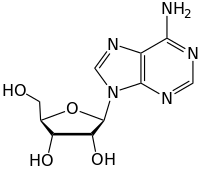
Nucleoside
Nucleosides are glycosylamines that can be thought of as nucleotides without a phosphate group. A nucleoside consists simply of a nucleobase (also termed a nitrogenous base) and a five-carbon sugar (ribose or 2'-deoxyribose) whereas a nucleotide is composed of a nucleobase, a five-carbon sugar, and one or more phosphate groups. In a nucleoside, the anomeric carbon is linked through a glycosidic bond to the N9 of a purine or the N1 of a pyrimidine. Nucleotides are the molecular building blocks of DNA and RNA.
List of nucleosides and corresponding nucleobases
This list does not include modified nucleobases and the corresponding nucleosides
The reason for 2 symbols, shorter and longer, is that the shorter ones are better for contexts where explicit disambiguation is superfluous (because context disambiguates) and the longer ones are for contexts where explicit disambiguation is judged to be needed or wise. For example, when discussing long nucleobase sequences in genomes, the CATG symbol system is much preferable to the Cyt-Ade-Thy-Gua symbol system (see Nucleic acid sequence § Notation for examples), but in discussions where confusion is likelier, the unambiguous symbols can be used.
| Nitrogenous base | Ribonucleoside | Deoxyribonucleoside |
|---|---|---|
 adenine symbol A or Ade |
 adenosine symbol A or Ado |
 deoxyadenosine symbol dA or dAdo |
 guanine symbol G or Gua |
 guanosine symbol G or Guo |
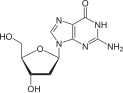 deoxyguanosine symbol dG or dGuo |
 thymine (5-methyluracil) symbol T or Thy |
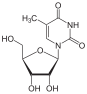 5-methyluridine (ribothymidine) symbol m⁵U |
 thymidine (deoxythymidine) symbol dT or dThd (dated: T or Thd) |
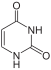 uracil symbol U or Ura |
 uridine symbol U or Urd |
 deoxyuridine symbol dU or dUrd |
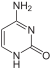 cytosine symbol C or Cyt |
 cytidine symbol C or Cyd |
 deoxycytidine symbol dC or dCyd |
Sources
Nucleosides can be produced from nucleotides de novo, particularly in the liver, but they are more abundantly supplied via ingestion and digestion of nucleic acids in the diet, whereby nucleotidases break down nucleotides (such as the thymidine monophosphate) into nucleosides (such as thymidine) and phosphate. The nucleosides, in turn, are subsequently broken down in the lumen of the digestive system by nucleosidases into nucleobases and ribose or deoxyribose. In addition, nucleotides can be broken down inside the cell into nitrogenous bases, and ribose-1-phosphate or deoxyribose-1-phosphate.
Use in medicine and technology
In medicine several nucleoside analogues are used as antiviral or anticancer agents.[1][2][3][4] The viral polymerase incorporates these compounds with non-canonical bases. These compounds are activated in the cells by being converted into nucleotides. They are administered as nucleosides since charged nucleotides cannot easily cross cell membranes.
In molecular biology, several analogues of the sugar backbone exist. Due to the low stability of RNA, which is prone to hydrolysis, several more stable alternative nucleoside/nucleotide analogues that correctly bind to RNA are used. This is achieved by using a different backbone sugar. These analogues include locked nucleic acids (LNA), morpholinos and peptide nucleic acids (PNA).
In sequencing, dideoxynucleotides are used. These nucleotides possess the non-canonical sugar dideoxyribose, which lacks 3' hydroxyl group (which accepts the phosphate). DNA polymerases cannot distinguish between these and regular deoxyribonucleotides, but when incorporated a dideoxynucleotide cannot bond with the next base and the chain is terminated.
Prebiotic synthesis of ribonucleosides
In order to understand how life arose, knowledge is required of the chemical pathways that permit formation of the key building blocks of life under plausible prebiotic conditions. According to the RNA world hypothesis free-floating ribonucleosides and ribonucleotides were present in the primitive soup. Molecules as complex as RNA must have arisen from small molecules whose reactivity was governed by physico-chemical processes. RNA is composed of purine and pyrimidine nucleotides, both of which are necessary for reliable information transfer, and thus Darwinian natural selection and evolution. Nam et al.[5] demonstrated the direct condensation of nucleobases with ribose to give ribonucleosides in aqueous microdroplets, a key step leading to RNA formation. Also, a plausible prebiotic process for synthesizing pyrimidine and purine ribonucleosides and ribonucleotides using wet-dry cycles was presented by Becker et al.[6]
See also
References
- ^ Ramesh, Deepthi; Vijayakumar, Balaji Gowrivel; Kannan, Tharanikkarasu (December 2020). "Therapeutic potential of uracil and its derivatives in countering pathogenic and physiological disorders". European Journal of Medicinal Chemistry. 207: 112801. doi:10.1016/j.ejmech.2020.112801. PMID 32927231. S2CID 221724578.
- ^ Galmarini, Carlos M.; MacKey, John R.; Dumontet, Charles (2002). "Nucleoside analogues and nucleobases in cancer treatment". The Lancet Oncology. 3 (7): 415–424. doi:10.1016/S1470-2045(02)00788-X. PMID 12142171.
- ^ Jordheim, Lars Petter; Durantel, David; Zoulim, Fabien; Dumontet, Charles (2013). "Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases". Nature Reviews Drug Discovery. 12 (6): 447–464. doi:10.1038/nrd4010. PMID 23722347. S2CID 39842610.
- ^ Ramesh, Deepthi; Vijayakumar, Balaji Gowrivel; Kannan, Tharanikkarasu (12 February 2021). "Advances in Nucleoside and Nucleotide Analogues in Tackling Human Immunodeficiency Virus and Hepatitis Virus Infections". ChemMedChem. 16 (9): 1403–1419. doi:10.1002/cmdc.202000849. PMID 33427377. S2CID 231576801. Archived from the original on 14 December 2021. Retrieved 13 March 2021.
- ^ Nam, Inho; Nam, Hong Gil; Zare, Richard N. (2018-01-02). "Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets". Proceedings of the National Academy of Sciences of the United States of America. 115 (1): 36–40. Bibcode:2018PNAS..115...36N. doi:10.1073/pnas.1718559115. PMC 5776833. PMID 29255025.
- ^ Becker, Sidney; Feldmann, Jonas; Wiedemann, Stefan; Okamura, Hidenori; Schneider, Christina; Iwan, Katharina; Crisp, Antony; Rossa, Martin; Amatov, Tynchtyk; Carell, Thomas (2019-10-04). "Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides" (PDF). Science. 366 (6461): 76–82. Bibcode:2019Sci...366...76B. doi:10.1126/science.aax2747. PMID 31604305. S2CID 203719976. Archived (PDF) from the original on 2022-10-09.
External links
 Media related to Nucleosides at Wikimedia Commons
Media related to Nucleosides at Wikimedia Commons