Genetic history of the Middle East: Difference between revisions
| Line 43: | Line 43: | ||
===DNA studies on modern Egyptians=== |
===DNA studies on modern Egyptians=== |
||
In general, various DNA studies have found that the gene frequencies of North African populations are intermediate between those of The Horn of Africa and Sub Saharan Africa,<ref>[http://books.google.com/books?id=FrwNcwKaUKoC&printsec=frontcover#PPA136,M1 Cavalli-Sforza, History and Geography of Human Genes], The intermediacy of North Africa and to a lesser extent Europe is apparent</ref> though Egypt’s NRY frequency distributions appear to be much more similar to those of the |
In general, various DNA studies have found that the gene frequencies of North African populations are intermediate between those of The Horn of Africa and Sub Saharan Africa,<ref>[http://books.google.com/books?id=FrwNcwKaUKoC&printsec=frontcover#PPA136,M1 Cavalli-Sforza, History and Geography of Human Genes], The intermediacy of North Africa and to a lesser extent Europe is apparent</ref> though Egypt’s NRY frequency distributions appear to be much more similar to those of the Near East than to any sub-Saharan African population, suggesting a much larger Near East genetic component.<ref name="luis">The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations – Luis; Rowold; Regueiro; Caeiro; Cinnioğlu; Roseman; Underhill; Cavalli-Sforza; and Herrera. – see http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1182266</ref><ref name="Cavalli-Sforza">Cavalli-Sforza, L.L., P. Menozzi, and A. Piazza. 1994. The History and Geography of Human Genes. Princeton: Princeton University Press.</ref><ref name="Cavalli-Sforza" /><ref name="Bosch1997">{{cite journal | last1 = Bosch | first1 = E. ''et al.'' | year = 1997 | title = Population history of north Africa: evidence from classical genetic markers | url = | journal = Human Biology | volume = 69 | issue = 3| pages = 295–311 }}</ref><ref>{{cite journal | author=Arredi B, Poloni E, Paracchini S, Zerjal T, Fathallah D, Makrelouf M, Pascali V, Novelletto A, Tyler-Smith C | title=A predominantly Nilo Saharan origin for Y-chromosomal DNA variation in North Africa. | journal=Am J Hum Genet | volume=75 | issue=2 | pages=338–45 | year=2004 | pmid=15202071 | doi = 10.1086/423147 | pmc=1216069}}</ref><ref>{{cite journal | author=Manni F, Leonardi P, Barakat A, Rouba H, Heyer E, Klintschar M, McElreavey K, Quintana-Murci L | title=Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa. | journal=[[Hum Biol]]| volume=74 | issue=5 | pages=645–58 | year=2002 | pmid=12495079 | doi = 10.1353/hub.2002.0054}}</ref><ref>{{cite book|title= The History and Geography of Human Genes|authorlink=Cavalli-Sforza|last=Cavalli-Sforza|chapter=Synthetic maps of Africa|chapterurl=http://books.google.com/books?id=FrwNcwKaUKoC&printsec=frontcover#PPA189,M1|isbn= 0691087504}}The present population of the Sahara is Sudan in the extreme north, with an increase of Negroid component as one goes south</ref> |
||
Luis, Rowold et al. found that the diverse NRY haplotypes observed in a population of mixed [[Arab]]ic speakers and [[Berbers]] found that the majority of haplogroups, about 61% were of [[North African]] origin.<ref name="luis">The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations – Luis; Rowold; Regueiro; Caeiro; Cinnioğlu; Roseman; Underhill; Cavalli-Sforza; and Herrera. – see http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1182266</ref> |
Luis, Rowold et al. found that the diverse NRY haplotypes observed in a population of mixed [[Arab]]ic speakers and [[Berbers]] found that the majority of haplogroups, about 61% were of [[North African]] origin.<ref name="luis">The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations – Luis; Rowold; Regueiro; Caeiro; Cinnioğlu; Roseman; Underhill; Cavalli-Sforza; and Herrera. – see http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1182266</ref> |
||
Revision as of 13:52, 14 March 2012
The archaeogenetics of the Near East involves the study of aDNA or ancient DNA, identifying haplogroups and haplotypes of ancient skeletal remains from both YDNA and mtDNA for populations of the Ancient Near East (the modern Middle East, i.e. Egypt, Arabia, the Levant, Mesopotamia, Anatolia and Persia).
To date, isolation of mtDNA has been most successful.
Developments in DNA sequencing in the 1970s and 1980s provided researchers with the tools needed to study human genetic variation and the genetics of human populations to discover founder populations of modern people groups and human migrations. In 2005, National Geographic launched The Genographic Project, led by 12 prominent scientists and researchers, to study and map historical human migration patterns by collecting and analyzing DNA samples from hundreds of thousands of people from around the world.
Iraq-Mesopotamia
Anthropologist Carleton S. Coon is quoted as saying The Iraqi population is without doubt much the same today as it was in Sumerian, Akkadian, Assyrian and Babylonian times.[1] The Iraqi people are a Caucasian people. It has been found that Y-DNA Haplogroup J2 originated in northern Iraq (Ancient Assyria).[2] In spite of the importance of this region, genetic studies on the Iraqi people are limited and generally restricted to analysis of classical markers due to Iraq's modern political instability,[2] although there have been several published studies displaying the genealogical connection between all Iraqi people and the neighbouring countries, regardless religious and linguistic barriers (Iraq contains Semitic peoples such as Arabs, Assyrians and Mandeans, Turkic peoples such as Turcomans,and Indo-European peoples such as Kurds, Armenians, Shabaks and Circassians). One such study reveals a close genetic relationship between all Iraqis, Kurds, Caspian Iranians and Svani Georgians.[3]
Iraqi mitochondrial DNA (mtDNA) haplogroup distribution is similar to that of Iran, Syria, Israel, Palestine, Georgia, and Armenia, whereas it substantially differs from that observed in the Arabian Peninsula.[2] Iraqi Y-chromosome DNA (Y-DNA) haplogroup distribution is similar to that of Lebanon, Turkey, and Syria.[2] No significant differences in Y-DNA variation were observed among Iraqi Arabs, Assyrians, or Kurds.[2]
For both mtDNA and Y-DNA variation, the large majority of the haplogroups observed in the Iraqi population (H, J, T, and U for the mtDNA, J2 and J1 for the Y-DNA) are those considered to have originated in Western Asia and to have later spread mainly in Western Eurasia.[2] The Eurasian haplogroups R1b and R1a represent the second most frequent component of the Iraqi Y-chromosome gene pool, the latter suggests that the population movements from Central Asia/Eastern Europe into modern Iran also influenced Iraq to some degree.[2]
Many historians and anthropologists provide strong circumstantial evidence to posit that Iraq's Maʻdān people share very strong links to the ancient Sumerians[4][5] - the most ancient inhabitants of southern Iraq,[4] and that Iraq's Mandaeans and Assyrians share the strongest links to the Babylonians.[6] The Arabic speaking Beni Delphi (sons of Delphi) tribe of Iraq is believed to have Greek origins, from the Macedonian soldiers of Alexander the Great and the colonists of the Seleucid Empire.
The Assyrian Christian population are fairly closely related to other Iraqis,[7][4] and also to modern Jordanians and some Near Eastern Jewish populations, yet due to religious and cultural endogamy have a very distinct genetic profile that distinguishes their population.[8] "The Assyrians are a fairly homogeneous group of people, believed to originate from the land of old Assyria in northern Iraq [..] they are Christians and are bona fide descendants of their ancient namesakes."[9] The relatively close genetic link with the indigenous Pre Arab Mesopotamian Assyrians and Mandeans indicates that many Iraqis who today speak Arabic are also to a great extent originally of Mesopotamian roots,as opposed to being ethnic Arabs.[10][11]
In a 2011 study focusing on the genetics of the Maʻdān people of Iraq, researchers identified Y chromosome haplotypes shared by Marsh Arabs, Arabic speaking Iraqis, Assyrians and Mandeans "supporting a common indigenous local background."[4]
Studies have reported that most Irish and Britons are descendants of farmers who left modern day Iraq and Syria 10,000 years ago.[12] Genetic researchers say they have found compelling evidence that four out of five (80% of) white Europeans can trace their roots to the Ancient Near East.[12] In another study, scientists analysed DNA from the 8,000 year-old remains of early farmers found at an ancient graveyard in Germany. They compared the genetic signatures to those of modern populations and found similarities with the DNA of people living in today's South Eastern Turkey and Iraq.[13]
Levant
Zalloua and Wells (2004), under the auspices of a grant from National Geographic Magazine examined the origins of the Phoenicians. The debate between Wells and Zalloua was whether haplogroup J2 (M172) should be identified as that of the Phoenicians or that of its "parent" haplogroup M89 on the YDNA phylogenetic tree.[14] Initial consensus suggested that J2 be identified with the Canaanite-Phoenician (Northwest Semitic) population, with avenues open for future research.[15] As Wells commented, "The Phoenicians were the Canaanites"[16] It was reported in the PBS description of the National Geographic TV Special on this study entitled "Quest for the Phoenicians" that ancient DNA was included in this study as extracted from the tooth of a 2500 year-old Phoenician mummy.[17]
Wells identified the haplogroup of the Canaanites as haplogroup J2.[18] The National Geographic Genographic Project linked haplogroup J2 to the site of Jericho, Tel el-Sultan, ca. 8500 BCE and indicated that in modern populations, haplogroup J2 is found in the Middle East, Southern Europe and North Africa, with especially high distribution among present-day Lebanese, Jewish populations (30%), Southern Italians (20%), and lower frequencies in Southern Spain (10%).[19]
In 2004, a team of geneticists from Stanford University, the Hebrew University of Jerusalem, Tartu University (Estonia), Barzilai Medical Center (Ashkelon, Israel), and the Assaf Harofeh Medical Center (Zerifin, Israel), studied the modern Samaritan community living in Israel in comparison with modern Israeli populations to explore the ancient genetic history of these people groups. The Samaritans or Shomronim (singular: Shomroni; Hebrew: שומרוני) trace their origins to the Assyrian province of Shomron (Samaria) in ancient Israel in the period after the Assyrian conquest circa 722 BCE. Shomron was the capital of the Northern Kingdom of Israel when it was conquered by the Assyrians and gave the name to the ancient province of Samaria and the Samaritan people group. Tradition holds that the Samaritans were a mixed group of Israelites who were not exiled or were sent back or returned from exile and non-Israelites relocated to the region by the Assyrians. The modern-day Samaritans are believed to be the direct descendants of the ancient Samaritans.
Their findings reported on four family lineages among the Samaritans: the Tsdaka family (tradition: tribe of Menasseh), the Joshua-Marhiv and Danfi families (tradition: tribe of Ephraim), and the Cohen family (tradition: tribe of Levi). All Samaritan families were found in haplogroups J1 and J2, except the Cohen family which was found in haplogroup E3b1a-M78.[20] This article predated the E3b1a subclades based on the research of Cruciani, et al.[21] The Samaritan Cohen family were Levites until the previous Cohen family died out around 1700, so the fact that they don't share CMH is expected. These findings may offer more proof that Near Eastern E1b1b was one of the founding lineages of the Levites.
Egypt
Attempts to extract ancient DNA or aDNA from Ancient Egyptian remains have yielded little or no success. Climatic conditions and the mummification process could hasten the deterioration of DNA. Contamination from handling and intrusion from microbes have also created obstacles to recovery of Ancient DNA.[22] Consequently most DNA studies have been carried out on modern Egyptian populations with the intent of learning about the influences of historical migrations on the population of Egypt.[23][24][25][26] However, there was one notable study of ancient mummies of the 12th Dynasty, performed by Paabo and Di Rienzo, which identified multiple lines of descent, including some from sub-Saharan Africa.[27] The other lineages were not identified but Keita (1996) speculates that they may also have been African in origin.[25]
DNA studies on modern Egyptians
In general, various DNA studies have found that the gene frequencies of North African populations are intermediate between those of The Horn of Africa and Sub Saharan Africa,[28] though Egypt’s NRY frequency distributions appear to be much more similar to those of the Near East than to any sub-Saharan African population, suggesting a much larger Near East genetic component.[29][30][30][31][32][33][34]
Luis, Rowold et al. found that the diverse NRY haplotypes observed in a population of mixed Arabic speakers and Berbers found that the majority of haplogroups, about 61% were of North African origin.[29]
A study by Krings et al. from 1999 on mitochondrial DNA clines along the Nile Valley found that a North African cline runs from Northern Egypt to Southern Sudan.[35] Another study based on maternal lineages links modern Egyptians with people from modern Eritrea/Ethiopia such as the Afro-Asiatic-speaking Tigre.[36] Similarly, an mtDNA study of modern Egyptians from the Gurna region near Thebes in Southern Egypt revealed that Eurasian haplogroups represented 61% of the population, with the remainder 39% being of Ethiopic and Mediterranean origin. The oral tradition of the Gurna people indicates that they descend from the ancient Egyptians [37]
A study using the Y-chromosome of modern Egyptian males found similar results, namely that North East African haplogroups are predominant in the South but the predominant haplogroups in the North are characteristic of North African populations.[38]
A study of Copts group in Sudan (as opposed to the ethnically distinct Egyptian Copts) found relatively high frequencies of Sub-Saharan Haplogroup B (Y-DNA). The Sudanese Copts are converts to Egyptian Christianity from the 6th Century AD. According to the study, the presence of Sub-Saharan haplogroups may also consistent with the historical record in which southern Egypt was colonized by Nilotic populations during the early state formation.[39] However, it is generally accepted that Egyptian Copts are not ethnically related to those of Sudan, as conversion of ethnic Nubian kings to Christianity occurred in the 6th century AD[40]. According to tradition, a missionary sent by Byzantine empress Theodora arrived in Nobatia and started preaching the gospel about 540 AD. It is possible that the conversion process began earlier, however, under the aegis of Coptic missionaries from Egypt. Egyptian Copts, in fact, have been shown to have B Haplogroup DNA types and NRY haplotypes.[41]
Other studies have shown that modern Egyptians have genetic affinities primarily with populations of the Near East, North and Northeast Africa,[42][43][44][45] and to a lesser extent Middle Eastern and European populations.[46] Studies done on ancient Egyptians' remains have shown uniformity and homogeneity among the samples, and cranial/limb ratio similarity with populations from North Africa, Somalia, Nubia, Southwest Asia and Europe.[47][48][49][50][51] Blood typing and DNA sampling on ancient Egyptian mummies is scant; however, blood typing of dynastic mummies found ABO frequencies to be most similar to modern Egyptians[52] and some also to Northern Haratin populations. ABO blood group distribution shows that the Egyptians form a sister group to North African populations, including Berbers, Nubians and Canary Islanders.[53]
Some genetic studies done on modern Egyptians suggest that most have close relations to other extant North Africans,[54] and other studies show that they have a distinct North African gene pool, with both the Sahara and Straits of Gibraltar acting as effective barriers to genetic migration[44] with only limited introductions from the Near East, Europe and the Horn of Africa .[45] A 2004 mtDNA study of upper Egyptians from Gurna found a genetic ancestral heritage to modern Northeast Africans, characterized by a high M1 haplotype frequency.[43] Though there has been much debate of the origins of haplogroup M1 a recent 2007 study had concluded that M1 has West Asia origins and not a Sub Saharan African origin[55] Origin A 2003 Y chromosome study was performed by Lucotte on modern Egyptians, with haplotypes V, XI, and IV being most common. Haplotype V is common in Berbers and has a low frequency outside North Africa. Haplotypes V, XI, and IV are all supra-Saharan haplotypes, and they are far more dominant in Egyptians than in Near Eastern or European groups.[56]
Historically there have been differing accounts of the appearance of ancient Egyptians as compared to people of other nations. Egyptologists generally consider the ancient Egyptians to have been a continuum from the lighter northern population of Lower Egypt to the generally darker Upper Egyptians.[57] A number of supporting studies have therefore been undertaken on craniometric patterns and skeletal remains. The results have varied, and interpretation has been complicated by conflict over the baselines to be used in analysing this data.[25][49][58][59][60][61][62][49][63][64][65]
Anatolia-Turkey
A genetic study based on modern male Y-chromosome DNA from Anatolia (also known as Asia Minor) has revealed gene flow from multiple geographic origins, which may correspond to various migrations over time. The predominant male lineages of Anatolian males are shared with European and neighboring Near Eastern populations (94.1%). Lineages related to Central Asia, India, and North Africa were far less prevalent among the males sampled. Y-chromosome haplogroup G-M201 was implied to have a possible association with the Hattians (Hittites).[66]
Research using HLA profiles stemming from Anatolian population groups suggests the possibility that most of the region's people are descendants of varying ethnic groups who lived in Anatolia during the Hittite empire. No genetic signals for intrusions during the Bronze Age collapse at around 1200 BC or Indo-Iranian invasions could be detected. It was concluded that subsequent invasions, if they occurred, had few invaders in comparison to populations already settled by 2000 BC, i.e. Anatolian Hittite and Hurrian (Mitanni) groups, and in the south east, Assyrians. The populations present in Anatolia from 2000 BC to 1200 BC is likely have given rise to the genetic make-up of the present-day Kurdish, Armenian and Turkish populations.[67]
According to Cinnioglu et al., (2004)[68] there are many Y-DNA haplogroups present in Turkey. The majority haplogroups are primarily shared with Middle Eastern, Caucasian, and European populations such as haplogroups E3b, G, J, I, R1a, R1b, K and T which form 78.5% from the Turkish Gene pool (without R1b, K, and which notably occur elsewhere, it is 59.3%) and contrast with a smaller share of haplogroups related to Central Asia (N and Q)- 5.7% (but it rises to 36% if K, R1a, R1b and L- which infrequently occur in Central Asia, but are notable in many other Western Turkic groups), India H, R2 - 1.5% and North Africa A, E3*, E3a - 1%. Some of the percentages identified were:
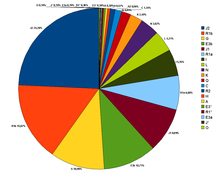
- J1=9% - Typical amongst people from the Arabian Peninsula and Dagestan in the Caucasus (ranging from 3% from Turks around Konya to 12% in Kurds).
- J2=24% - J2 (M172) Typical of populations of the Near East, Southeast Europe, Southwest Asia and the Caucasus, with a moderate distribution through much of Central Asia, South Asia, and North Africa
- R1a=6.9% - Typical of Central Asian, Caucasus, Eastern Europeans and Indo-Aryan people.
- I=5.3% - Typical of Central Europeans, Western Caucasian and Balkan populations.
- R1b=14.7% -Typical of Western Europeans and Eurasian People [69]
- E1b1b1=10.7% - Typical of people from the Mediterranean
- G=10.9% - Typical of people from the Caucasus and to a lesser extent the Middle East.
- N=3.8% - Typical of Uralic, Siberian and Altaic populations.
- T=2.5% - Typical of Mediterranean, North African and South Asian/Indian populations
- K=4.5% - Typical of Near Eastern populations and Caucasian populations.
- L=4.2% - Typical of Indian Subcontinent and Khorasan populations.
- Q=1.9% - Typical of Northern Altaic populations.
See also
- Ancient DNA
- Ancient Near East
- Archaeogenetics
- Ethnic groups of the Middle East
- Genetic history of indigenous peoples of the Americas
- Genetic history of Europe
- Genetic origins of the Turkish people
- Origin of Egyptians
- Origins of the Kurds
- Y-chromosomal Aaron
- Origin of the Nilotic peoples
References
- ^ Carlton S Coon
- ^ a b c d e f g Cite error: The named reference
Zaherywas invoked but never defined (see the help page). - ^ Cite error: The named reference
CSwas invoked but never defined (see the help page). - ^ a b c d Cite error: The named reference
BMCwas invoked but never defined (see the help page). - ^ Spencer, William (2000). Iraq: Old Land, New Nation in Conflict. Twenty-First Century Books. p. 17. ISBN 9780761313564.
But one writer has suggested after a visit to the marshes near the site of ancient Sumer that "some Iraqis still have a touch of the Sumerian in them."
- ^ "Iraq's Marsh Arabs". Retrieved 2010-12-10.
- ^ Cite error: The named reference
Geneticwas invoked but never defined (see the help page). - ^ Dr. Joel J. Elias, Emeritus, University of California, The Genetics of Modern Assyrians and their Relationship to Other People of the Middle East
- ^ Luigi Luca Cavalli-Sforza, Paolo Menozzi, Alberto Piazza, The History and Geography of Human Genes, p. 243
- ^ Kjeilen, Tore. "Assyrians". LookLex Encyclopaedia. Retrieved 16 February 2012.
- ^ Kjeilen, Tore. "Iraq / Peoples". LookLex Encyclopaedia. Retrieved 29 February 2012.
- ^ a b Derbyshire, David (2010-01-20). "Most Britons descended from male farmers who left Iraq and Syria 10,000 years ago". London: Daily Mail. Retrieved 2010-12-10.
- ^ "Migrants from the Near East 'brought farming to Europe'". BBC. 2010-11-10. Retrieved 2010-12-10.
- ^ C:\Documents and Settings\agellon\My Documents\GATC Files\YCC Web\2002_genres\Fig 1.htm
- ^ National Geographic Magazine, October 2004. Available online: http://ngm.nationalgeographic.com/features/world/asia/lebanon/phoenicians-text/1; and http://www.independent.com.mt/news.asp?newsitemid=57215 [accessed: March 10, 2008]
- ^ Who Were the Phoenicians? - National Geographic Magazine
- ^ http://www.pbs.org/previews/phoenicians/ [Accessed April 6, 2008]
- ^ http://ngm.nationalgeographic.com/features/world/asia/lebanon/phoenicians-text/5; and http://www.independent.com.mt/news.asp?newsitemid=57215 [Accessed April 11, 2008]
- ^ The Atlas of the Human Journey-Genetic Markers-Haplogroup J2 (M172): https://www3.nationalgeographic.com/genographic/atlas.html [Accessed April 11, 2008]
- ^ Shen, P (2004). "Reconstruction of Patrilineages and Matrilineages of Samaritans and Other Israeli Populations From Y-Chromosome and Mitochondrial DNA Sequence Variation" (PDF). Human Mutation. 24 (3): 248–260. doi:10.1002/humu.20077. PMID 15300852.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Cruciani, F. (2006). "Molecular Dissection of the Y Chromosome Haplogroup E-M78 (E3b1a): A Posteriori Evaluation of a Microsatellite-Network-Based Approach Through Six New Biallelic Markers" (PDF). Human Mutation. 27: 831–2. doi:10.1002/humu.9445. PMID 16835895.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Encyclopedia of the Archaeology of Ancient Egypt By Kathryn A. Bard, Steven Blake Shubert pp 278-279
- ^ S.O.Y. Keita & A. J. Boyce, S. O. Y.; Boyce, A. J. (Anthony J.) (2005). "Genetics, Egypt, and History: Interpreting Geographical Patterns of Y Chromosome Variation" (PDF). History in Africa. 32: 221. doi:10.1353/hia.2005.0013.
- ^ Shomarka Keita (2005), S. O. Y. (2005). "Y-Chromosome Variation in Egypt" (PDF). African Archaeological Review. 22 (2): 61. doi:10.1007/s10437-005-4189-4. ISBN 1043700541894.
{{cite journal}}: Check|isbn=value: invalid prefix (help)CS1 maint: numeric names: authors list (link) - ^ a b c Keita, S.O.Y. (2005). "History in the Interpretation of the Pattern of p49a,f TaqI RFLP Y-Chromosome Variation in Egypt" (PDF). American Journal of Human Biology. 17 (5): 559–67. doi:10.1002/ajhb.20428. PMID 16136533. Cite error: The named reference "Keita" was defined multiple times with different content (see the help page).
- ^ Shomarka Keita: What genetics can tell us
- ^ Paabo, S., and A. Di Rienzo, A molecular approach to the study of Egyptian history. In Biological Anthropology and the Study of Ancient Egypt. V. Davies and R. Walker, eds. pp. 86-90. London: British Museum Press. 1993
- ^ Cavalli-Sforza, History and Geography of Human Genes, The intermediacy of North Africa and to a lesser extent Europe is apparent
- ^ a b The Levant versus the Horn of Africa: Evidence for Bidirectional Corridors of Human Migrations – Luis; Rowold; Regueiro; Caeiro; Cinnioğlu; Roseman; Underhill; Cavalli-Sforza; and Herrera. – see http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1182266
- ^ a b Cavalli-Sforza, L.L., P. Menozzi, and A. Piazza. 1994. The History and Geography of Human Genes. Princeton: Princeton University Press.
- ^ Bosch, E.; et al. (1997). "Population history of north Africa: evidence from classical genetic markers". Human Biology. 69 (3): 295–311.
{{cite journal}}: Explicit use of et al. in:|first1=(help) - ^ Arredi B, Poloni E, Paracchini S, Zerjal T, Fathallah D, Makrelouf M, Pascali V, Novelletto A, Tyler-Smith C (2004). "A predominantly Nilo Saharan origin for Y-chromosomal DNA variation in North Africa". Am J Hum Genet. 75 (2): 338–45. doi:10.1086/423147. PMC 1216069. PMID 15202071.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Manni F, Leonardi P, Barakat A, Rouba H, Heyer E, Klintschar M, McElreavey K, Quintana-Murci L (2002). "Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa". Hum Biol. 74 (5): 645–58. doi:10.1353/hub.2002.0054. PMID 12495079.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Cavalli-Sforza. "Synthetic maps of Africa". The History and Geography of Human Genes. ISBN 0691087504.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help)The present population of the Sahara is Sudan in the extreme north, with an increase of Negroid component as one goes south - ^ Krings, T; Salem, AE; Bauer, K; Geisert, H; Malek, AK; Chaix, L; Simon, C; Welsby, D; Di Rienzo, A (1992). "mtDNA Analysis of Nile River Valley Populations: Genetic Corridor or a Barrier to Migration?" (PDF). Am J Hum Genet. 64 (4): 1116–76. doi:10.1086/302314. PMC 1377841. PMID 10090902.
- ^ Kivisild T, Reidla M, Metspalu E, Rosa A, Brehm A, Pennarun E, Parik J, Geberhiwot T, Usanga E, Villems R (2004). "Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears". Am J Hum Genet. 75 (5): 752–70. doi:10.1086/425161. PMC 1182106. PMID 15457403.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Stevanovitch, A.; Gilles, A.; Bouzaid, E.; Kefi, R.; Paris, F.; Gayraud, R. P.; Spadoni, J. L.; El-Chenawi, F.; Beraud-Colomb, E. (2004). "Mitochondrial DNA Sequence Diversity in a Sedentary Population from Egypt". Annals of Human Genetics. 68 (Pt 1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
- ^ Lucotte, G.; Mercier, G. (2001). "Brief communication: Y-chromosome haplotypes in Egypt" (PDF). American Journal of Physical Anthropology. 121 (1): 63–6. doi:10.1002/ajpa.10190. PMID 12687584.
- ^ Hassan, Hisham Y.; Underhill, Peter A.; Cavalli-Sforza, Luca L.; Ibrahim, Muntaser E. (2008). "Y-Chromosome Variation Among Sudanese:Restricted Gene Flow, Concordance With Language, Geography, and History" (PDF). American Journal of Physical Anthropology. 137 (3): 316–23. doi:10.1002/ajpa.20876. PMID 18618658.
- ^ http://atheism.about.com/library/FAQs/islam/countries/bl_SudanChristianNubia.htm
- ^ Frank Yurco, "An Egyptological Review" in Mary R. Lefkowitz and Guy MacLean Rogers, eds. Black Athena Revisited. Chapel Hill: University of North Carolina Press, 1996. p. 62-100
- ^ Kivisild T, Reidla M, Metspalu E; et al. (2004). "Ethiopian mitochondrial DNA heritage: tracking gene flow across and around the gate of tears". Am. J. Hum. Genet. 75 (5): 752–70. doi:10.1086/425161. PMC 1182106. PMID 15457403.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ a b Stevanovitch A, Gilles A, Bouzaid E; et al. (2004). "Mitochondrial DNA sequence diversity in a sedentary population from Egypt". Ann. Hum. Genet. 68 (Pt 1): 23–39. doi:10.1046/j.1529-8817.2003.00057.x. PMID 14748828.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) Cite error: The named reference "pmid14748828" was defined multiple times with different content (see the help page). - ^ a b Arredi B, Poloni E, Paracchini S, Zerjal T, Fathallah D, Makrelouf M, Pascali V, Novelletto A, Tyler-Smith C (2004). "A predominantly neolithic origin for Y-chromosomal DNA variation in North Africa". Am J Hum Genet. 75 (2): 338–45. doi:10.1086/423147. PMC 1216069. PMID 15202071.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Manni F, Leonardi P, Barakat A, Rouba H, Heyer E, Klintschar M, McElreavey K, Quintana-Murci L (2002). "Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa". Hum Biol. 74 (5): 645–58. doi:10.1353/hub.2002.0054. PMID 12495079.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Luca Cavalli-Sforza, Luigi (1996-08-05). The History and Geography of Human Genes. Princeton University Press. ISBN 978-0691029054.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Zakrzewski, S.R. (2003). "Variation in ancient Egyptian stature and body proportions". American Journal of Physical Anthropology. 121 (3): 219–229. doi:10.1002/ajpa.10223. PMID 12772210.
- ^ The questionable contribution of the Neolithic and the Bronze Age
- ^ a b c Irish, J.D. (2006). "Who were the ancient Egyptians? Dental affinities among Neolithic through postdynastic peoples" (PDF). Am J Phys Anthropol. 129 (4): 529–543. doi:10.1002/ajpa.20261. PMID 16331657. Cite error: The named reference "Irish2006" was defined multiple times with different content (see the help page).
- ^ Keita, S. (1992). "Further Studies of Crania From Ancient Northern Africa: An Analysis of Crania From First Dynasty Egyptian Tombs, Using Multiple Discriminant Functions". American Journal of Physical Anthropology. 87 (3): 245–54. doi:10.1002/ajpa.1330870302. PMID 1562056.
- ^ Brace CL, Tracer DP, Yaroch LA, Robb J, Brandt K, Nelson AR (1993). Clines and clusters versus "race:" a test in ancient Egypt and the case of a death on the Nile. Yrbk Phys Anthropol 36:1–31.
- ^ Borgognini Tarli S.M., Paoli G. 1982. Survey on paleoserological studies. Homo, 33: 69-89
- ^ Cavalli-Sforza, L.L., P. Menozzi, and A. Piazza. 1994. The History and Geography of Human Genes. Princeton: Princeton University Press, 169-174
- ^ Cavalli-Sforza, L.L., P. Menozzi, and A. Piazza. 1994, The History and Geography of Human Genes. Princeton:Princeton University Press.
- ^ Mitochondrial lineage M1 traces an early human backflow to Africa
- ^ Keita, S.O. (2005). "History in the interpretation of the pattern of p49a, f TaqI RFLP Y-chromosome variation in Egypt: a consideration of multiple lines of evidence" (PDF). Am J Hum Biol. 17 (5): 559–67. doi:10.1002/ajhb.20428. PMID 16136533. Retrieved 2007-10-12.
- ^ Yurco, Frank. "Were the Ancient Egyptians Black or White?" (PDF). BAR magazine. Retrieved 2007-10-03.
- ^ Zakrzewski, S.R. (2007). "Population continuity or population change: Formation of the ancient Egyptian state". American Journal of Physical Anthropology. 132 (4): 501–509. doi:10.1002/ajpa.20569. PMID 17295300.
- ^ Brace et al., 'Clines and clusters versus "race"' (1993)
- ^ S.O.Y. Keita and Rick A. Kittles,' The Persistence of Racial Thinking and the Myth of Racial Divergence', American Anthropologist (Vol. 99, no. 3, 1997), pp. 534-44; pp. 534, 540.
- ^ S.O.Y. Keita. "Early Nile Valley Farmers from El-Badari: Aboriginals or "European" Agro-Nostratic Immigrants? Craniometric Affinities Considered With Other Data". Journal of Black Studies, Vol. 36 No. 2, pp. 191-208 (2005)
- ^ S.O.Y. Keita, American Journal of Physical Anthropology, 87: 245-254 (1992)
- ^ Frank Yurco, "An Egyptological Review" in Mary R. Lefkowitz and Guy MacLean Rogers, eds. Black Athena Revisited. Chapel Hill: University of North Carolina Press, 1996. p. 62-100
- ^ Guns Germs and Steel
- ^ Guns Germs and Steel, pages 101-102
- ^ Cinnioğlu, C.; King, Roy; Kivisild, Toomas; Atasoy, S; Cavalleri, GL; Lillie, AS; Roseman, CC; Lin, AA; et al. (2004). "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics. 114 (2): 127–148. doi:10.1007/s00439-003-1031-4. PMID 14586639.
{{cite journal}}: Explicit use of et al. in:|last4=(help). - ^ http://www3.interscience.wiley.com/journal/118967462/abstract
- ^ a b Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet (2004) 114 : 127–148, Springer-Verlag 2003
- ^ Klyosov A.A. "The principal mystery in the relationship of Indo-European and Türkic linguistic families, and an attempt to solve it with the help of DNA genealogy: reflections of a non-linguist"//Proceedings of Russian Academy of DNA Genealogy (ISSN 1942-7484), Vol. 3, No 1, pp. 3 - 58
Bibliography
- Cruciani, F. (2006). "Molecular Dissection of the Y Chromosome Haplogroup E-M78 (E3b1a): A Posteriori Evaluation of a Microsatellite-Network-Based Approach Through Six New Biallelic Markers" (PDF). Human Mutation. 27: 831–2. doi:10.1002/humu.9445. PMID 16835895.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - King, R. J.; Ozcan, S. S., Carter, T., Kalfoglu, E., Atasoy, S., Triantaphyllidis, C., Kouvatsi, A., Lin, A. A., Chow, C-E. T., Zhivotovsky, L. A., Michalodimitrakis, M., Underhill, P. A., (2008). "Differential Y-chromosome Anatolian Influences on the Greek and Cretan Neolithic." Annals of Human Genetics 72 Issue 2 March 2008: 205-214.
- Shen, P; Lavi, T; Kivisild, T; Chou, V; Sengun, D; Gefel, D; Shpirer, I; Woolf, E; Hillel, J; et al. (2004). "Reconstruction of Patrilineages and Matrilineages of Samaritans and Other Israeli Populations From Y-Chromosome and Mitochondrial DNA Sequence Variation". Human Mutation. 24 (3): 248–260. doi:10.1002/humu.20077. PMID 15300852.
{{cite journal}}: Explicit use of et al. in:|first9=(help) - Zalloua, P., Wells, S. (2004) “Who Were the Phoenicians?” National Geographic Magazine, October 2004.
