Haplogroup I-M253
This article needs attention from an expert in Human Genetic History. The specific problem is: Nomenclature of haplogroup(s) and subclades. |
| Haplogroup I-M253 | |
|---|---|
 | |
| Possible time of origin | 11-21,000[1] or 28-33,000 years ago[2] |
| Possible place of origin | Europe |
| Ancestor | I-M170 |
| Defining mutations | M253, M307, P30, P40 |
In human genetics, Haplogroup I-M253 is a Y chromosome haplogroup which occurs at greatest frequency in Fenno-Scandia. The mutations identified with Haplogroup I-M253 (Y-DNA) are M253, M307, P30, and P40. These are known as single nucleotide polymorphisms (SNPs). It is a subclade of Haplogroup I. Before a reclassification in 2008,[3] the group was known as Haplogroup I1a.
The group displays a very clear frequency gradient, with a peak of approximately 40 percent among the populations of western Finland and more than 50 percent in the province of Satakunta,[4] and around 38 percent in Sweden as a whole, with a peak of 52 percent in Västra Götaland County in central Sweden.[5]
Origins
Haplogroup I-M253 arose from haplogroup I-M170, which appears ancient in Europe. The haplogroup was previously[year needed] thought to have originated 15,000 years ago in Iberia, but as of 2010 it was estimated to have originated between 4,000 - 5,000 years ago, in Chalcolithic Europe.[6] A new study in 2015 estimated the origin as between 3,470 to 5,070 years ago or between 3,180 to 3,760 years ago, using two different techniques.[7] It is suggested that it initially dispersed from the area that is now Denmark.[8] A 2014 study in Hungary uncovered remains of nine individuals from the Linear Pottery culture, one of whom was found to have carried the M253 SNP which defines Haplogroup I1. This culture is thought to have been present between 6,500 to 7,500 years ago.[9]
Subclades
Note: The systematic subclade names have changed several times in recent years,[year needed] and are likely to change again, as new markers which clarify the sequence of branchings of the tree are discovered.[10]
- I-M253 (M253,[11] M307.2/P203.2,[12] L64, L75, L80, L81, L118, L121/S62, L123, L124/S64, L125/S65, L157.1, L186, L187, L840, M450/S109, P30, P40, S63, S66, S107, S108, S110, S111.[10]
- I-DF29 (DF29/S438)
- I-CTS6364 (CTS6364/Z2336)
- I-L22 (L22/S142)
- I-P109 (P109)
- I-L205 (L205)
- I-L300 (L300/S241)
- I-Z74 (Z74)
- I-L287 (L287)
- I-L258 (L258/S335)
- I-L813 (L813)
- I-L287 (L287)
- I-M227 (M227)
- I-L22 (L22/S142)
- I-PF49 (PF49)
- I-Z58 (S244/Z58)
- I-Z59 (S246/Z59)
- I-Z60(S337/Z60, S439/Z61, Z62)
- I-Z140 (Z140, Z141)
- I-L338 (L338)
- I-F2642 (F2642)
- I-Z73 (Z73)
- I-L1302 (L1302)
- I-L573 (L573)
- I-L803 (L803)
- I-Z140 (Z140, Z141)
- I-Z382 (Z382)
- I-Z60(S337/Z60, S439/Z61, Z62)
- I-Z138 (S296/Z138, Z139)
- I-Z2541 (Z2541)
- I-Z59 (S246/Z59)
- I-Z63 (S243/Z63)
- I-BY151 (BY151)
- I-BY351 (BY351)
- I-CTS10345 (CTS10345)
- I-Y10994 (Y10994)
- I-Y7075 (Y7075)
- I-CTS10345 (CTS10345)
- I-S2078 (S2078)
- I-S2077 (S2077)
- I-Y2245 (Y2245/PR683)
- I-L1237 (L1237)
- I-FGC9550 (FGC9550)
- I-S10360 (S10360)
- I-S15301 (S15301)
- I-Y7234 (Y7234)
- I-L1237 (L1237)
- I-Y2245 (Y2245/PR683)
- I-S2077 (S2077)
- I-Y8331 (Y8331/BY62)
- I-BY351 (BY351)
- I-Y13952 (Y13952)
- I-BY151 (BY151)
- I-CTS6364 (CTS6364/Z2336)
- I-Z131 (Z131)
- I-DF29 (DF29/S438)
Distribution by population
I-M253 is found at its highest density in Northern Europe and other countries that experienced extensive migration from Northern Europe, either in the Migration Period, the Viking period or modern times. It is found in all places invaded by the ancient Germanic peoples and the Vikings. In the modern era, significant I-M253 populations have also taken root in immigrant nations and former European colonies such as United States, Australia and Canada.
| Population | Sample size | I | I-M253 (M253) | I-M227 (M227) | Source |
|---|---|---|---|---|---|
| Austria | 43 | 9.3 | 2.3 | 0 | Underhill et al. 2007 |
| Belarusians: Vitbsk | 100 | 15 | 1 | 0 | Underhill et al. 2007 |
| Belarusians: Brest | 97 | 20.6 | 1 | 0 | Underhill et al. 2007 |
| Bosnia | 100 | 42 | 2 | 0 | Rootsi et al. 2004 |
| Bulgaria | 808 | 26.6 | 4.3 | 0 | Karachanak et al. 2013 |
| Czech Republic | 47 | 31.9 | 8.5 | 0 | Underhill et al. 2007 |
| Czech | 53 | 17.0 | 1.9 | 0 | Rootsi et al. 2004 |
| Denmark | 122 | 39.3 | 32.8 | 0 | Underhill et al. 2007 |
| England | 104 | 19.2 | 15.4 | 0 | Underhill et al. 2007 |
| Estonia | 210 | 18.6 | 14.8 | 0.5 | Rootsi et al. 2004 |
| Estonia | 118 | - | 11.9 | - | Lappalainen et al. 2008 |
| Finland (west) | 230 | - | 40 | - | Lappalainen et al. 2008 |
| Finland (east) | 306 | - | 19 | - | Lappalainen et al. 2008 |
| France | 58 | 17.2 | 8.6 | 1.7 | Underhill et al. 2007 |
| France | 12 | 16.7 | 16.7 | 0 | Cann et al. 2002 |
| France (Low Normandy) | 42 | 21.4 | 11.9 | 0 | Rootsi et al. 2004 |
| Germany | 125 | 24 | 15.2 | 0 | Underhill et al. 2007 |
| Greece | 171 | 15.8 | 2.3 | 0 | Underhill et al. 2007 |
| Hungary | 113 | 25.7 | 13.3 | 0 | Rootsi et al. 2004 |
| Ireland | 100 | 11 | 6 | 0 | Underhill et al. 2007 |
| Latvia | 113 | - | 3.5 | - | Lappalainen et al. 2008 |
| Lithuania | 164 | - | 4.9 | - | Lappalainen et al. 2008 |
| Netherlands | 93 | 20.4 | 14 | 0 | Underhill et al. 2007 |
| Russians | 16 | 25 | 12.5 | 0 | Cann et al. 2002 |
| Russia: Pskov | 130 | 16.9 | 5.4 | 0 | Underhill et al. 2007 |
| Russia: Kostroma | 53 | 26.4 | 11.3 | 0 | Underhill et al. 2007 |
| Russia: Smolensk | 103 | 12.6 | 1.9 | 0 | Underhill et al. 2007 |
| Russia: Voronez | 96 | 19.8 | 3.1 | 0 | Underhill et al. 2007 |
| Russia: Arkhangelsk | 145 | 15.8 | 7.6 | 0 | Underhill et al. 2007 |
| Russia: Cossac | 89 | 24.7 | 4.5 | 0 | Underhill et al. 2007 |
| Russia: Karelians | 140 | 10 | 8.6 | 0 | Underhill et al. 2007 |
| Russia: Karelians | 132 | - | 15.2 | - | Lappalainen et al. 2008 |
| Russia: Vepsa | 39 | 5.1 | 2.6 | 0 | Underhill et al. 2007 |
| Slovakia | 70 | 14.3 | 4.3 | 0 | Rootsi et al. 2004 |
| Slovenia | 95 | 26.3 | 7.4 | 0 | Underhill et al. 2007 |
| Sweden | 160 | - | 35.6 | - | Lappalainen et al. 2008 |
| Swiss | 144 | 7.6 | 5.6 | 0 | Rootsi et al. 2004 |
| Turkey | 523 | 5.4 | 1.1 | 0 | Underhill et al. 2007 |
| Ukrainians: Lvov | 101 | 23.8 | 4.9 | 0 | Underhill et al. 2007 |
| Ukrainians: Ivanovo-Frankov | 56 | 21.4 | 1.8 | 0 | Underhill et al. 2007 |
| Ukrainians: Hmelnitz | 176 | 26.2 | 6.1 | 0 | Underhill et al. 2007 |
| Ukrainians: Cherkasso | 114 | 28.1 | 4.3 | 0 | Underhill et al. 2007 |
| Ukrainians: Belgorod | 56 | 26.8 | 5.3 | 0 | Underhill et al. 2007 |
Publicly accessible databases
There are several public access databases where I-M253 populations can be found:
- http://www.eupedia.com/europe/european_y-dna_haplogroups.shtml
- http://www.semargl.me/
- http://www.ysearch.org/
- http://www.yhrd.org/
- http://www.yfull.com/tree/I1/
Britain
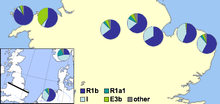
In 2002 a paper was published by Michael E. Weale and colleagues showing genetic evidence for population differences between the English and Welsh populations, including a markedly higher level of Y-DNA haplogroup I in England than in Wales. They saw this as convincing evidence of Anglo-Saxon mass invasion of eastern Great Britain from northern Germany and Denmark during the Migration Period.[13] The authors assumed that populations with large proportions of haplogroup I originated from northern Germany or southern Scandinavia, particularly Denmark, and that their ancestors had migrated across the North Sea with Anglo-Saxon migrations and Danish Vikings. The main claim by the researchers was
that an Anglo-Saxon immigration event affecting 50–100% of the Central English male gene pool at that time is required. We note, however, that our data do not allow us to distinguish an event that simply added to the indigenous Central English male gene pool from one where indigenous males were displaced elsewhere or one where indigenous males were reduced in number … This study shows that the Welsh border was more of a genetic barrier to Anglo-Saxon Y chromosome gene flow than the North Sea … These results indicate that a political boundary can be more important than a geophysical one in population genetic structuring.

In 2003 a paper was published by Christian Capelli and colleagues which supported, but modified, the conclusions of Weale and colleagues.[14] This paper, which sampled Great Britain and Ireland on a grid, found a smaller difference between Welsh and English samples, with a gradual decrease in Haplogroup I frequency moving westwards in southern Great Britain. The results suggested to the authors that Norwegian Vikings invaders had heavily influenced the northern area of the British Isles, but that both English and mainland Scottish samples all have German/Danish influence.
Famous figures
Alexander Hamilton, through genealogy and the testing of his descendants (assuming actual paternity matching his genealogy), has been placed within Y-DNA haplogroup I-M253.[15]
Mutations

The following are the technical specifications for known I-M253 haplogroup SNP and STR mutations.
Name: M253[16]
- Type: SNP
- Source: M (Peter Underhill, Ph.D. of Stanford University)
- Position: ChrY:13532101..13532101 (+ strand)
- Position (base pair): 283
- Total size (base pairs): 400
- Length: 1
- ISOGG HG: I1
- Primer F (Forward 5′→ 3′): GCAACAATGAGGGTTTTTTTG
- Primer R (Reverse 5′→ 3′): CAGCTCCACCTCTATGCAGTTT
- YCC HG: I1
- Nucleotide alleles change (mutation): C to T
Name: M307[17]
- Type: SNP
- Source: M (Peter Underhill, Ph.D. of Stanford University)
- Position: ChrY:21160339..21160339 (+ strand)
- Length: 1
- ISOGG HG: I1
- Primer F: TTATTGGCATTTCAGGAAGTG
- Primer R: GGGTGAGGCAGGAAAATAGC
- YCC HG: I1
- Nucleotide alleles change (mutation): G to A
Name: P30[18]
- Type: SNP
- Source: PS (Michael Hammer, Ph.D. of the University of Arizona and James F. Wilson, D.Phil. at the University of Edinburgh)
- Position: ChrY:13006761..13006761 (+ strand)
- Length: 1
- ISOGG HG: I1
- Primer F: GGTGGGCTGTTTGAAAAAGA
- Primer R: AGCCAAATACCAGTCGTCAC
- YCC HG: I1
- Nucleotide alleles change (mutation): G to A
- Region: ARSDP
Name: P40[19]
- Type: SNP
- Source: PS (Michael Hammer, Ph.D. of the University of Arizona and James F. Wilson, D.Phil. at the University of Edinburgh)
- Position: ChrY:12994402..12994402 (+ strand)
- Length: 1
- ISOGG HG: I1
- Primer F: GGAGAAAAGGTGAGAAACC
- Primer R: GGACAAGGGGCAGATT
- YCC HG: I1
- Nucleotide alleles change (mutation): C to T
- Region: ARSDP
References
- ^ Rootsi, Siiri; et al. (2004). "Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe" (PDF). American Journal of Human Genetics. 75: 128–137. doi:10.1086/422196. PMC 1181996. PMID 15162323.
- ^ P.A. Underhill, N.M. Myres, S. Rootsi, C.T. Chow, A.A. Lin, R.P. Otillar, R. King, L.A. Zhivotovsky, O. Balanovsky, A. Pshenichnov, K.H. Ritchie, L.L. Cavalli-Sforza, T. Kivisild, R. Villems, S.R. Woodward, New Phylogenetic Relationships for Y-chromosome Haplogroup I: Reappraising its Phylogeography and Prehistory, in P. Mellars, K. Boyle, O. Bar-Yosef and C. Stringer (eds.), Rethinking the Human Evolution (2007), pp. pp. 33-42.
- ^ Karafet, Tatiana M.; Mendez, F. L.; Meilerman, M. B.; Underhill, P. A.; Zegura, S. L.; Hammer, M. F. (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research. 18 (5): 830–8. doi:10.1101/gr.7172008. PMC 2336805. PMID 18385274.
- ^ Lappalainen, T.; Laitinen, V.; Salmela, E.; Andersen, P.; Huoponen, K.; Savontaus, M.-L.; Lahermo, P. (2008). "Migration Waves to the Baltic Sea Region". Annals of Human Genetics. 72 (3): 337–348. doi:10.1111/j.1469-1809.2007.00429.x. PMID 18294359.
- ^ Lappalainen, T.; Hannelius, U.; Salmela, E.; von Döbeln, U.; Lindgren, C. M.; Huoponen, K.; Savontaus, M.-L.; Kere, J.; Lahermo, P. (2009). "Population Structure in Contemporary Sweden: A Y-Chromosomal and Mitochondrial DNA Analysis". Annals of Human Genetics. 73 (1): 61–73. doi:10.1111/j.1469-1809.2008.00487.x. PMID 19040656.
- ^ Pedro Soares, Alessandro Achilli, Ornella Semino, William Davies, Vincent Macaulay, Hans-Jürgen Bandelt, Antonio Torroni, and Martin B. Richards, The Archaeogenetics of Europe, Current Biology, vol. 20 (February 23, 2010), R174–R183. yDNA Haplogroup I: Subclade I1, Family Tree DNA,
- ^ "TMRCAs of major haplogroups in Europe estimated using two methods. : Large-scale recent expansion of European patrilineages shown by population resequencing : Nature Communications : Nature Publishing Group". www.nature.com. Retrieved 2015-05-19.
- ^ Peter A. Underhill et al., New Phylogenetic Relationships for Y-chromosome Haplogroup I: Reappraising its Phylogeography and Prehistory, in Rethinking the Human Revolution (2007), pp. 33-42. P. Mellars, K. Boyle, O. Bar-Yosef, C. Stringer (Eds.) McDonald Institute for Archaeological Research, Cambridge, UK.
- ^ "Tracing the genetic origin of Europe's first farmers reveals insights into their social organization". biorxiv.org.
- ^ a b Copyright 2015 ISOGG. "ISOGG 2015 Y-DNA Haplogroup I". isogg.org.
{{cite web}}: CS1 maint: numeric names: authors list (link) - ^ Cinnioglu, Cengiz; King, Roy; Kivisild, Toomas; Kalfoglu, Ersi; Atasoy, Sevil; Cavalleri, Gianpiero L.; Lillie, Anita S.; Roseman, Charles C.; Lin, Alice A.; Prince, Kristina; Oefner, Peter J.; Shen, Peidong; Semino, Ornella; Cavalli-Sforza, L. Luca; Underhill, Peter A.; et al. (2004). "Excavating Y-chromosome haplotype strata in Anatolia". Human Genetics. 114 (2): 127–148. doi:10.1007/s00439-003-1031-4. PMID 14586639.
{{cite journal}}: Explicit use of et al. in:|first9=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ P, Shen et al., Reconstruction of Patrilineages and Matrilineages of Samaritans and Other Israeli Populations From Y-Chromosome and Mitochondrial DNA Sequence Variation, Human Mutations, vol. 24, no. 3 (Sep 2004), pp.248-60.
- ^ Weale, Michael E.; Weiss, Deborah A.; Jager, Rolf F.; Bradman, Neil; Thomas, Mark G. (2002). "Y chromosome Evidence for Anglo-Saxon Mass Migration". Molecular Biology and Evolution. 19 (7): 1008–1021. doi:10.1093/oxfordjournals.molbev.a004160. PMID 12082121.
- ^ Capelli, Cristian; Redhead, Nicola; Abernethy, Julia K.; Gratrix, Fiona; Wilson, James F.; Moen, Torolf; Hervig, Tor; Richards, Martin; Stumpf, Michael P.H.; Underhill, Peter A.; Bradshaw, Paul; Shaha, Alom; Thomas, Mark G.; Bradman, Neal; Goldstein, David B.; et al. (2003). "A Y Chromosome Census of the British Isles" (PDF). Current Biology. 13 (11): 979–984. doi:10.1016/S0960-9822(03)00373-7. PMID 12781138.
{{cite journal}}: Explicit use of et al. in:|first9=(help); Unknown parameter|displayauthors=ignored (|display-authors=suggested) (help) - ^ "Founding Father DNA". isogg.org.
- ^ snpdev. "Reference SNP (refSNP) Cluster Report: rs9341296". nih.gov.
- ^ snpdev. "Reference SNP (refSNP) Cluster Report: rs13447354". nih.gov.
- ^ P30
- ^ P40
