Haplogroup F-M89
| Haplogroup F-M89 | |
|---|---|
 | |
| Possible time of origin | 57,500–62,500;(Raghavan 2014);[1] 45,000–55,700 BP (Karafet 2008);[2] 43,000–56,800 BP (Hammer & Zegura 2002).[3] |
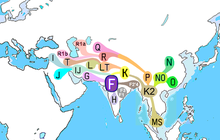
| Possible place of origin | West Asia or South Asia [4][5][6][7] or Southeast Asia[8] |
| Ancestor | CF |
| Descendants | Primary: F1, F2, F3, GHIJK. |
| Defining mutations | M89/PF2746, L132.1, M213/P137/Page38, M235/Page80, P14, P133, P134, P135, P136, P138, P139, P140, P141, P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187, P316 |
Haplogroup F, also known as F-M89 and previously as Haplogroup FT, is a very common Y-chromosome haplogroup. The clade and its subclades constitute over 90% of paternal lineages outside of Africa.
The vast majority of individual males with F-M89 fall into its direct descendant Haplogroup GHIJK (F1329/M3658/PF2622/YSC0001299).[9] in addition to GHIJK, haplogroup F has three other immediate descendant subclades: F1 (P91/P104), F2 (M427/M428), and F3 (M481). These three, with F* (M89*), constitute the paragroup F(xGHIJK). They are primarily found throughout South Asia, Southeast Asia and parts of East Asia.
Haplogroup GHIJK branches subsequently split into two direct descendants: G (M201/PF2957) and HIJK (F929/M578/PF3494/S6397). HIJK in turn splits into H (L901/M2939) and IJK (F-L15). The descendants of the haplogroup IJK include the clades I, J, K, and, ultimately, several major haplogroups descended from Haplogroup K, namely: haplogroups M, N, O, P, Q, R, S, L, and T.
Origins
[edit]It is estimated that the SNP M89 appeared 38,700–55,700 years ago, and most likely originated in West Asia[2][10] or Southeast Asia[8] It has also been suggested by previous research that F-M89 most likely first appeared in the Arabian Peninsula, Levant or North Africa, about 43,800–56,800 years ago.[3] It has also been speculated that the possible location of this lineage's first expansion and rise to prevalence appears to have been in the Indian Subcontinent, or somewhere close to it, and most of the descendant subclades and haplogroups appear to have radiated outward from The Persian Gulf and/or neighbouring parts of the Middle East and Southeast Asia.
Some lineages derived from Haplogroup F-M89 appear to have back-migrated into Africa from West Asia, during prehistory. For example, subclades of F-M89 were discovered in ancient DNA samples from Sudan, which were associated with both Meroitic and Post-Meroitic [1] Archived 2022-08-25 at the Wayback Machine burials.
Distribution
[edit]The vast majority of living individuals carrying F-M89 belong to subclades of GHIJK. By comparison, cases of the paragroup F(xG,H,I,J,K) – that is, either basal F* (M89) or the primary subclades F1 (P91; P104), F2 (M427; M428) and F3 (M481) – are relatively rare worldwide.
F(xG,H,I,J,K)
[edit]A lack of precise, high resolution testing in the past makes it difficult to discuss F*, F1, F2* and F3* separately. ISOGG states that F(xG,H,I,J,K) has not been well studied, occurs "infrequently" in modern populations and peaks in South Asia, especially Sri Lanka.[9] It also appears to have long been present in South East Asia. However, the possibility of misidentification is considered to be relatively high and some cases may in fact belong to misidentified subclades of Haplogroup GHIJK. This was, for instance, the case with the subclade Haplogroup H2 (P96), which was originally named "F3", i.e. – a name that has since been reassigned to F-M481.
F(xF1,F2,F3) has been reported among 10% of males in Sri Lanka, 5.2% of males across India (including up to 10% of males in South India), 5% in Pakistan, as well as lower levels among the Tamang people (Nepal), and in Iran.
Men originating in Indonesia have also been reported to carry F(xG,H,I,J,K) – especially F-M89* – at relatively significant levels. It has been reported at rates of 4-5% in Sulawesi and Lembata. One study, which did not comprehensively screen for other subclades of F-M89 (including some subclades of GHIJK), found that Indonesian men with the SNP P14/PF2704 (which is equivalent to M89), comprise 1.8% of men in West Timor, 1.5% of Flores 5.4% of Lembata 2.3% of Sulawesi and 0.2% in Sumatra.[11][12] F1 (P91), F2 (M427) and F3 (M481; previously F5) are all highly rare and virtually exclusive to regions/ethnic minorities in Sri Lanka, India, Nepal, South China, Thailand, Burma, and Vietnam.
In Central Asia, examples of F(xG,H,I,J,K) have been reported in individuals from Turkmenistan and Uzbekistan.[13]
Kutanan et al. (2020) have found F*-M89 in 50.0% (8/16) of a sample of Red Lahu, 47.1% (8/17) of a sample of Black Lahu, and 6.7% (1/15) of a sample of Lisu in Mae Hong Son Province of Thailand. All these Loloish-speaking members of F*-M89 in northwestern Thailand have been found to be quite closely related in the paternal line, with the TMRCA of their Y-DNA estimated to be 584 years before present. However, the aforementioned Y-chromosomes are only distantly related to instances of F*-M89 observed in samples of other populations of Thailand, including 5.6% (1/18) of a sample of Phuan from Central Thailand, 11.8% (2/17) of a sample of Soa from Northeast Thailand, and 29% (2/7) of a sample of Saek from Northeast Thailand. The TMRCA of the Loloish cluster from North Thailand and the Y-DNA of the Phuan individual from Central Thailand has been estimated to be 12,675 years before present. The TMRCA of the F*-M89 cluster from Northeast Thailand has been estimated to be 6,492 years before present. The TMRCA of all these F*-M89 individuals from Thailand has been estimated to be 16,006 years before present.[14]
There is also evidence of westward Paleolithic back-migration of F(xG,H,I,J,K) from South Asia, to Iran, Arabia and North East Africa,[15][16] as well as subclades of haplogroup K to South-East Europe.[17]
Neolithic migration into Europe from Southwest Asia, by first wave of farmers in Europe has been put forward as the source of F and G2a found in European Neolithic remains, dating from circa 4000 BCE. These remains, according to Herrerra et al. (2012) showed a "greater genetic similarity" to "individuals from the modern Near East" than to modern Europeans.[18] F(xG,H,I,J,K) may have been found in Bronze Age remains from Europe, namely the individuals known as DEB 20 and DEB 38, who lived about 7,000–7,210 BP, and were found at the Derenburg Meerenstieg II site in Germany.[19]
Three less certain cases, which have not been tested for all subclades of GHIJK, have been found among Neolithic remains in Europe. I0411 (Troc 4), who lived 7,195–7,080 years BP, was found in the Els Trocs cave, near Bisaurri (modern Spain) – while haplogroups G, I1 (I-M450; I-S247) J, L1b2, Q1b1, Q1a2a, R1a1a1 (R-L449), R1b1a2b1a (R-M35) and T were ruled out,[19] I2a1b1 and R1b1a2 were found in other remains from the same site (Troc 5 and Troc 2). Similarly, three sets of remains from Hungary were not tested for all subclades of GHIJK: BAM 17, BAM 26 (both from Alsónyék Bátaszék, circa 7,850–7,675 years BP) and TOLM 3 (7,030–7,230 BP, found in Tolna-Mözs).[19] (An individual known to scholars as "Oase 1", who lived circa 37,800 years BP in Eastern Europe, was initially classified as belonging either to paragroup F(xGHIJK) or within K.[17] However, subsequent research has revealed that Oase 1 belonged to K2a*.[20])
- found higher in Korea, Philippines, and Indonesia
Some cases reported amongst modern populations of Europeans, Native Americans and Pacific Islanders may be due to migration and admixture of F(xG,H,I,J,K), as a result of contact with South and/or South East Asia, during the early modern era (16th–19th Century).[9] Such examples include:
- low levels in Polynesia;[21]
- higher among Seminole and Boruca Native Americans;[22]
- rare cases in the Netherlands;[2]
- two cases in Portugal.[9]
F* (M89*)
[edit]Basal F-M89* has been reported among 5.2% of males in India.[10] A regional breakdown was provided by Chiaroni et al. (2009): 10% in South India; 8% in Central India; about 1.0% in North India and Western India, as well as 5% in Pakistan; 10% in Sri Lanka; 4% among the Tamang people of Nepal; 2% in Borneo and Java; 4-5% in Sulawesi and Lembata in Southeast Asia.[11]
In Iran, 2.3% of Bandari males from Hormozgan Province have been found to carry basal F-M89.[15]
Haplogorup F* has been found in only 11.67% of Yunnan Han Chinese tested.[23] Xi'an (1/34),[23]
Haplogroup F-M89 has also been observed in Northeast Africa among two Christian period individuals, who were excavated on the Nile's Fourth Cataract and on Meroe Island.[24] The remains of the Bacho-Kiro cave prehistoric individual F6-620 / AA7-738, from Initial Upper Paleolithic, dated to between 45,930 and 42,580 calibrated years before present, carry also the basal lineage of the Y chromosome haplogroup F-M89.[25]
F1 (P91)
[edit]This subclade is defined by the SNP P91. It is most common in Sri Lanka.[2][26]
F2 (M427)
[edit]F2 Y-chromosomes have been reported among minorities from the borderlands of South China (Yunnan and Guizhou), Thailand, Burma, and Vietnam, namely the Yi and Kucong or Lahu Shi ("Yellow Lahu"), a subgroup of the Lahu.[27]
F3 (M481)
[edit]The newly defined and rare subclade F3 (M481; previously F5) has been found in India and Nepal, among the Tharu people and in Andhra Pradesh.[28] F-M481 should not be confused with Haplogroup H2 (L279, L281, L284, L285, L286, M282, P96), which was previously misclassified under F-M89, as "F3".
Haplogroup GHIJK
[edit]Basal GHIJK has never been found, either in living males or ancient remains.
Subclades – including some major haplogroups – are widespread in modern populations of the Central Asia, Caucasus, Middle East, South Asia, Europe, South East Asia, Pacific Islands and Native Americans.
Phylogenetics
[edit]In Y-chromosome phylogenetics, subclades are the branches of haplogroups. These subclades are also defined by single nucleotide polymorphisms (SNPs) or unique event polymorphisms (UEPs).
Phylogenetic trees
[edit]There are several confirmed and proposed phylogenetic trees available for haplogroup F-M89. The scientifically accepted one is the Y-Chromosome Consortium (YCC) one published in Karafet 2008[2] and subsequently updated. A draft tree that shows emerging science is provided by Thomas Krahn at the Genomic Research Center in Houston, Texas. The International Society of Genetic Genealogy (ISOGG) also provides an amateur tree.
The Genomic Research Center draft tree
[edit]The Genomic Research Center's draft tree for haplogroup F-M89 is as follows.[29] (Only the first three levels of subclades are shown.)
- F-M89 P14, M89, M213, P133, P134, P135, P136, P138, P139, P140, P141, P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187, P316, L132.1, L313, L498
- F-P91 P91, P104
- F-M427 M427, M428
- F-P96 P96, M282, L279, L281, L284, L285, L286
- F-L280 L280
- G-M201 M201, P257, L116, L154, L204, L240, L269, L402, L605, L769, L770, L836, L837, L1258, U2, U3, U6, U7, U12, U17, U20, U21, U23, U33
- H-M69 M69, M370, PAGES00049
- IJK L15, L16
YCC/ISOGG tree
[edit]This is the official scientific tree produced by the Y-Chromosome Consortium (YCC). The last major update was in 2008.[2] Subsequent updates have been quarterly and biannual. The current version is a revision of the 2010 update.[citation needed]
- CF
- F (L132.1, M89/PF2746).
- F1 (P91, P104)
- F2 (M427, M428)
- F3 (M481)
- Macrohaplogroup GHIJK (F1329/M3658/PF2622/YSC0001299).
- F (L132.1, M89/PF2746).
Phylogenetic history
[edit]Prior to 2002, there were in academic literature at least seven naming systems for the Y-Chromosome Phylogenetic tree. This led to considerable confusion. In 2002, the major research groups came together and formed the Y-Chromosome Consortium (YCC). They published a joint paper that created a single new tree that all agreed to use. Later, a group of citizen scientists with an interest in population genetics and genetic genealogy formed a working group to create an amateur tree aiming at being above all timely. The table below brings together all of these works at the point of the landmark 2002 YCC Tree. This allows a researcher reviewing older published literature to quickly move between nomenclatures.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F-M89 | 2 | VI | 1R | 20 | Eu10 | H4 | B | F* | F | F | F | F | F | F | F | F | F | F |
| F-APT/H-APT | 15 | VI | 1R | 20 | Eu10 | H4 | B | F1 | H2 | H2 | H2 | H2 | H2 | H2 | H2 | H2 | H2 | H2 |
See also
[edit]Genetics
[edit]- Genetics and archaeogenetics of South Asia
- Genetic history of the Middle East
- Conversion table for Y chromosome haplogroups
- Genetic Genealogy
- Haplogroup
- Haplotype
- Human Y-chromosome DNA haplogroup
- Molecular phylogenetics
- Paragroup
- Subclade
- Y-chromosomal Aaron
- Y-chromosome haplogroups in populations of the world
- Y-DNA haplogroups by ethnic group
- Y-DNA haplogroups in populations of South Asia
Backbone Tree
[edit]References
[edit]- ^ Estimated time that F split from C = 70,000–75,000 BP; estimated time when G split from HIJK = 45,000-50,000 Raghavan, M.; et al. (2014). "Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans". Nature. 505 (7481): 87–91. Bibcode:2014Natur.505...87R. doi:10.1038/nature12736. PMC 4105016. PMID 24256729.
- ^ a b c d e f Karafet, Tatiana; et al. (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research. 18 (5): 830–8. doi:10.1101/gr.7172008. PMC 2336805. PMID 18385274.
- ^ a b Hammer, M.F.; Zegura, S.L. (2002). "The human Y chromosome haplogroup tree: Nomenclature and phylogeography of its major divisions". Annual Review of Anthropology. 31: 303–321. doi:10.1146/annurev.anthro.31.040402.085413.
- ^ Kivisild et al 2003, The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations
- ^ Sengupta et al 2005, Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists
- ^ Sanghamitra Sahoo et al 2006, A prehistory of Indian Y chromosomes: Evaluating demic diffusion scenarios
- ^ Arunkumar et al 2012
- ^ a b Hallast, Pille; Agdzhoyan, Anastasia; Balanovsky, Oleg; Xue, Yali; Tyler-Smith, Chris (2020-07-14). "A Southeast Asian origin for present-day non-African human Y chromosomes". Human Genetics. 140 (2): 299–307. doi:10.1007/s00439-020-02204-9. ISSN 1432-1203. PMC 7864842. PMID 32666166.
- ^ a b c d ISOGG, 2015, Y-DNA Haplogroup F and its Subclades - 2015 (8 September 2015).
- ^ a b Sengupta, Sanghamitra; et al. (2006). "Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists". The American Journal of Human Genetics. 78 (2): 202–21. doi:10.1086/499411. PMC 1380230. PMID 16400607.
- ^ a b Chiaroni, Jacques; Underhill, Peter A.; Cavalli-Sforza, Luca L. (1 December 2009). "Y chromosome diversity, human expansion, drift, and cultural evolution". Proceedings of the National Academy of Sciences of the United States of America. 106 (48): 20174–9. Bibcode:2009PNAS..10620174C. doi:10.1073/pnas.0910803106. PMC 2787129. PMID 19920170.
- ^ Tumonggor, MK; Karafet, TM; Downey, S; Lansing, JS; Norquest, P; Sudoyo, H; Hammer, MF; Cox, MP (2014). "Isolation, contact and social behavior shaped genetic diversity in West Timor". Journal of Human Genetics. 59 (9): 494–503. doi:10.1038/jhg.2014.62. PMC 4521296. PMID 25078354.
- ^ Balaresque et al. (2015), Y-chromosome descent clusters and male differential reproductive success: young lineage expansions dominate Asian pastoral nomadic populations, Supplementary Table 2
- ^ Wibhu Kutanan, Rasmi Shoocongdej, Metawee Srikummool, et al. (2020), "Cultural variation impacts paternal and maternal genetic lineages of the Hmong-Mien and Sino-Tibetan groups from Thailand." European Journal of Human Genetics. https://doi.org/10.1038/s41431-020-0693-x
- ^ a b Grugni, V; Battaglia, V; Hooshiar Kashani, B; et al. (2012). "Ancient migratory events in the Middle East: new clues from the Y-chromosome variation of modern Iranians". PLOS ONE. 7 (7): e41252. Bibcode:2012PLoSO...741252G. doi:10.1371/journal.pone.0041252. PMC 3399854. PMID 22815981.
- ^ Yousif, Hisham; Eltayeb, Muntaser. "Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan" (PDF). University of Khartoum. Retrieved 17 July 2016.
- ^ a b Qiaomei Fu; Mateja Hajdinjak; Oana Teodora Moldovan; Silviu Constantin; Swapan Mallick; Pontus Skoglund; Nick Patterson; Nadin Rohland; Iosif Lazaridis; Birgit Nickel; Bence Viola; Kay Prüfer; Matthias Meyer; Janet Kelso; David Reich; Svante Pääbo (13 August 2015). "An early modern human from Romania with a recent Neanderthal ancestor". Nature. 524 (7564): 216–219. Bibcode:2015Natur.524..216F. doi:10.1038/nature14558. PMC 4537386. PMID 26098372.
- ^ Herrera, KJ; Lowery, RK; Hadden, L; et al. (March 2012). "Neolithic patrilineal signals indicate that the Armenian plateau was repopulated by agriculturalists". Eur. J. Hum. Genet. 20 (3): 313–20. doi:10.1038/ejhg.2011.192. PMC 3286660. PMID 22085901.
- ^ a b c Jean Manco, 2016, DNA from the European Neolithic (1 March 2016).
- ^ G. David Poznik et al., 2016, "Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences" Nature Genetics, no. 48, pp. 593–599.
- ^ "Melanesian and Asian Origins of Polynesians: mtDNA and Y Chromosome Gradients Across the Pacific". Archived from the original on 2011-05-13.
- ^ Phillip Edward Melton, 2008, Genetic History and Pre-Columbian Diaspora of Chibchan Speaking Populations: Molecular Genetic Evidence; Ann Arbor, Michigan; ProQuest, p. 29.
- ^ a b Soon-Hee Kim 2011, High frequencies of Y-chromosome haplogroup O2b-SRY465 lineages in Korea: a genetic perspective on the peopling of Korea
- ^ Mohamed, Hisham Yousif Hassan. "Genetic Patterns of Y-chromosome and Mitochondrial DNA Variation, with Implications to the Peopling of the Sudan". University of Khartoum. p. 76. Archived from the original (PDF) on 10 November 2020. Retrieved 22 August 2016.
- ^ Hajdinjak, M., Mafessoni, F., Skov, L. et al. "Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry." Nature 592, 253–257 (2021). https://doi.org/10.1038/s41586-021-03335-3
- ^ "PhyloTree y - Minimal y tree".
- ^ Black, M.L.; Wise, C.A.; Wang, W.; Bittles, A.H. (June 2006). "Combining Genetics and Population History in the Study of Ethnic Diversity in the People's Republic of China". Human Biology. 78 (3): 277–293. doi:10.1353/hub.2006.0041. PMID 17216801. S2CID 42002729.
- ^ Fornarino, S; Pala, M; Battaglia, V; et al. (2009). "Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation". BMC Evol. Biol. 9: 154. doi:10.1186/1471-2148-9-154. PMC 2720951. PMID 19573232.
- ^ "Archived copy". Archived from the original on 2013-05-26. Retrieved 2013-01-06.
{{cite web}}: CS1 maint: archived copy as title (link)
External links
[edit]- "GENO 2.0: The Greated Journey Ever Told". National Geographic. Archived from the original on 2011-07-26.
- "Y-DNA Haplogroup Tree 2013, Version: 8.72". International Society of Genetic Genealogy. 25 August 2013.
- McDonald, J. D. (2005). "World Haplogroups Maps" (PDF). Archived from the original (PDF) on 2004-07-28. Retrieved 2007-02-06.
