Jul 2001). "Genome annotation: from sequence to biology". Nature Reviews Genetics. 2 (7): 493–503. doi:10.1038/35080529. PMID 11433356.
- ^ Brent MR (Jan 2008). "Steady progress and recent breakthroughs in the accuracy of automated genome annotation" (PDF). Nature Reviews Genetics. 9 (1): 62–73. doi:10.1038/nrg2220. PMID 18087260.
- ^ Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, García-Girón C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kähäri AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM (Jan 2013). "Ensembl 2013". Nucleic Acids Research. 41 (Database issue): D48-55. doi:10.1093/nar/gks1236. PMC 3531136. PMID 23203987.
{{cite journal}}: Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ Keith, Jonathan M, ed. (2008). "Bioinformatics". Methods in Molecular Biology. 453. doi:10.1007/978-1-60327-429-6. ISBN 978-1-60327-428-9.
{{cite journal}}: Cite journal requires|journal=(help); Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Marsden RL, Lewis TA, Orengo CA (2007). "Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint". BMC Bioinformatics. 8: 86. doi:10.1186/1471-2105-8-86. PMC 1829165. PMID 17349043.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Brenner SE, Levitt M (Jan 2000). "Expectations from structural genomics". Protein Science. 9 (1): 197–200. doi:10.1110/ps.9.1.197. PMC 2144435. PMID 10739263.
- ^ Brenner SE (Oct 2001). "A tour of structural genomics" (PDF). Nature Reviews Genetics. 2 (10): 801–9. doi:10.1038/35093574. PMID 11584296.
- ^ a b Francis, Richard C (2011). Epigenetics : the ultimate mystery of inheritance. New York: W.W. Norton. ISBN 9780393070057.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Laird PW (Mar 2010). "Principles and challenges of genomewide DNA methylation analysis". Nature Reviews Genetics. 11 (3): 191–203. doi:10.1038/nrg2732. PMID 20125086.
- ^ Hugenholtz P, Goebel BM, Pace NR (Sep 1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". Journal of Bacteriology. 180 (18): 4765–74. PMC 107498. PMID 9733676.
- ^ Eisen JA (Mar 2007). "Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes". PLoS Biology. 5 (3): e82. doi:10.1371/journal.pbio.0050082. PMC 1821061. PMID 17355177.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Marco, D, ed. (2010). Metagenomics: Theory, Methods and Applications. Caister Academic Press. ISBN 978-1-904455-54-7.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Marco, D, ed. (2011). Metagenomics: Current Innovations and Future Trends. Caister Academic Press. ISBN 978-1-904455-87-5.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H (Jun 2003). "Prophage genomics". Microbiology and Molecular Biology Reviews. 67 (2): 238–76, table of contents. doi:10.1128/MMBR.67.2.238-276.2003. PMC 156470. PMID 12794192.
- ^ McGrath, S; van Sinderen, D, eds. (2007). Bacteriophage: Genetics and Molecular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-14-1.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Fouts DE (November 2006). "Phage_Finder: automated identification and classification of prophage regions in complete bacterial genome sequences". Nucleic Acids Research. 34 (20): 5839–51. doi:10.1093/nar/gkl732. PMC 1635311. PMID 17062630.
- ^ Herrero, A; Flores, E, eds. (2008). The Cyanobacteria: Molecular Biology, Genomics and Evolution (1st ed.). Caister Academic Press. ISBN 978-1-904455-15-8.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Hudson KL (Sep 2011). "Genomics, health care, and society". The New England Journal of Medicine. 365 (11): 1033–41. doi:10.1056/NEJMra1010517. PMID 21916641.
- ^ O'Donnell CJ, Nabel EG (Dec 2011). "Genomics of cardiovascular disease". The New England Journal of Medicine. 365 (22): 2098–109. doi:10.1056/NEJMra1105239. PMID 22129254.
- ^ Lu YF, Goldstein DB, Angrist M, Cavalleri G (Sep 2014). "Personalized medicine and human genetic diversity". Cold Spring Harbor Perspectives in Medicine. 4 (9): a008581. doi:10.1101/cshperspect.a008581. PMID 25059740.
- ^ Church, George M; Regis, Edward (2012). Regenesis : how synthetic biology will reinvent nature and ourselves. New York: Basic Books. ISBN 9780465021758.
{{cite book}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ Baker M (May 2011). "Synthetic genomes: The next step for the synthetic genome". Nature. 473 (7347): 403, 405–8. Bibcode:2011Natur.473..403B. doi:10.1038/473403a. PMID 21593873.
External links
- Annual Review of Genomics and Human Genetics
- BMC Genomics: A BMC journal on Genomics
- Genomics journal
- Genomics.org: An openfree genomics portal.
- NHGRI: US government's genome institute
- JCVI Comprehensive Microbial Resource
- KoreaGenome.org: The first Korean Genome published and the sequence is available freely.
- GenomicsNetwork: Looks at the development and use of the science and technologies of genomics.
- Institute for Genome Sciences: Genomics research.
- MIT OpenCourseWare HST.512 Genomic Medicine A free, self-study course in genomic medicine. Resources include audio lectures and selected lecture notes.
- ENCODE threads explorer Machine learning approaches to genomics. Nature (journal)
- Global map of genomics laboratories
- Genomics: Scitable by nature education