Bacterial phyla: Difference between revisions
m added phyla |
added more phyla |
||
| Line 52: | Line 52: | ||
* '''[[Thermotogae]]''' (''[[Thermotoga maritima]]'') |
* '''[[Thermotogae]]''' (''[[Thermotoga maritima]]'') |
||
== <!-- If adding a newly proposed phylum, if the phylum was detected through 16S rRNA gene sequences alone, citation should be the study that found the first sequences as well as the first study to confirm that the phylum exists through more rigorous analyses (e.g.: first genome) --> == |
|||
== List of bacterial phyla == |
|||
The following is a list of bacterial phyla that have been proposed. |
The following is a list of bacterial phyla that have been proposed. |
||
{| class="wikitable sortable" |
{| class="wikitable sortable" |
||
| Line 110: | Line 110: | ||
| |
| |
||
|- |
|- |
||
|[[Aerophobota]] |
|[[Aerophobota]] / Aerophobetes |
||
|CD12, BHI80-139 |
|CD12, BHI80-139 |
||
| |
| |
||
| Line 176: | Line 176: | ||
| |
| |
||
|- |
|- |
||
|[[Balneolaeota]]<ref>{{Cite journal|last=Hahnke|first=Richard L.|last2=Meier-Kolthoff|first2=Jan P.|last3=García-López|first3=Marina|last4=Mukherjee|first4=Supratim|last5=Huntemann|first5=Marcel|last6=Ivanova|first6=Natalia N.|last7=Woyke|first7=Tanja|last8=Kyrpides|first8=Nikos C.|last9=Klenk|first9=Hans-Peter|last10=Göker|first10=Markus|date=2016|title=Genome-Based Taxonomic Classification of Bacteroidetes|url=https://www.ncbi.nlm.nih.gov/pubmed/28066339|journal=Frontiers in Microbiology|volume=7|pages=2003|doi=10.3389/fmicb.2016.02003|issn=1664-302X|pmc=5167729|pmid=28066339}}</ref> |
|||
|[[Balneolaeota]] |
|||
| |
| |
||
| |
| |
||
| Line 224: | Line 224: | ||
| |
| |
||
|- |
|- |
||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
|OP5<ref name=":9">{{Cite journal|last=Dunfield|first=Peter F.|last2=Tamas|first2=Ivica|last3=Lee|first3=Kevin C.|last4=Morgan|first4=Xochitl C.|last5=McDonald|first5=Ian R.|last6=Stott|first6=Matthew B.|date=2012|title=Electing a candidate: a speculative history of the bacterial phylum OP10|url=https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1111/j.1462-2920.2012.02742.x|journal=Environmental Microbiology|language=en|volume=14|issue=12|pages=3069–3080|doi=10.1111/j.1462-2920.2012.02742.x|issn=1462-2920}}</ref> |
|OP5<ref name=":9">{{Cite journal|last=Dunfield|first=Peter F.|last2=Tamas|first2=Ivica|last3=Lee|first3=Kevin C.|last4=Morgan|first4=Xochitl C.|last5=McDonald|first5=Ian R.|last6=Stott|first6=Matthew B.|date=2012|title=Electing a candidate: a speculative history of the bacterial phylum OP10|url=https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1111/j.1462-2920.2012.02742.x|journal=Environmental Microbiology|language=en|volume=14|issue=12|pages=3069–3080|doi=10.1111/j.1462-2920.2012.02742.x|issn=1462-2920}}</ref> |
||
| |
| |
||
| Line 238: | Line 232: | ||
|[[Calditrichaeota]]<ref>{{Cite journal|last=Kublanov|first=Ilya V.|last2=Sigalova|first2=Olga M.|last3=Gavrilov|first3=Sergey N.|last4=Lebedinsky|first4=Alexander V.|last5=Rinke|first5=Christian|last6=Kovaleva|first6=Olga|last7=Chernyh|first7=Nikolai A.|last8=Ivanova|first8=Natalia|last9=Daum|first9=Chris|last10=Reddy|first10=T.B.K.|last11=Klenk|first11=Hans-Peter|date=2017-02-20|title=Genomic Analysis of Caldithrix abyssi, the Thermophilic Anaerobic Bacterium of the Novel Bacterial Phylum Calditrichaeota|url=http://dx.doi.org/10.3389/fmicb.2017.00195|journal=Frontiers in Microbiology|volume=8|doi=10.3389/fmicb.2017.00195|issn=1664-302X}}</ref> |
|[[Calditrichaeota]]<ref>{{Cite journal|last=Kublanov|first=Ilya V.|last2=Sigalova|first2=Olga M.|last3=Gavrilov|first3=Sergey N.|last4=Lebedinsky|first4=Alexander V.|last5=Rinke|first5=Christian|last6=Kovaleva|first6=Olga|last7=Chernyh|first7=Nikolai A.|last8=Ivanova|first8=Natalia|last9=Daum|first9=Chris|last10=Reddy|first10=T.B.K.|last11=Klenk|first11=Hans-Peter|date=2017-02-20|title=Genomic Analysis of Caldithrix abyssi, the Thermophilic Anaerobic Bacterium of the Novel Bacterial Phylum Calditrichaeota|url=http://dx.doi.org/10.3389/fmicb.2017.00195|journal=Frontiers in Microbiology|volume=8|doi=10.3389/fmicb.2017.00195|issn=1664-302X}}</ref> |
||
|Caldithrix |
|Caldithrix |
||
|FCB group |
|FCB group<ref name="MDM" /> |
||
| |
| |
||
| |
| |
||
| Line 254: | Line 248: | ||
| |
| |
||
|- |
|- |
||
|[[Chlamydiae]]<ref>{{Cite journal|date=2001|editor-last=Boone|editor-first=David R.|editor2-last=Castenholz|editor2-first=Richard W.|editor3-last=Garrity|editor3-first=George M.|title=Bergey’s Manual® of Systematic Bacteriology|url=http://dx.doi.org/10.1007/978-0-387-21609-6|doi=10.1007/978-0-387-21609-6}}</ref> |
|||
|[[Chlamydiae]] |
|||
| |
| |
||
| |
| |
||
| Line 286: | Line 280: | ||
|[[Cloacimonetes]]<ref>{{Cite journal|last=Chouari|first=Rakia|last2=Le Paslier|first2=Denis|last3=Dauga|first3=Catherine|last4=Daegelen|first4=Patrick|last5=Weissenbach|first5=Jean|last6=Sghir|first6=Abdelghani|date=2005-04|title=Novel Major Bacterial Candidate Division within a Municipal Anaerobic Sludge Digester|url=http://dx.doi.org/10.1128/aem.71.4.2145-2153.2005|journal=Applied and Environmental Microbiology|volume=71|issue=4|pages=2145–2153|doi=10.1128/aem.71.4.2145-2153.2005|issn=0099-2240}}</ref> |
|[[Cloacimonetes]]<ref>{{Cite journal|last=Chouari|first=Rakia|last2=Le Paslier|first2=Denis|last3=Dauga|first3=Catherine|last4=Daegelen|first4=Patrick|last5=Weissenbach|first5=Jean|last6=Sghir|first6=Abdelghani|date=2005-04|title=Novel Major Bacterial Candidate Division within a Municipal Anaerobic Sludge Digester|url=http://dx.doi.org/10.1128/aem.71.4.2145-2153.2005|journal=Applied and Environmental Microbiology|volume=71|issue=4|pages=2145–2153|doi=10.1128/aem.71.4.2145-2153.2005|issn=0099-2240}}</ref> |
||
|WWE1 |
|WWE1 |
||
| ⚫ | |||
| ⚫ | |||
| |
| |
||
| |
| |
||
| Line 351: | Line 345: | ||
|- |
|- |
||
|[[Delongbacteria]]<ref name=":1" /> |
|[[Delongbacteria]]<ref name=":1" /> |
||
|RIF26 |
|RIF26, H-178 |
||
| |
| |
||
|No |
|No |
||
| Line 374: | Line 368: | ||
| |
| |
||
|- |
|- |
||
|Dictyoglomi<ref>{{Citation|last=Patel|first=Bharat K. C.|title=Phylum XX. Dictyoglomi phyl. nov.|date=2010|url=http://dx.doi.org/10.1007/978-0-387-68572-4_9|work=Bergey’s Manual® of Systematic Bacteriology|pages=775–780|publisher=Springer New York|isbn=978-0-387-95042-6|access-date=2020-05-15}}</ref> |
|||
|Dictyoglomi |
|||
| |
| |
||
| |
| |
||
| Line 386: | Line 380: | ||
| |
| |
||
|- |
|- |
||
|[[Dormibacteraeota]]<ref>{{Cite journal|last=Ji|first=Mukan|last2=Greening|first2=Chris|last3=Vanwonterghem|first3=Inka|last4=Carere|first4=Carlo R.|last5=Bay|first5=Sean K.|last6=Steen|first6=Jason A.|last7=Montgomery|first7=Kate|last8=Lines|first8=Thomas|last9=Beardall|first9=John|last10=van Dorst|first10=Josie|last11=Snape|first11=Ian|date=2017-12|title=Atmospheric trace gases support primary production in Antarctic desert surface soil|url=https://www.nature.com/articles/nature25014|journal=Nature|language=en|volume=552|issue=7685|pages=400–403|doi=10.1038/nature25014|issn=1476-4687}}</ref> |
|[[Dormibacteraeota]]<ref name=":13">{{Cite journal|last=Ji|first=Mukan|last2=Greening|first2=Chris|last3=Vanwonterghem|first3=Inka|last4=Carere|first4=Carlo R.|last5=Bay|first5=Sean K.|last6=Steen|first6=Jason A.|last7=Montgomery|first7=Kate|last8=Lines|first8=Thomas|last9=Beardall|first9=John|last10=van Dorst|first10=Josie|last11=Snape|first11=Ian|date=2017-12|title=Atmospheric trace gases support primary production in Antarctic desert surface soil|url=https://www.nature.com/articles/nature25014|journal=Nature|language=en|volume=552|issue=7685|pages=400–403|doi=10.1038/nature25014|issn=1476-4687}}</ref> |
||
|AD3 |
|AD3 |
||
| |
| |
||
| Line 398: | Line 392: | ||
| |
| |
||
|- |
|- |
||
|[[Edwardsbacteria]]<ref name=":1" /> |
|[[Edwardsbacteria]]<ref name=":12" /><ref name=":1" /> |
||
|RIF29, UBP-2 <ref name=":14">{{Cite journal|last=Youssef|first=Noha H.|last2=Farag|first2=Ibrahim F.|last3=Hahn|first3=C. Ryan|last4=Premathilake|first4=Hasitha|last5=Fry|first5=Emily|last6=Hart|first6=Matthew|last7=Huffaker|first7=Krystal|last8=Bird|first8=Edward|last9=Hambright|first9=Jimmre|last10=Hoff|first10=Wouter D.|last11=Elshahed|first11=Mostafa S.|date=2019-01-01|title=Candidatus Krumholzibacterium zodletonense gen. nov., sp nov, the first representative of the candidate phylum Krumholzibacteriota phyl. nov. recovered from an anoxic sulfidic spring using genome resolved metagenomics|url=http://www.sciencedirect.com/science/article/pii/S072320201830331X|journal=Systematic and Applied Microbiology|series=Taxonomy of uncultivated Bacteria and Archaea|language=en|volume=42|issue=1|pages=85–93|doi=10.1016/j.syapm.2018.11.002|issn=0723-2020}}</ref> |
|||
|RIF29 |
|||
| |
| |
||
|No |
|No |
||
| Line 410: | Line 404: | ||
| |
| |
||
|- |
|- |
||
|[[Elusimicrobia]] |
|[[Elusimicrobia]]<ref name=":8" /> |
||
|Termite Group 1 |
|OP7, Termite Group 1 (TG1)<ref name=":9" /> |
||
| ⚫ | |||
| |
| |
||
|Yes<ref>{{Cite journal|last=Herlemann|first=D. P. R.|last2=Geissinger|first2=O.|last3=Ikeda-Ohtsubo|first3=W.|last4=Kunin|first4=V.|last5=Sun|first5=H.|last6=Lapidus|first6=A.|last7=Hugenholtz|first7=P.|last8=Brune|first8=A.|date=2009-05-01|title=Genomic Analysis of “Elusimicrobium minutum,” the First Cultivated Representative of the Phylum “Elusimicrobia” (Formerly Termite Group 1)|url=https://aem.asm.org/content/75/9/2841|journal=Applied and Environmental Microbiology|language=en|volume=75|issue=9|pages=2841–2849|doi=10.1128/AEM.02698-08|issn=0099-2240|pmid=19270133}}</ref> |
|||
| |
| |
||
|- |
|- |
||
|[[Eremiobacteraeota]]<ref>{{Cite journal|last=Nogales|first=Balbina|last2=Moore|first2=Edward R. B.|last3=Llobet-Brossa|first3=Enrique|last4=Rossello-Mora|first4=Ramon|last5=Amann|first5=Rudolf|last6=Timmis|first6=Kenneth N.|date=2001-04-01|title=Combined Use of 16S Ribosomal DNA and 16S rRNA To Study the Bacterial Community of Polychlorinated Biphenyl-Polluted Soil|url=https://aem.asm.org/content/67/4/1874|journal=Applied and Environmental Microbiology|language=en|volume=67|issue=4|pages=1874–1884|doi=10.1128/AEM.67.4.1874-1884.2001|issn=0099-2240|pmid=11282645}}</ref><ref name=":13" /> |
|||
|[[Eremiobacteraeota]] |
|||
|WPS-2, Palusbacterota<ref>{{Cite web|title=Evolutionary Implications of Anoxygenic Phototrophy in the Bacterial Phylum Candidatus Palusbacterota (WPS-2)|url=http://dx.doi.org/10.1101/534180|last=Ward|first=Lewis M.|last2=Cardona|first2=Tanai|date=2019-01-29|website=dx.doi.org|access-date=2020-05-15|last3=Holland-Moritz|first3=Hannah}}</ref> |
|||
|WPS-2 |
|||
| |
| |
||
|No |
|No |
||
| Line 484: | Line 478: | ||
|[[Gemmatimonadetes]]<ref name=":11">{{Cite journal|last=Zhang|first=Hui|last2=Sekiguchi|first2=Yuji|last3=Hanada|first3=Satoshi|last4=Hugenholtz|first4=Philip|last5=Kim|first5=Hongik|last6=Kamagata|first6=Yoichi|last7=Nakamura|first7=Kazunori|date=2003|title=Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov.|url=https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.02520-0|journal=International Journal of Systematic and Evolutionary Microbiology,|volume=53|issue=4|pages=1155–1163|doi=10.1099/ijs.0.02520-0|issn=1466-5026}}</ref> |
|[[Gemmatimonadetes]]<ref name=":11">{{Cite journal|last=Zhang|first=Hui|last2=Sekiguchi|first2=Yuji|last3=Hanada|first3=Satoshi|last4=Hugenholtz|first4=Philip|last5=Kim|first5=Hongik|last6=Kamagata|first6=Yoichi|last7=Nakamura|first7=Kazunori|date=2003|title=Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov.|url=https://www.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.02520-0|journal=International Journal of Systematic and Evolutionary Microbiology,|volume=53|issue=4|pages=1155–1163|doi=10.1099/ijs.0.02520-0|issn=1466-5026}}</ref> |
||
| |
| |
||
|FCB group<ref name="MDM" /> |
|||
| ⚫ | |||
|Yes<ref name=":11" /> |
|Yes<ref name=":11" /> |
||
| |
| |
||
| Line 506: | Line 500: | ||
| |
| |
||
|- |
|- |
||
|[[Gracilibacteria]]<ref name=":15">{{Cite journal|last=Ley|first=Ruth E.|last2=Harris|first2=J. Kirk|last3=Wilcox|first3=Joshua|last4=Spear|first4=John R.|last5=Miller|first5=Scott R.|last6=Bebout|first6=Brad M.|last7=Maresca|first7=Julia A.|last8=Bryant|first8=Donald A.|last9=Sogin|first9=Mitchell L.|last10=Pace|first10=Norman R.|date=2006-05-01|title=Unexpected Diversity and Complexity of the Guerrero Negro Hypersaline Microbial Mat|url=https://aem.asm.org/content/72/5/3685|journal=Applied and Environmental Microbiology|language=en|volume=72|issue=5|pages=3685–3695|doi=10.1128/AEM.72.5.3685-3695.2006|issn=0099-2240|pmid=16672518}}</ref> |
|||
|[[Gracilibacteria]] |
|||
|GN02, BD1-5 |
|GN02, BD1-5 |
||
|CPR |
|CPR |
||
| Line 563: | Line 557: | ||
| |
| |
||
|CPR |
|CPR |
||
|No |
|||
| ⚫ | |||
| ⚫ | |||
|KD3-62 |
|||
| ⚫ | |||
| ⚫ | |||
|No |
|No |
||
| |
| |
||
| Line 594: | Line 582: | ||
| |
| |
||
|No |
|No |
||
| ⚫ | |||
| ⚫ | |||
|[[Krumholzibacteriota]]<ref name=":14" /> |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| |
| |
||
|- |
|- |
||
| Line 610: | Line 604: | ||
|[[Latescibacteria]] |
|[[Latescibacteria]] |
||
|WS3 |
|WS3 |
||
|FCB group |
|FCB group<ref name="MDM" /> |
||
|No |
|No |
||
| ⚫ | |||
| ⚫ | |||
|LCP-89<ref>{{Cite journal|last=Youssef|first=Noha H.|last2=Farag|first2=Ibrahim F.|last3=Hahn|first3=C. Ryan|last4=Jarett|first4=Jessica|last5=Becraft|first5=Eric|last6=Eloe-Fadrosh|first6=Emiley|last7=Lightfoot|first7=Jorge|last8=Bourgeois|first8=Austin|last9=Cole|first9=Tanner|last10=Ferrante|first10=Stephanie|last11=Truelock|first11=Mandy|date=2019-05-15|title=Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities|url=https://aem.asm.org/content/85/10/e00110-19|journal=Applied and Environmental Microbiology|language=en|volume=85|issue=10|doi=10.1128/AEM.00110-19|issn=0099-2240|pmid=30902854}}</ref> |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| |
| |
||
|- |
|- |
||
| Line 658: | Line 658: | ||
|Marinimicrobia |
|Marinimicrobia |
||
|SAR406, Marine Group A |
|SAR406, Marine Group A |
||
|FCB group<ref name="MDM" /> |
|||
| ⚫ | |||
|Yes |
|Yes |
||
| |
| |
||
| Line 674: | Line 674: | ||
|Superphylum |
|Superphylum |
||
|- |
|- |
||
|[[Modulibacteria]]<ref name=":15" /><ref>{{Cite journal|last=Sekiguchi|first=Yuji|last2=Ohashi|first2=Akiko|last3=Parks|first3=Donovan H.|last4=Yamauchi|first4=Toshihiro|last5=Tyson|first5=Gene W.|last6=Hugenholtz|first6=Philip|date=2015-01-27|title=First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4312070/|journal=PeerJ|volume=3|doi=10.7717/peerj.740|issn=2167-8359|pmc=4312070|pmid=25650158}}</ref> |
|||
|[[Modulibacteria]] |
|||
|KSB3 |
|KSB3, GN06 |
||
| |
| |
||
|No |
|No |
||
| Line 692: | Line 692: | ||
| |
| |
||
|- |
|- |
||
|NC10<ref>{{Cite journal|last=Holmes|first=Andrew J.|last2=Tujula|first2=Niina A.|last3=Holley|first3=Marita|last4=Contos|first4=Annalisa|last5=James|first5=Julia M.|last6=Rogers|first6=Peter|last7=Gillings|first7=Michael R.|date=2001|title=Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia|url=https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1046/j.1462-2920.2001.00187.x|journal=Environmental Microbiology|language=en|volume=3|issue=4|pages=256–264|doi=10.1046/j.1462-2920.2001.00187.x|issn=1462-2920}}</ref><ref name="Rappe" /> |
|||
|NC10 |
|||
| |
| |
||
| |
| |
||
| Line 710: | Line 710: | ||
| |
| |
||
|- |
|- |
||
|Nitrospinae<ref>{{Cite journal|last=Luecker|first=Sebastian|last2=Nowka|first2=Boris|last3=Rattei|first3=Thomas|last4=Spieck|first4=Eva|last5=Daims|first5=Holger|date=2013|title=The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer|url=https://www.frontiersin.org/articles/10.3389/fmicb.2013.00027/full|journal=Frontiers in Microbiology|language=English|volume=4|doi=10.3389/fmicb.2013.00027|issn=1664-302X}}</ref> |
|||
|Nitrospinae |
|||
| |
| |
||
| |
| |
||
| Line 745: | Line 745: | ||
|No |
|No |
||
|Superphylum |
|Superphylum |
||
| ⚫ | |||
|[[PAUC34f]]<ref>{{Cite journal|last=Astudillo‐García|first=Carmen|last2=Slaby|first2=Beate M.|last3=Waite|first3=David W.|last4=Bayer|first4=Kristina|last5=Hentschel|first5=Ute|last6=Taylor|first6=Michael W.|date=2018|title=Phylogeny and genomics of SAUL, an enigmatic bacterial lineage frequently associated with marine sponges|url=https://sfamjournals.onlinelibrary.wiley.com/doi/abs/10.1111/1462-2920.13965|journal=Environmental Microbiology|language=en|volume=20|issue=2|pages=561–576|doi=10.1111/1462-2920.13965|issn=1462-2920}}</ref> |
|||
|sponge‐associated unclassified lineage (SAUL) |
|||
|FCB group |
|||
| ⚫ | |||
| ⚫ | |||
|- |
|- |
||
|[[Perigrinibacteria]]<ref>{{Cite journal|last=Wrighton|first=K. C.|last2=Thomas|first2=B. C.|last3=Sharon|first3=I.|last4=Miller|first4=C. S.|last5=Castelle|first5=C. J.|last6=VerBerkmoes|first6=N. C.|last7=Wilkins|first7=M. J.|last8=Hettich|first8=R. L.|last9=Lipton|first9=M. S.|last10=Williams|first10=K. H.|last11=Long|first11=P. E.|date=2012-09-27|title=Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla|url=http://dx.doi.org/10.1126/science.1224041|journal=Science|volume=337|issue=6102|pages=1661–1665|doi=10.1126/science.1224041|issn=0036-8075}}</ref> |
|[[Perigrinibacteria]]<ref>{{Cite journal|last=Wrighton|first=K. C.|last2=Thomas|first2=B. C.|last3=Sharon|first3=I.|last4=Miller|first4=C. S.|last5=Castelle|first5=C. J.|last6=VerBerkmoes|first6=N. C.|last7=Wilkins|first7=M. J.|last8=Hettich|first8=R. L.|last9=Lipton|first9=M. S.|last10=Williams|first10=K. H.|last11=Long|first11=P. E.|date=2012-09-27|title=Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla|url=http://dx.doi.org/10.1126/science.1224041|journal=Science|volume=337|issue=6102|pages=1661–1665|doi=10.1126/science.1224041|issn=0036-8075}}</ref> |
||
| Line 758: | Line 764: | ||
| |
| |
||
|- |
|- |
||
|[[Poribacteria]]<ref>{{Cite journal|last=Fieseler|first=Lars|last2=Horn|first2=Matthias|last3=Wagner|first3=Michael|last4=Hentschel|first4=Ute|date=2004-6|title=Discovery of the Novel Candidate Phylum “Poribacteria” in Marine Sponges|url=https://www.ncbi.nlm.nih.gov/pmc/articles/PMC427773/|journal=Applied and Environmental Microbiology|volume=70|issue=6|pages=3724–3732|doi=10.1128/AEM.70.6.3724-3732.2004|issn=0099-2240|pmid=15184179}}</ref> |
|||
|[[Poribacteria]] |
|||
| |
| |
||
| |
| |
||
| Line 840: | Line 846: | ||
|CPR |
|CPR |
||
|No |
|No |
||
| ⚫ | |||
| ⚫ | |||
|[[Sumerlaeota]]<ref>{{Cite journal|last=Derakshani|first=Manigee|last2=Lukow|first2=Thomas|last3=Liesack|first3=Werner|date=2001-02-01|title=Novel Bacterial Lineages at the (Sub)Division Level as Detected by Signature Nucleotide-Targeted Recovery of 16S rRNA Genes from Bulk Soil and Rice Roots of Flooded Rice Microcosms|url=http://dx.doi.org/10.1128/aem.67.2.623-631.2001|journal=Applied and Environmental Microbiology|volume=67|issue=2|pages=623–631|doi=10.1128/aem.67.2.623-631.2001|issn=1098-5336}}</ref><ref>{{Cite journal|last=Kadnikov|first=Vitaly V.|last2=Mardanov|first2=Andrey V.|last3=Beletsky|first3=Alexey V.|last4=Rakitin|first4=Andrey L.|last5=Frank|first5=Yulia A.|last6=Karnachuk|first6=Olga V.|last7=Ravin|first7=Nikolai V.|date=2019-01|title=Phylogeny and physiology of candidate phylum BRC1 inferred from the first complete metagenome-assembled genome obtained from deep subsurface aquifer|url=https://www.ncbi.nlm.nih.gov/pubmed/30201528|journal=Systematic and Applied Microbiology|volume=42|issue=1|pages=67–76|doi=10.1016/j.syapm.2018.08.013|issn=1618-0984|pmid=30201528}}</ref> |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| |
| |
||
|- |
|- |
||
| Line 854: | Line 866: | ||
| |
| |
||
|- |
|- |
||
|TA06 |
|TA06<ref name=":4" /> |
||
| |
| |
||
| |
| |
||
| Line 872: | Line 884: | ||
| |
| |
||
|- |
|- |
||
|[[Tectomicrobia]]<ref>{{Cite journal|last=Wilson|first=Micheal C.|last2=Mori|first2=Tetsushi|last3=Rückert|first3=Christian|last4=Uria|first4=Agustinus R.|last5=Helf|first5=Maximilian J.|last6=Takada|first6=Kentaro|last7=Gernert|first7=Christine|last8=Steffens|first8=Ursula A. E.|last9=Heycke|first9=Nina|last10=Schmitt|first10=Susanne|last11=Rinke|first11=Christian|date=2014-02|title=An environmental bacterial taxon with a large and distinct metabolic repertoire|url=https://www.nature.com/articles/nature12959|journal=Nature|language=en|volume=506|issue=7486|pages=58–62|doi=10.1038/nature12959|issn=1476-4687}}</ref> |
|||
|[[Tectomicrobia]] |
|||
| |
| |
||
| |
| |
||
| Line 909: | Line 921: | ||
|- |
|- |
||
|UBP-1<ref name=":12">{{Cite journal|last=Parks|first=Donovan H.|last2=Rinke|first2=Christian|last3=Chuvochina|first3=Maria|last4=Chaumeil|first4=Pierre-Alain|last5=Woodcroft|first5=Ben J.|last6=Evans|first6=Paul N.|last7=Hugenholtz|first7=Philip|last8=Tyson|first8=Gene W.|date=2017-11|title=Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life|url=https://www.nature.com/articles/s41564-017-0012-7|journal=Nature Microbiology|language=en|volume=2|issue=11|pages=1533–1542|doi=10.1038/s41564-017-0012-7|issn=2058-5276}}</ref> |
|UBP-1<ref name=":12">{{Cite journal|last=Parks|first=Donovan H.|last2=Rinke|first2=Christian|last3=Chuvochina|first3=Maria|last4=Chaumeil|first4=Pierre-Alain|last5=Woodcroft|first5=Ben J.|last6=Evans|first6=Paul N.|last7=Hugenholtz|first7=Philip|last8=Tyson|first8=Gene W.|date=2017-11|title=Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life|url=https://www.nature.com/articles/s41564-017-0012-7|journal=Nature Microbiology|language=en|volume=2|issue=11|pages=1533–1542|doi=10.1038/s41564-017-0012-7|issn=2058-5276}}</ref> |
||
| ⚫ | |||
| ⚫ | |||
|No |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
| |
| |
||
| |
| |
||
Revision as of 00:04, 16 May 2020
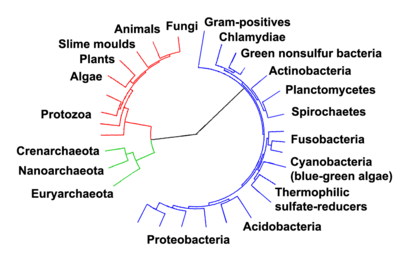
The bacterial phyla are the major lineages of the domain Bacteria. When bacterial nomenclature was controlled under the Botanical Code, the term division was used, but now that bacterial nomenclature (with the exception of cyanobacteria) is controlled under the Bacteriological Code, the term phylum is preferred.
While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.[2]
It has been estimated that ~1,300 bacterial phyla exist.[2] As of May 2020, only 41 bacterial phyla are formally accepted by the LPSN[3] while 89 bacterial phyla are recognized on the Silva database although many more have been proposed [4][5] and many more undoubtedly remain to be discovered. As of 2017, approximately 72% of widely recognized bacterial phyla were candidate phyla.[6]
There are no fixed rules to the nomenclature of bacterial phyla. It was proposed that the suffix "-bacteria" be used for phyla.
Molecular phylogenetics
Traditionally, phylogeny was inferred and taxonomy established based on studies of morphology. Recently molecular phylogenetics has been used to allow better elucidation of the evolutionary relationship of species by analysing their DNA and protein sequences, for example their ribosomal DNA.[7] The lack of easily accessible morphological features, such as those present in animals and plants, hampered early efforts of classification and resulted in erroneous, distorted and confused classification, an example of which, noted Carl Woese, is Pseudomonas whose etymology ironically matched its taxonomy, namely "false unit".[8]
Initial sub-division
In 1987, Carl Woese, regarded as the forerunner of the molecular phylogeny revolution, divided Eubacteria into 11 divisions based on 16S ribosomal RNA (SSU) sequences, listed below[8][10] . Many new phyla have been proposed since then.
- Purple Bacteria and their relatives (later renamed Proteobacteria[11])
- alpha subdivision (purple non-sulfur bacteria, rhizobacteria, Agrobacterium, Rickettsiae, Nitrobacter)
- beta subdivision (Rhodocyclus, (some) Thiobacillus, Alcaligenes, Spirillum, Nitrosovibrio)
- gamma subdivision (enterics, fluorescent pseudomonads, purple sulfur bacteria, Legionella, (some) Beggiatoa)
- delta subdivision (Sulfur and sulfate reducers (Desulfovibrio), Myxobacteria, Bdellovibrio)
- Gram-positive Eubacteria[Note 1]
- High-G+C species (later renamed Actinobacteria[15]) (Actinomyces, Streptomyces, Arthrobacter, Micrococcus, Bifidobacterium)
- Low-G+C species (later renamed Firmicutes[15]) (Clostridium, Peptococcus, Bacillus, Mycoplasma)
- Photosynthetic species (Heliobacteria)
- Species with Gram-negative walls (Megasphaera, Sporomusa)
- Cyanobacteria and chloroplasts (Aphanocapsa, Oscillatoria, Nostoc, Synechococcus, Gloeobacter, Prochloron)
- Spirochetes and relatives
- Spirochetes (Spirochaeta, Treponema, Borrelia)
- Leptospiras (Leptospira, Leptonema)
- Green sulfur bacteria (Chlorobium, Chloroherpeton)
- Bacteroides, Flavobacteria and relatives (later renamed Bacteroidetes
- Bacteroides (Bacteroides, Fusobacterium)
- Flavobacterium group (Flavobacterium, Cytophaga, Saprospira, Flexibacter)
- Planctomyces and relatives (later renamed Planctomycetes)
- Planctomyces group (Planctomyces, Pasteuria [sic][Note 2])
- Thermophiles (Isocystis pallida)
- Chlamydiae (Chlamydia psittaci, Chlamydia trachomatis)
- Radioresistant micrococci and relatives (now commonly referred to as Deinococcus–Thermus[16] or Thermi)[Note 3]
- Deinococcus group (Deinococcus radiodurans)
- Thermophiles (Thermus aquaticus)
- Green non-sulfur bacteria and relatives (later renamed Chloroflexi[20])
- Chloroflexus group (Chloroflexus, Herpetosiphon)
- Thermomicrobium group (Thermomicrobium roseum)
- Thermotogae (Thermotoga maritima)
The following is a list of bacterial phyla that have been proposed.
Uncultivated phyla and metagenomics
This section needs additional citations for verification. (January 2018) |
With the advent of methods to analyse environmental DNA (metagenomics), the 16S rRNA of an extremely large number of undiscovered species have been found, showing that there are several whole phyla which have no known cultivable representative and that some phyla lack in culture major subdivisions as is the case for Verrucomicrobia and Chloroflexi.[54] The term Candidatus is used for proposed species for which the lack of information prevents it to be validated, such as where the only evidence is DNA sequence data, even if the whole genome has been sequenced.[65][66] When the species are members of a whole phylum it is called a candidate division (or candidate phylum)[67] and in 2003 there were 26 candidate phyla out of 52.[54] A candidate phylum was defined by Hugenholtz and Pace in 1998, as a set of 16S ribosomal RNA sequences with less than 85% similarity to already-described phyla.[24] More recently an even lower threshold of 75% was proposed.[68] Three candidate phyla were known before 1998, prior to the 85% threshold definition by Hugenholtz and Pace:
- OS-K group (from Octopus spring, Yellowstone National Park)
- Marine Group A (from Pacific ocean)
- Termite Group 1 (from a Reticulitermes speratus termite gut, now Elusimicrobia)[69]
Since then several other candidate phyla have been identified:[54])
- OP1, OP3, OP5 (now Caldiserica), OP8, OP9 (now Atribacteria),[70] OP10 (now Armatimonadetes), OP11 (obsidian pool, Yellowstone National Park)[54]
- WS2, WS3, WS5, WS6 (Wurtsmith contaminated aquifer)[54]
- SC3 and SC4 (from arid soil)[54]
- vadinBE97 (now Lentisphaerae)[54]
- NC10 (from flooded caves, paddy fields, intertidal zones, etc)[54]
- BRC1 (from bulk soil and rice roots)[54]
- ABY1 (from sediment)[54]
- Guyamas1 (from hydrothermal vents)[54]
- GN01, GN02, GN04 (from a Guerrero Negro hypersaline microbial mat)[54]
- NKB19 (from activated sludge)[54]
- SBR1093 (from activated sludge)[54]
- TM6 and TM7 (Torf, Mittlere Schicht lit. "peat, middle layer")[54]
Since then a candidate phylum called Poribacteria was discovered, living in symbiosis with sponges and extensively studied.[71] (Note: the divergence of the major bacterial lineages predates sponges) Another candidate phylum, called Tectomicrobia, was also found living in symbiosis with sponges.[72] And Nitrospina gracilis, which had long eluded phylogenetic placement, was proposed to belong to a new phylum, Nitrospinae.[73]
Other candidate phyla that have been the center of some studies are TM7,[67] the genomes of organisms of which have even been sequenced (draft),[74] WS6[75] and Marine Group A.[54]
Two species of the candidate phylum OP10, which is now called Armatimonadetes, where recently cultured: Armatimonas rosea isolated from the rhizoplane of a reed in a lake in Japan[76] and Chthonomonas calidirosea from an isolate from geothermally heated soil at Hell's Gate, Tikitere, New Zealand.[77]
One species, Caldisericum exile, of the candidate phylum OP5 was cultured, leading to it being named Caldiserica.[78]
The candidate phylum VadinBE97 is now known as Lentisphaerae after Lentisphaera araneosa and Victivallis vadensis were cultured.[79]
More recently several candidate phyla have been given provisional names despite the fact that they have no cultured representatives:[33]
- Candidate phylum ACD58 was renamed Berkelbacteria[80]
- Candidate phylum CD12 (also known as candidate phylum BHI80-139) was renamed Aerophobetes
- Candidate phylum EM19 was renamed Calescamantes
- Candidate phylum GN02 (also known as candidate phylum BD1-5) was renamed Gracilibacteria
- Candidate phylum KSB3 was renamed Modulibacteria[81]
- Candidate phylum NKB19 was renamed Hydrogenedentes
- Candidate phylum OctSpa1-106 was renamed Fervidibacteria
- Candidate phylum OD1 was renamed Parcubacteria
- Candidate phylum OP1 was renamed Acetothermia
- Candidate phylum OP3 was renamed Omnitrophica
- Candidate phylum OP8 was renamed Aminicenantes
- Candidate phylum OP9 (also known as candidate phylum JS1) was renamed Atribacteria
- Candidate phylum OP11 was renamed Microgenomates
- Candidate phylum PER was renamed Perigrinibacteria[27]
- Candidate phylum SAR406 (also known as candidate phylum Marine Group A) was renamed Marinimicrobia
- Candidate phylum SR1 was renamed Absconditabacteria[70]
- Candidate phylum TM6 was renamed Dependentiae[82]
- Candidate phylum TM7 was renamed Saccharibacteria[83]
- Candidate phylum WS3 was renamed Latescibacteria
- Candidate phylum WWE1 was renamed Cloacimonetes
- Candidate phylum WWE3 was renamed Katanobacteria[70]
- Candidate phylum ZB1 was renamed Ignavibacteriae[84]
Despite these lineages not being officially recognised (due to the ever-increasing number of sequences belonging to undescribed phyla) the ARB-Silva database lists 67 phyla, including 37 candidate phyla (Acetothermia, Aerophobetes, Aminicenantes, aquifer1, aquifer2, Atribacteria, Calescamantes, CKC4, Cloacimonetes, GAL08, GOUTA4, Gracilibacteria, Fermentibacteria (Hyd24-12),[85] Hydrogenedentes, JL-ETNP-Z39, Kazan-3B-09, Latescibacteria, LCP-89, LD1-PA38, Marinimicrobia, Microgenomates, OC31, Omnitrophica, Parcubacteria, PAUC34f, RsaHF231, S2R-29, Saccharibacteria, SBYG-2791, SHA-109, SM2F11, SR1, TA06, TM6, WCHB1-60, WD272, and WS6),[21] the Ribosomal Database Project 10, lists 49 phyla, including 20 candidate phyla (Acetothermia, Aminicenantes, Atribacteria, BRC1, Calescamantes, Cloacimonetes, Hydrogenedentes, Ignavibacteriae, Latescibacteria, Marinimicrobia, Microgenomates, Nitrospinae, Omnitrophica, Parcubacteria, Poribacteria, SR1, Saccharibacteria, WPS-1, WPS-2, and ZB3),[86] and NCBI lists 120 phyla, including 90 candidate phyla (AC1, Acetothermia, Aerophobetes, Aminicenantes, Atribacteria, Berkelbacteria, BRC1, CAB-I, Calescamantes, CPR1, CPR2, CPR3, EM 3, Fervidibacteria, GAL15, GN01, GN03, GN04, GN05, GN06, GN07, GN08, GN09, GN10, GN11, GN12, GN13, GN14, GN15, Gracilibacteria, Fermentibacteria (Hyd24-12),[85] Hydrogenedentes, JL-ETNP-Z39, KD3-62, kpj58rc, KSA1, KSA2, KSB1, KSB2, KSB3, KSB4, Latescibacteria, marine group A, Marinimicrobia, Microgenomates, MSBL2, MSBL3, MSBL4, MSBL5, MSBL6, NC10, Nitrospinae, NPL-UPA2, NT-B4, Omnitrophica, OP2, OP4, OP6, OP7, OS-K, Parcubacteria, Peregrinibacteria, Poribacteria, RF3, Saccharibacteria, SAM, SBR1093, Sediment-1, Sediment-2, Sediment-3, Sediment-4, SPAM, SR1, TA06, TG2, TM6, VC2, WOR-1, WOR-3, WPS-1, WPS-2, WS1, WS2, WS4, WS5, WS6, WWE3, WYO, ZB3, and Zixibacteria).[87]
Many phyla of this type forms a larger clade distinct from other bacteria called the candidate phyla radiation.[88]
Superphyla
Despite the unclear branching order for most bacterial phyla, several groups of phyla consistently cluster together and are referred to as superphyla.
The FCB Group
The FCB group (now called Sphingobacteria) includes Bacteroidetes, the unplaced genus Caldithrix, Chlorobi, candidate phylum Cloacimonetes, Fibrobacteres, Gemmatimonadates, candidate phylum Ignavibacteriae, candidate phylum Latescibacteria, candidate phylum Marinimicrobia, and candidate phylum Zixibacteria.[33][81]
The PVC Group
The PVC group (now called Planctobacteria) includes Chlamydiae, Lentisphaerae, candidate phylum Omnitrophica, Planctomycetes, candidate phylum Poribacteria, and Verrucomicrobia.[33][81]
Patescibacteria
The superphylum Patescibacteria was originally proposed to encompass the phyla Microgenomates (OP11), Parcubacteria (OD1), and Gracilibacteria (GNO2 / BD1-5)[33]. Unfortunately, the meaning of the term 'Patescibacteria' has become confused and is sometimes erroneously used interchangeably with the term Candidate Phyla Radiation (CPR)[89] . To complicate matters, it has been suggested that the Microgenomates and Parcubacteria groups within the Patescibacteria are themselves actually superphyla (see the section on cryptic superphyla below).
Terrabacteria
The proposed superphylum, Terrabacteria,[90] includes Actinobacteria, Cyanobacteria, Deinococcus–Thermus, Chloroflexi, Firmicutes, and candidate phylum OP10.[90][91][33][81]
Proteobacteria
It has been proposed that some classes of the phylum Proteobacteria may be phyla in their own right, which would make Proteobacteria a superphylum.[68] For example, the Deltaproteobacteria group does not consistently form a monophyletic lineage with the other Proteobacteria classes [70].
Candidate Phyla Radiation (CPR)
The CPR is a descriptive term referring to a massive monophyletic radiation of candidate phyla that exists within the Bacterial domain[88]. It includes the Patescibacteria group as well as dozens of additional phyla and superphyla.
Cryptic Superphyla
Several candidate phyla (Microgenomates, Omnitrophica, Parcubacteria, and Saccharibacteria) and several accepted phyla (Elusimicrobia, Caldiserica, and Armatimonadetes) have been suggested to actually be superphyla that were incorrectly described as phyla because rules for defining a bacterial phylum are lacking. For example, it is suggested that candidate phylum Microgenomates is actually a superphylum that encompasses 28 subordinate phyla and that phylum Elusimocrobia is actually a superphylum that encompasses 7 subordinate phyla.[68]
Overview of phyla
As of January 2016[update], there are 30 phyla in the domain "Bacteria" accepted by LPSN.[3] There are no fixed rules to the nomenclature of bacterial phyla. It was proposed that the suffix "-bacteria" be used for phyla,[92] but generally the name of the phylum is generally the plural of the type genus, with the exception of the Firmicutes, Cyanobacteria, and Proteobacteria, whose names do not stem from a genus name (Actinobacteria instead is from Actinomyces).
Acidobacteria
The Acidobacteria (diderm Gram negative) is the most abundant bacterial phylum in many soils, but its members are mostly uncultured. Additionally, they are phenotypically diverse and include not only acidophiles, but also many non-acidophiles.[93] Generally its members divide slowly, exhibit slow metabolic rates under low-nutrient conditions and can tolerate fluctuations in soil hydration.[94]
Actinobacteria
The Actinobacteria is a phylum of monoderm Gram positive bacteria, many of which are notable secondary metabolite producers. There are only two phyla of monoderm Gram positive bacteria, the other being the Firmicutes; the actinobacteria generally have higher GC content so are sometimes called "high-CG Gram positive bacteria". Notable genera/species include Streptomyces (antibiotic production), Cutibacterium acnes (odorous skin commensal) and Propionibacterium freudenreichii (holes in Emmental)
Aquificae
The Aquificae (diderm Gram negative) contains only 14 genera (including Aquifex and Hydrogenobacter). The species are hyperthermophiles and chemolithotrophs (sulphur). According to some studies, this may be one of the most deep branching phyla.
Armatimonadetes
Bacteroidetes
The Bacteroidetes (diderm Gram negative) is a member of the FBC superphylum. Some species are opportunistic pathogens, while other are the most common human gut commensal bacteria. Gained notoriety in the non-scientific community with the urban myth of a bacterial weight loss powder.[95]
Caldiserica
This phylum was formerly known as candidate phylum OP5, Caldisericum exile is the sole representative.
Chlamydiae
The Chlamydiae (diderms, weakly Gram negative) is a phylum of the PVC superphylum. It is composed of only 6 genera of obligate intracellular pathogens with a complex life cycle. Species include Chlamydia trachomatis (chlamydia infection).
Chlorobi
Chlorobi is a member of the FBC superphylum. It contains only 7 genera of obligately anaerobic photoautotrophic bacteria, known colloquially as Green sulfur bacteria. The reaction centre for photosynthesis in Chlorobi and Chloroflexi (another photosynthetic group) is formed by a structures called the chlorosome as opposed to phycobilisomes of cyanobacteria (another photosynthetic group).[96]
Chloroflexi
Chloroflexi, a diverse phylum including thermophiles and halorespirers, are known colloquially as Green non-sulfur bacteria.
Chrysiogenetes
Chrysiogenetes, only 3 genera (Chrysiogenes arsenatis, Desulfurispira natronophila, Desulfurispirillum alkaliphilum)
Cyanobacteria
Cyanobacteria, major photosynthetic clade believed to have caused Earth's oxygen atmosphere, also known as the blue-green algae
Deferribacteres
Deinococcus–Thermus
Deinococcus–Thermus, Deinococcus radiodurans and Thermus aquaticus are "commonly known" species of this phylum.
Dictyoglomi
Elusimicrobia
Elusimicrobia, formerly candidate phylum Termite Group 1
Fibrobacteres
Fibrobacteres, member of the FBC superphylum.
Firmicutes
Firmicutes, Low-G+C Gram positive species most often spore-forming, in two/three classes: the class Bacilli such as the Bacillus spp. (e.g. B. anthracis, a pathogen, and B. subtilis, biotechnologically useful), lactic acid bacteria (e.g. Lactobacillus casei in yoghurt, Oenococcus oeni in malolactic fermentation, Streptococcus pyogenes, pathogen), the class Clostridia of mostly anaerobic sulphite-reducing saprophytic species, includes the genus Clostridium (e.g. the pathogens C. dificile, C. tetani, C. botulinum and the biotech C. acetobutylicum)
Fusobacteria
Gemmatimonadetes
Gemmatimonadetes, member of the FBC superphylum.
Lentisphaerae
Lentisphaerae, formerly clade VadinBE97, member of the PVC superphylum.
Nitrospirae
Planctomycetes
Planctomycetes, member of the PVC superphylum.
Proteobacteria
Proteobacteria, contains most of the "commonly known" species, such as Escherichia coli and Pseudomonas aeruginosa.
Spirochaetes
Spirochaetes, notable for compartmentalisation and species include Borrelia burgdorferi, which causes Lyme disease.
Synergistetes
The Synergistetes is a phylum whose members are diderm Gram negative, rod-shaped obligate anaerobes, some of which are human commensals.[97]
Tenericutes
The Tenericutes includes the class Mollicutes, formerly/debatedly of the phylum Firmicutes (sister clades). Despite their monoderm Gram-positive relatives, they lack peptidoglycan. Notable genus: Mycoplasma.
Thermodesulfobacteria
The Thermodesulfobacteria is a phylum composed of only three genera in the same family (Thermodesulfobacteriaceae: Caldimicrobium, Thermodesulfatator and Thermodesulfobacterium). The members of the phylum are thermophilic sulphate-reducers.
Thermomicrobia
The Thermomicrobia is a group of thermophilic green non-sulfur bacteria.
Thermotogae
The Thermotogae is a phylum of whose members possess an unusual outer envelope called the toga and are mostly hyperthermophilic obligate anaerobic fermenters.
Verrucomicrobia
Verrucomicrobia is a phylum of the PVC superphylum. Like the Planctomycetes species, its members possess a compartmentalised cell plan with a condensed nucleoid and the ribosomes pirellulosome (enclosed by the intracytoplasmic membrane) and paryphoplasm compartment between the intracytoplasmic membrane and cytoplasmic membrane.[98]
Branching order
The branching order of the phyla of bacteria is unclear.[99] Different studies arrive at different results due to different datasets and methods. For example, in studies using 16S and few other sequences Thermotogae and Aquificae appear as the most basal phyla, whereas in several phylogenomic studies, Firmicutes are the most basal.
- Branching order of bacterial phyla (Woese, 1987)
- Branching order of bacterial phyla (Rappe and Giovanoni, 2004)
- Branching order of bacterial phyla after ARB Silva Living Tree
- Branching order of bacterial phyla (Ciccarelli et al., 2006)
- Branching order of bacterial phyla (Battistuzzi et al., 2004)
- Branching order of bacterial phyla (Gupta, 2001)
- Branching order of bacterial phyla (Cavalier-Smith, 2002)
See also
- Bacterial taxonomy#Phyla endings
- International Code of Nomenclature of Bacteria
- List of Bacteria genera
- List of bacterial orders
- List of sequenced bacterial genomes
Footnotes
- ^ Until recently, it was believed than only Firmicutes and Actinobacteria were Gram-positive. However, the candidate phylum TM7 may also be Gram positive.[12] Chloroflexi however possess a single bilayer, but stain negative (with some exceptions[13]).[14]
- ^ Pasteuria is now assigned to phylum Bacilli, not to phylum Planctomycetes.
- ^ It has been proposed to call the clade Xenobacteria[17] or Hadobacteria[18] (the latter is considered an illegitimate name[19]).
References
- ^ Ciccarelli FD; et al. (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–7. Bibcode:2006Sci...311.1283C. CiteSeerX 10.1.1.381.9514. doi:10.1126/science.1123061. PMID 16513982.
- ^ a b Yarza, Pablo; Yilmaz, Pelin; Pruesse, Elmar; Glöckner, Frank Oliver; Ludwig, Wolfgang; Schleifer, Karl-Heinz; Whitman, William B.; Euzéby, Jean; Amann, Rudolf; Rosselló-Móra, Ramon (September 2014). "Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences". Nature Reviews Microbiology. 12 (9): 635–645. doi:10.1038/nrmicro3330. ISSN 1740-1534.
- ^ a b Bacterial phyla in LPSN; Parte, Aidan C.; Sardà Carbasse, Joaquim; Meier-Kolthoff, Jan P.; Reimer, Lorenz C.; Göker, Markus (1 November 2020). "List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ". International Journal of Systematic and Evolutionary Microbiology. 70 (11): 5607–5612. doi:10.1099/ijsem.0.004332.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av Anantharaman, Karthik; Brown, Christopher T.; Hug, Laura A.; Sharon, Itai; Castelle, Cindy J.; Probst, Alexander J.; Thomas, Brian C.; Singh, Andrea; Wilkins, Michael J.; Karaoz, Ulas; Brodie, Eoin L. (24 October 2016). "Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system". Nature Communications. 7 (1): 1–11. doi:10.1038/ncomms13219. ISSN 2041-1723.
- ^ Parks, Donovan H.; Rinke, Christian; Chuvochina, Maria; Chaumeil, Pierre-Alain; Woodcroft, Ben J.; Evans, Paul N.; Hugenholtz, Philip; Tyson, Gene W. (November 2017). "Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life". Nature Microbiology. 2 (11): 1533–1542. doi:10.1038/s41564-017-0012-7. ISSN 2058-5276.
- ^ a b c Dudek, Natasha K.; Sun, Christine L.; Burstein, David; Kantor, Rose S.; Aliaga Goltsman, Daniela S.; Bik, Elisabeth M.; Thomas, Brian C.; Banfield, Jillian F.; Relman, David A. (18 December 2017). "Novel Microbial Diversity and Functional Potential in the Marine Mammal Oral Microbiome". Current biology: CB. 27 (24): 3752–3762.e6. doi:10.1016/j.cub.2017.10.040. ISSN 1879-0445. PMID 29153320.
- ^ Olsen GJ, Woese CR, Overbeek R (1994). "The winds of (evolutionary) change: breathing new life into microbiology". Journal of Bacteriology. 176 (1): 1–6. doi:10.2172/205047. PMC 205007. PMID 8282683.
- ^ a b c Woese, CR (1987). "Bacterial evolution". Microbiological Reviews. 51 (2): 221–71. PMC 373105. PMID 2439888.
- ^ Schluenzen F; et al. (2000). "Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution". Cell. 102 (5): 615–23. doi:10.1016/S0092-8674(00)00084-2. PMID 11007480.
- ^ Holland L (22 May 1990). "Carl Woese in forefront of bacterial evolution revolution". The Scientist. 3 (10).
- ^ Stackebrandt; et al. (1988). "Proteobacteria classis nov., a name for the phylogenetic taxon that includes the "purple bacteria and their relatives"". Int. J. Syst. Bacteriol. 38 (3): 321–325. doi:10.1099/00207713-38-3-321.
- ^ Hugenholtz, P.; Tyson, G. W.; Webb, R. I.; Wagner, A. M.; Blackall, L. L. (2001). "Investigation of Candidate Division TM7, a Recently Recognized Major Lineage of the Domain Bacteria with No Known Pure-Culture Representatives". Applied and Environmental Microbiology. 67 (1): 411–9. doi:10.1128/AEM.67.1.411-419.2001. PMC 92593. PMID 11133473.
- ^ Yabe, S.; Aiba, Y.; Sakai, Y.; Hazaka, M.; Yokota, A. (2010). "Thermogemmatispora onikobensis gen. nov., sp. nov. and Thermogemmatispora foliorum sp. nov., isolated from fallen leaves on geothermal soils, and description of Thermogemmatisporaceae fam. nov. and Thermogemmatisporales ord. nov. within the class Ktedonobacteria". International Journal of Systematic and Evolutionary Microbiology. 61 (4): 903–910. doi:10.1099/ijs.0.024877-0. PMID 20495028.
- ^ Sutcliffe, I. C. (2011). "Cell envelope architecture in the Chloroflexi: A shifting frontline in a phylogenetic turf war". Environmental Microbiology. 13 (2): 279–282. doi:10.1111/j.1462-2920.2010.02339.x. PMID 20860732.
- ^ a b Stackebrandt, E.; Rainey, F. A.; Ward-Rainey, N. L. (1997). "Proposal for a New Hierarchic Classification System, Actinobacteria classis nov". International Journal of Systematic Bacteriology. 47 (2): 479–491. doi:10.1099/00207713-47-2-479.
- ^ J.P. Euzéby. "List of Prokaryotic names with Standing in Nomenclature: classification of Deinococcus–Thermus". Archived from the original on 27 January 2013. Retrieved 30 December 2010.
- ^ Bergey's Manual of Systematic Bacteriology 1st Ed.
- ^ Cavalier-Smith, T (2002). "The neomuran origin of Archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification". International Journal of Systematic and Evolutionary Microbiology. 52 (Pt 1): 7–76. doi:10.1099/00207713-52-1-7. PMID 11837318.
- ^ "List of Prokaryotic names with Standing in Nomenclature—Class Hadobacteria". LPSN. Archived from the original on 19 April 2012. Retrieved 30 December 2010. Euzéby, J.P. (1997). "List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet". Int J Syst Bacteriol. 47 (2): 590–2. doi:10.1099/00207713-47-2-590. ISSN 0020-7713. PMID 9103655.
- ^ Boone DR; Castenholz RW (18 May 2001) [1984 (Williams & Wilkins)]. Garrity GM (ed.). The Archaea and the Deeply Branching and Phototrophic Bacteria. Bergey's Manual of Systematic Bacteriology. Vol. 1 (2nd ed.). New York: Springer. pp. 721. ISBN 978-0-387-98771-2. British Library no. GBA561951.
- ^ a b c d e "ARB-Silva: comprehensive ribosomal RNA database". The ARB development Team. Retrieved 2 January 2016.
- ^ a b Tahon, Guillaume; Tytgat, Bjorn; Lebbe, Liesbeth; Carlier, Aurélien; Willems, Anne (1 July 2018). "Abditibacterium utsteinense sp. nov., the first cultivated member of candidate phylum FBP, isolated from ice-free Antarctic soil samples". Systematic and Applied Microbiology. 41 (4): 279–290. doi:10.1016/j.syapm.2018.01.009. ISSN 0723-2020.
- ^ a b Harris, J. Kirk; Kelley, Scott T.; Pace, Norman R. (2004-2). "New Perspective on Uncultured Bacterial Phylogenetic Division OP11". Applied and Environmental Microbiology. 70 (2): 845–849. doi:10.1128/AEM.70.2.845-849.2004. ISSN 0099-2240. PMID 14766563.
{{cite journal}}: Check date values in:|date=(help) - ^ a b c d e f g h Hugenholtz P; et al. (1998). "Novel division level bacterial diversity in a Yellowstone hot spring". Journal of Bacteriology. 180 (2): 366–76. PMC 106892. PMID 9440526.
- ^ Thrash, J. Cameron; Coates, John D. (2010), "Phylum XVII. Acidobacteria phyl. nov.", Bergey’s Manual® of Systematic Bacteriology, Springer New York, pp. 725–735, ISBN 978-0-387-95042-6, retrieved 15 May 2020
- ^ Goodfellow, Michael (2012), "Phylum XXVI. Actinobacteria phyl. nov.", Bergey’s Manual® of Systematic Bacteriology, Springer New York, pp. 33–2028, ISBN 978-0-387-95043-3, retrieved 15 May 2020
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa Brown CT; et al. (2015). "Unusual biology across a group comprising more than 15% of domain Bacteria". Nature. 523 (7559): 208–11. Bibcode:2015Natur.523..208B. doi:10.1038/nature14486. PMID 26083755.
- ^ Tamaki, Hideyuki; Tanaka, Yasuhiro; Matsuzawa, Hiroaki; Muramatsu, Mizuho; Meng, Xian-Ying; Hanada, Satoshi; Mori, Kazuhiro; Kamagata, Yoichi (2011-06). "Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10". International Journal of Systematic and Evolutionary Microbiology. 61 (Pt 6): 1442–1447. doi:10.1099/ijs.0.025643-0. ISSN 1466-5034. PMID 20622056.
{{cite journal}}: Check date values in:|date=(help) - ^ Hahnke, Richard L.; Meier-Kolthoff, Jan P.; García-López, Marina; Mukherjee, Supratim; Huntemann, Marcel; Ivanova, Natalia N.; Woyke, Tanja; Kyrpides, Nikos C.; Klenk, Hans-Peter; Göker, Markus (2016). "Genome-Based Taxonomic Classification of Bacteroidetes". Frontiers in Microbiology. 7: 2003. doi:10.3389/fmicb.2016.02003. ISSN 1664-302X. PMC 5167729. PMID 28066339.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c Dunfield, Peter F.; Tamas, Ivica; Lee, Kevin C.; Morgan, Xochitl C.; McDonald, Ian R.; Stott, Matthew B. (2012). "Electing a candidate: a speculative history of the bacterial phylum OP10". Environmental Microbiology. 14 (12): 3069–3080. doi:10.1111/j.1462-2920.2012.02742.x. ISSN 1462-2920.
- ^ Mori, K.; Yamaguchi, K.; Sakiyama, Y.; Urabe, T.; Suzuki, K.-i. (23 July 2009). "Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov". INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY. 59 (11): 2894–2898. doi:10.1099/ijs.0.010033-0. ISSN 1466-5026.
- ^ Kublanov, Ilya V.; Sigalova, Olga M.; Gavrilov, Sergey N.; Lebedinsky, Alexander V.; Rinke, Christian; Kovaleva, Olga; Chernyh, Nikolai A.; Ivanova, Natalia; Daum, Chris; Reddy, T.B.K.; Klenk, Hans-Peter (20 February 2017). "Genomic Analysis of Caldithrix abyssi, the Thermophilic Anaerobic Bacterium of the Novel Bacterial Phylum Calditrichaeota". Frontiers in Microbiology. 8. doi:10.3389/fmicb.2017.00195. ISSN 1664-302X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b c d e f g h i j Rinke C; et al. (2013). "Insights into the phylogeny and coding potential of microbial dark matter". Nature. 499 (7459): 431–7. Bibcode:2013Natur.499..431R. doi:10.1038/nature12352. PMID 23851394.
- ^ Boone, David R.; Castenholz, Richard W.; Garrity, George M., eds. (2001). "Bergey's Manual® of Systematic Bacteriology". doi:10.1007/978-0-387-21609-6.
{{cite journal}}: Cite journal requires|journal=(help) - ^ Chouari, Rakia; Le Paslier, Denis; Dauga, Catherine; Daegelen, Patrick; Weissenbach, Jean; Sghir, Abdelghani (2005-04). "Novel Major Bacterial Candidate Division within a Municipal Anaerobic Sludge Digester". Applied and Environmental Microbiology. 71 (4): 2145–2153. doi:10.1128/aem.71.4.2145-2153.2005. ISSN 0099-2240.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Hug, Laura A.; Thomas, Brian C.; Sharon, Itai; Brown, Christopher T.; Sharma, Ritin; Hettich, Robert L.; Wilkins, Michael J.; Williams, Kenneth H.; Singh, Andrea; Banfield, Jillian F. (2016). "Critical biogeochemical functions in the subsurface are associated with bacteria from new phyla and little studied lineages". Environmental Microbiology. 18 (1): 159–173. doi:10.1111/1462-2920.12930. ISSN 1462-2920.
- ^ a b Rheims, H; Rainey, F A; Stackebrandt, E (1996-09). "A molecular approach to search for diversity among bacteria in the environment". Journal of Industrial Microbiology & Biotechnology. 17 (3–4): 159–169. doi:10.1007/bf01574689. ISSN 0169-4146.
{{cite journal}}: Check date values in:|date=(help) - ^ Patel, Bharat K. C. (2010), "Phylum XX. Dictyoglomi phyl. nov.", Bergey’s Manual® of Systematic Bacteriology, Springer New York, pp. 775–780, ISBN 978-0-387-95042-6, retrieved 15 May 2020
- ^ a b Ji, Mukan; Greening, Chris; Vanwonterghem, Inka; Carere, Carlo R.; Bay, Sean K.; Steen, Jason A.; Montgomery, Kate; Lines, Thomas; Beardall, John; van Dorst, Josie; Snape, Ian (2017-12). "Atmospheric trace gases support primary production in Antarctic desert surface soil". Nature. 552 (7685): 400–403. doi:10.1038/nature25014. ISSN 1476-4687.
{{cite journal}}: Check date values in:|date=(help) - ^ a b c d e f g h i j k l m n o p q Parks, Donovan H.; Rinke, Christian; Chuvochina, Maria; Chaumeil, Pierre-Alain; Woodcroft, Ben J.; Evans, Paul N.; Hugenholtz, Philip; Tyson, Gene W. (2017-11). "Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life". Nature Microbiology. 2 (11): 1533–1542. doi:10.1038/s41564-017-0012-7. ISSN 2058-5276.
{{cite journal}}: Check date values in:|date=(help) - ^ a b Youssef, Noha H.; Farag, Ibrahim F.; Hahn, C. Ryan; Premathilake, Hasitha; Fry, Emily; Hart, Matthew; Huffaker, Krystal; Bird, Edward; Hambright, Jimmre; Hoff, Wouter D.; Elshahed, Mostafa S. (1 January 2019). "Candidatus Krumholzibacterium zodletonense gen. nov., sp nov, the first representative of the candidate phylum Krumholzibacteriota phyl. nov. recovered from an anoxic sulfidic spring using genome resolved metagenomics". Systematic and Applied Microbiology. Taxonomy of uncultivated Bacteria and Archaea. 42 (1): 85–93. doi:10.1016/j.syapm.2018.11.002. ISSN 0723-2020.
- ^ Herlemann, D. P. R.; Geissinger, O.; Ikeda-Ohtsubo, W.; Kunin, V.; Sun, H.; Lapidus, A.; Hugenholtz, P.; Brune, A. (1 May 2009). "Genomic Analysis of "Elusimicrobium minutum," the First Cultivated Representative of the Phylum "Elusimicrobia" (Formerly Termite Group 1)". Applied and Environmental Microbiology. 75 (9): 2841–2849. doi:10.1128/AEM.02698-08. ISSN 0099-2240. PMID 19270133.
- ^ Nogales, Balbina; Moore, Edward R. B.; Llobet-Brossa, Enrique; Rossello-Mora, Ramon; Amann, Rudolf; Timmis, Kenneth N. (1 April 2001). "Combined Use of 16S Ribosomal DNA and 16S rRNA To Study the Bacterial Community of Polychlorinated Biphenyl-Polluted Soil". Applied and Environmental Microbiology. 67 (4): 1874–1884. doi:10.1128/AEM.67.4.1874-1884.2001. ISSN 0099-2240. PMID 11282645.
- ^ Ward, Lewis M.; Cardona, Tanai; Holland-Moritz, Hannah (29 January 2019). "Evolutionary Implications of Anoxygenic Phototrophy in the Bacterial Phylum Candidatus Palusbacterota (WPS-2)". dx.doi.org. Retrieved 15 May 2020.
- ^ a b Zhang, Hui; Sekiguchi, Yuji; Hanada, Satoshi; Hugenholtz, Philip; Kim, Hongik; Kamagata, Yoichi; Nakamura, Kazunori (2003). "Gemmatimonas aurantiaca gen. nov., sp. nov., a Gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov". International Journal of Systematic and Evolutionary Microbiology,. 53 (4): 1155–1163. doi:10.1099/ijs.0.02520-0. ISSN 1466-5026.
{{cite journal}}: CS1 maint: extra punctuation (link) - ^ a b Ley, Ruth E.; Harris, J. Kirk; Wilcox, Joshua; Spear, John R.; Miller, Scott R.; Bebout, Brad M.; Maresca, Julia A.; Bryant, Donald A.; Sogin, Mitchell L.; Pace, Norman R. (1 May 2006). "Unexpected Diversity and Complexity of the Guerrero Negro Hypersaline Microbial Mat". Applied and Environmental Microbiology. 72 (5): 3685–3695. doi:10.1128/AEM.72.5.3685-3695.2006. ISSN 0099-2240. PMID 16672518.
- ^ Guermazi, Sonda; Daegelen, Patrick; Dauga, Catherine; Rivière, Delphine; Bouchez, Théodore; Godon, Jean Jacques; Gyapay, Gábor; Sghir, Abdelghani; Pelletier, Eric; Weissenbach, Jean; Le Paslier, Denis (2008-8). "Discovery and characterization of a new bacterial candidate division by an anaerobic sludge digester metagenomic approach". Environmental Microbiology. 10 (8): 2111–2123. doi:10.1111/j.1462-2920.2008.01632.x. ISSN 1462-2912. PMC 2702496. PMID 18459975.
{{cite journal}}: Check date values in:|date=(help) - ^ Eloe-Fadrosh, Emiley A.; Paez-Espino, David; Jarett, Jessica; Dunfield, Peter F.; Hedlund, Brian P.; Dekas, Anne E.; Grasby, Stephen E.; Brady, Allyson L.; Dong, Hailiang; Briggs, Brandon R.; Li, Wen-Jun (27 January 2016). "Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs". Nature Communications. 7 (1): 1–10. doi:10.1038/ncomms10476. ISSN 2041-1723.
- ^ Youssef, Noha H.; Farag, Ibrahim F.; Hahn, C. Ryan; Jarett, Jessica; Becraft, Eric; Eloe-Fadrosh, Emiley; Lightfoot, Jorge; Bourgeois, Austin; Cole, Tanner; Ferrante, Stephanie; Truelock, Mandy (15 May 2019). "Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities". Applied and Environmental Microbiology. 85 (10). doi:10.1128/AEM.00110-19. ISSN 0099-2240. PMID 30902854.
- ^ Di Rienzi, Sara C; Sharon, Itai; Wrighton, Kelly C; Koren, Omry; Hug, Laura A; Thomas, Brian C; Goodrich, Julia K; Bell, Jordana T; Spector, Timothy D; Banfield, Jillian F; Ley, Ruth E (1 October 2013). "The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria". eLife. 2. doi:10.7554/eLife.01102. ISSN 2050-084X. PMC 3787301. PMID 24137540.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Hugenholtz, Philip; Goebel, Brett M.; Pace, Norman R. (15 December 1998). "Impact of Culture-Independent Studies on the Emerging Phylogenetic View of Bacterial Diversity". Journal of Bacteriology. 180 (24): 6793–6793. doi:10.1128/jb.180.24.6793-6793.1998. ISSN 1098-5530.
- ^ Sekiguchi, Yuji; Ohashi, Akiko; Parks, Donovan H.; Yamauchi, Toshihiro; Tyson, Gene W.; Hugenholtz, Philip (27 January 2015). "First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking". PeerJ. 3. doi:10.7717/peerj.740. ISSN 2167-8359. PMC 4312070. PMID 25650158.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Holmes, Andrew J.; Tujula, Niina A.; Holley, Marita; Contos, Annalisa; James, Julia M.; Rogers, Peter; Gillings, Michael R. (2001). "Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia". Environmental Microbiology. 3 (4): 256–264. doi:10.1046/j.1462-2920.2001.00187.x. ISSN 1462-2920.
- ^ a b c d e f g h i j k l m n o p q Rappe, M. S.; Giovannoni, S. J. (2003). "The Uncultured Microbial Majority". Annual Review of Microbiology. 57: 369–94. doi:10.1146/annurev.micro.57.030502.090759. PMID 14527284.
- ^ Luecker, Sebastian; Nowka, Boris; Rattei, Thomas; Spieck, Eva; Daims, Holger (2013). "The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer". Frontiers in Microbiology. 4. doi:10.3389/fmicb.2013.00027. ISSN 1664-302X.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Astudillo‐García, Carmen; Slaby, Beate M.; Waite, David W.; Bayer, Kristina; Hentschel, Ute; Taylor, Michael W. (2018). "Phylogeny and genomics of SAUL, an enigmatic bacterial lineage frequently associated with marine sponges". Environmental Microbiology. 20 (2): 561–576. doi:10.1111/1462-2920.13965. ISSN 1462-2920.
- ^ Wrighton, K. C.; Thomas, B. C.; Sharon, I.; Miller, C. S.; Castelle, C. J.; VerBerkmoes, N. C.; Wilkins, M. J.; Hettich, R. L.; Lipton, M. S.; Williams, K. H.; Long, P. E. (27 September 2012). "Fermentation, Hydrogen, and Sulfur Metabolism in Multiple Uncultivated Bacterial Phyla". Science. 337 (6102): 1661–1665. doi:10.1126/science.1224041. ISSN 0036-8075.
- ^ Fieseler, Lars; Horn, Matthias; Wagner, Michael; Hentschel, Ute (2004-6). "Discovery of the Novel Candidate Phylum "Poribacteria" in Marine Sponges". Applied and Environmental Microbiology. 70 (6): 3724–3732. doi:10.1128/AEM.70.6.3724-3732.2004. ISSN 0099-2240. PMID 15184179.
{{cite journal}}: Check date values in:|date=(help) - ^ Derakshani, Manigee; Lukow, Thomas; Liesack, Werner (1 February 2001). "Novel Bacterial Lineages at the (Sub)Division Level as Detected by Signature Nucleotide-Targeted Recovery of 16S rRNA Genes from Bulk Soil and Rice Roots of Flooded Rice Microcosms". Applied and Environmental Microbiology. 67 (2): 623–631. doi:10.1128/aem.67.2.623-631.2001. ISSN 1098-5336.
- ^ Kadnikov, Vitaly V.; Mardanov, Andrey V.; Beletsky, Alexey V.; Rakitin, Andrey L.; Frank, Yulia A.; Karnachuk, Olga V.; Ravin, Nikolai V. (2019-01). "Phylogeny and physiology of candidate phylum BRC1 inferred from the first complete metagenome-assembled genome obtained from deep subsurface aquifer". Systematic and Applied Microbiology. 42 (1): 67–76. doi:10.1016/j.syapm.2018.08.013. ISSN 1618-0984. PMID 30201528.
{{cite journal}}: Check date values in:|date=(help) - ^ a b c d Baker, Brett J.; Lazar, Cassandre Sara; Teske, Andreas P.; Dick, Gregory J. (13 April 2015). "Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria". Microbiome. 3 (1): 14. doi:10.1186/s40168-015-0077-6. ISSN 2049-2618. PMC 4411801. PMID 25922666.
{{cite journal}}: CS1 maint: PMC format (link) CS1 maint: unflagged free DOI (link) - ^ Wilson, Micheal C.; Mori, Tetsushi; Rückert, Christian; Uria, Agustinus R.; Helf, Maximilian J.; Takada, Kentaro; Gernert, Christine; Steffens, Ursula A. E.; Heycke, Nina; Schmitt, Susanne; Rinke, Christian (2014-02). "An environmental bacterial taxon with a large and distinct metabolic repertoire". Nature. 506 (7486): 58–62. doi:10.1038/nature12959. ISSN 1476-4687.
{{cite journal}}: Check date values in:|date=(help) - ^ Reysenbach, Anna-Louise; Huber, Robert; Stetter, Karl O.; Davey, Mary Ellen; MacGregor, Barbara J.; Stahl, David A. (2001), "Phylum BII. Thermotogae phy. nov.", Bergey’s Manual® of Systematic Bacteriology, Springer New York, pp. 369–387, ISBN 978-1-4419-3159-7, retrieved 15 May 2020
- ^ Castelle, Cindy J.; Hug, Laura A.; Wrighton, Kelly C.; Thomas, Brian C.; Williams, Kenneth H.; Wu, Dongying; Tringe, Susannah G.; Singer, Steven W.; Eisen, Jonathan A.; Banfield, Jillian F. (27 August 2013). "Extraordinary phylogenetic diversity and metabolic versatility in aquifer sediment". Nature Communications. 4 (1): 1–10. doi:10.1038/ncomms3120. ISSN 2041-1723.
- ^ Murray, R. G. E.; Schleifer, K. H. (1994). "Taxonomic Notes: A Proposal for Recording the Properties of Putative Taxa of Procaryotes". International Journal of Systematic Bacteriology. 44 (1): 174–6. doi:10.1099/00207713-44-1-174. PMID 8123559.
- ^ Frederiksen, W (1995). "Judicial commission of the international committee on systematic bacteriology: Minutes of the meetings, 2 and 6 July 1994, Prague, Czech Republic" (PDF). Int. J. Syst. Bacteriol. 45 (1): 195–196. doi:10.1099/00207713-45-1-195.
- ^ a b Hugenholtz P; et al. (1998). "Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity". Journal of Bacteriology. 180 (18): 4765–74. doi:10.1128/JB.180.18.4765-4774.1998. PMC 107498. PMID 9733676.
- ^ a b c Yarza P; et al. (2014). "Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences". Nature Reviews Microbiology. 12 (9): 635–645. doi:10.1038/nrmicro3330. hdl:10261/123763. PMID 25118885.
- ^ Geissinger O; et al. (2009). "The Ultramicrobacterium Elusimicrobium minutum gen. nov., sp. nov., the first cultivated representative of the Termite Group 1 Phylum". Applied and Environmental Microbiology. 75 (9): 2831–40. doi:10.1128/AEM.02697-08. PMC 2681718. PMID 19270135.
- ^ a b c d Hug LA; et al. (2016). "A new view of the tree of life". Nature Microbiology. Article 16048 (5): 16048. doi:10.1038/nmicrobiol.2016.48. PMID 27572647.
- ^ Fieseler L; et al. (2004). "Discovery of the Novel Candidate Phylum "Poribacteria" in Marine Sponges". Applied and Environmental Microbiology. 70 (6): 3724–32. doi:10.1128/AEM.70.6.3724-3732.2004. PMC 427773. PMID 15184179.
- ^ Wilson MC; et al. (2014). "An environmental bacterial taxon with a large and distinct metabolic repertoire" (PDF). Nature. 506 (7486): 58–62. Bibcode:2014Natur.506...58W. doi:10.1038/nature12959. PMID 24476823.
- ^ Lucker S; et al. (2013). "The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer". Front. Microbiol. 4: 27. doi:10.3389/fmicb.2013.00027. PMC 3578206. PMID 23439773.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR (July 2007). "Dissecting biological "dark matter" with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth". Proceedings of the National Academy of Sciences of the United States of America. 104 (29): 11889–94. doi:10.1073/pnas.0704662104. PMC 1924555. PMID 17620602.
- ^ Dojka MA; et al. (2000). "Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria". Applied and Environmental Microbiology. 66 (4): 1617–21. doi:10.1128/AEM.66.4.1617-1621.2000. PMC 92031. PMID 10742250.
- ^ Tamaki H; et al. (2010). "Armatimonas rosea gen. nov., sp. nov., a Gram-negative, aerobic, chemoheterotrophic bacterium of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10". International Journal of Systematic and Evolutionary Microbiology. 61 (Pt 6): 1442–7. doi:10.1099/ijs.0.025643-0. PMID 20622056.
- ^ Lee KCY; et al. (2010). "Chthonomonas calidirosea gen. nov., sp. nov., an aerobic, pigmented, thermophilic microorganism of a novel bacterial class, Chthonomonadetes classis. nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10". International Journal of Systematic and Evolutionary Microbiology. 61 (Pt 10): 2482–90. doi:10.1099/ijs.0.027235-0. PMID 21097641.
- ^ Mori K; et al. (2009). "Caldisericum exile gen. nov., sp. nov., an anaerobic, thermophilic, filamentous bacterium of a novel bacterial phylum, Caldiserica phyl. nov., originally called the candidate phylum OP5, and description of Caldisericaceae fam. nov., Caldisericales ord. nov. and Caldisericia classis nov". International Journal of Systematic and Evolutionary Microbiology. 59 (Pt 11): 2894–8. doi:10.1099/ijs.0.010033-0. PMID 19628600.
- ^ Cho JC; et al. (2004). "Lentisphaera araneosa gen. nov., sp. nov, a transparent exopolymer producing marine bacterium, and the description of a novel bacterial phylum, Lentisphaerae". Environmental Microbiology. 6 (6): 611–21. doi:10.1111/j.1462-2920.2004.00614.x. PMID 15142250.
- ^ Wrighton KC; et al. (2014). "Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer". The ISME Journal. 8 (7): 1452–1463. doi:10.1038/ismej.2013.249. PMC 4069391. PMID 24621521.
- ^ a b c d Sekiguchi Y; et al. (2015). "First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking". PeerJ. 3: e740. doi:10.7717/peerj.740. PMC 4312070. PMID 25650158.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Yeoh YK; et al. (2015). "Comparative Genomics of Candidate Phylum TM6 Suggests That Parasitism Is Widespread and Ancestral in This Lineage". Mol Biol Evol. 33 (4): 915–927. doi:10.1093/molbev/msv281. PMC 4776705. PMID 26615204.
- ^ Albertsen M; et al. (2013). "Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes". Nat. Biotechnol. 31 (6): 533–8. doi:10.1038/nbt.2579. PMID 23707974.
- ^ Podosokorskaya OA; et al. (2013). "Characterization of Melioribacter roseus gen. nov., sp. nov., a novel facultatively anaerobic thermophilic cellulolytic bacterium from the class Ignavibacteria, and a proposal of a novel bacterial phylum Ignavibacteriae". Environ. Microbiol. 15 (6): 1759–71. doi:10.1111/1462-2920.12067. PMID 23297868.
- ^ a b Kirkegaard, Rasmus Hansen; Dueholm, Morten Simonsen; McIlroy, Simon Jon; Nierychlo, Marta; Karst, Søren Michael; Albertsen, Mads; Nielsen, Per Halkjær (1 October 2016). "Genomic insights into members of the candidate phylum Hyd24-12 common in mesophilic anaerobic digesters". The ISME Journal. 10 (10): 2352–2364. doi:10.1038/ismej.2016.43. ISSN 1751-7370. PMC 5030696. PMID 27058503.
- ^ "Hierarchy Browser". Ribosomal database project. Retrieved 2 January 2016.
- ^ "Taxonomy Browser". NCBI. Retrieved 2 January 2016.
- ^ a b Castelle CJ, Banfield JF (March 2018). "Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life". Cell. 172 (6): 1181–1197. doi:10.1016/j.cell.2018.02.016. PMID 29522741.
- ^ Castelle, Cindy J.; Banfield, Jillian F. (8 March 2018). "Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life". Cell. 172 (6): 1181–1197. doi:10.1016/j.cell.2018.02.016. ISSN 0092-8674.
- ^ a b c Battistuzzi FU, Feijao A, Hedges SB (November 2004). "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evolutionary Biology. 4: 44. doi:10.1186/1471-2148-4-44. PMC 533871. PMID 15535883.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Battistuzzi, F. U.; Hedges, S. B. (6 November 2008). "A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land". Molecular Biology and Evolution. 26 (2): 335–343. doi:10.1093/molbev/msn247. PMID 18988685.
- ^ Murray, R. G. E. 1984. The higher taxa, or, a place for everything…?, p. 33. In N. R. Krieg and J. G. Holt (ed.), Bergey’s manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore.
- ^ Kielak, A.; Pijl, A. S.; Van Veen, J. A.; Kowalchuk, G. A. (2008). "Phylogenetic diversity of Acidobacteria in a former agricultural soil". The ISME Journal. 3 (3): 378–382. doi:10.1038/ismej.2008.113. PMID 19020558.
- ^ Ward NL; et al. (2009). "Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils". Applied and Environmental Microbiology. 75 (7): 2046–2056. doi:10.1128/AEM.02294-08. PMC 2663196. PMID 19201974.
- ^ Duncan SH; et al. (2008). "Human colonic microbiota associated with diet, obesity and weight loss". International Journal of Obesity. 32 (11): 1720–1724. doi:10.1038/ijo.2008.155. PMID 18779823.
- ^ Oostergetel GT, Amerongen H, Boekema EJ (2010). "The chlorosome: A prototype for efficient light harvesting in photosynthesis". Photosynthesis Research. 104 (2–3): 245–255. doi:10.1007/s11120-010-9533-0. PMC 2882566. PMID 20130996.
- ^ Marchandin HLN; et al. (2010). "Phylogeny, diversity and host specialization in the phylum Synergistetes with emphasis on strains and clones of human origin". Research in Microbiology. 161 (2): 91–100. doi:10.1016/j.resmic.2009.12.008. PMID 20079831.
- ^ Lee, K. C.; Webb, R. I.; Janssen, P. H.; Sangwan, P.; Romeo, T.; Staley, J. T.; Fuerst, J. A. (2009). "Phylum Verrucomicrobia representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes". BMC Microbiology. 9: 5. doi:10.1186/1471-2180-9-5. PMC 2647929. PMID 19133117.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Pace, N. R. (2009). "Mapping the Tree of Life: Progress and Prospects". Microbiology and Molecular Biology Reviews. 73 (4): 565–576. doi:10.1128/MMBR.00033-09. PMC 2786576. PMID 19946133.
- ^ Lapage, S. P.; Sneath PHA; Lessel, E. F.; Skerman VBD; Seeliger HPR; Clark, W. A. (1992). Lapage SP; Sneath PHA; Lessel EF; Skerman VBD; Seeliger HPR; Clark WA (eds.). International Code of Nomenclature of Bacteria, 1990 Revision. Washington (DC): American Society for Microbiology. ISBN 978-1-55581-039-9. PMID 21089234.
- ^ Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzéby J, Tindall BJ (2007). "Taxonomic Outline of the Bacteria and Archaea, Release 7.7". Taxonomic Outline of Bacteria and Archaea. doi:10.1601/TOBA7.7.
- ^ Euzéby JP. "List of Prokaryotic names with Standing in Nomenclature". Retrieved 30 December 2010.
Cite error: A list-defined reference named "woese90" is not used in the content (see the help page).
