Haplogroup F-M89: Difference between revisions
Brewcrewer (talk | contribs) rv, should probably be removed in its entirety, per this talk page and RSN. also please cease with personal attacks. |
→Paragroup F*: as per talk: these 2 sources seem to please everyone if combined, and having 2 sources is neater than having 1 source and 1 tag |
||
| Line 32: | Line 32: | ||
The '''F*''' haplogroup is paraphyletic and often rare, except in '''[[India]]''', which codistributes with [[Haplogroup H (Y-DNA)|H]].<ref name=Chiaroni>Chiaroni J. et al 2009, [http://www.pnas.org/content/106/48/20174.full.pdf+html Y chromosome diversity, human expansion, drift, and cultural evolution]</ref> India has a frequency of 12.5%, being the most frequent haplogroup in [[Indian tribes (India)|tribal groups]] after that of H: among tribal groups is at 18.1% with a moderate distribution among [[Caste system in India|caste groups]] (approx. 9.6%),<ref name=Cordaux/> so the gene flow between castes was low but it is important in [[dalit|panchamas]] and [[sudras]].<ref>Zerjal Tatiana et al 2007, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2590678/?tool=pubmed Y-chromosomal insights into the genetic impact of the caste system in India]</ref> So it is important in [[Tamil Nadu]] and [[Andhra Pradesh]],<ref name="Sengupta"/> in [[Koya (Mappila Subdivision)|Koya]] tribes is at 27% and in [[Sinhalese people]] 10%.<ref name="Kivisild"/> |
The '''F*''' haplogroup is paraphyletic and often rare, except in '''[[India]]''', which codistributes with [[Haplogroup H (Y-DNA)|H]].<ref name=Chiaroni>Chiaroni J. et al 2009, [http://www.pnas.org/content/106/48/20174.full.pdf+html Y chromosome diversity, human expansion, drift, and cultural evolution]</ref> India has a frequency of 12.5%, being the most frequent haplogroup in [[Indian tribes (India)|tribal groups]] after that of H: among tribal groups is at 18.1% with a moderate distribution among [[Caste system in India|caste groups]] (approx. 9.6%),<ref name=Cordaux/> so the gene flow between castes was low but it is important in [[dalit|panchamas]] and [[sudras]].<ref>Zerjal Tatiana et al 2007, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2590678/?tool=pubmed Y-chromosomal insights into the genetic impact of the caste system in India]</ref> So it is important in [[Tamil Nadu]] and [[Andhra Pradesh]],<ref name="Sengupta"/> in [[Koya (Mappila Subdivision)|Koya]] tribes is at 27% and in [[Sinhalese people]] 10%.<ref name="Kivisild"/> |
||
It is found in two of the north [[Portuguese people|Portuguese]] populations (0.5%) and it may have possibly occurred as a result of admixture since Portugal had significant contacts with India about 500 years ago.<ref>T. Whit Athey 2005, [http://www.jogg.info/12/Pitfalls.pdf Pitfalls in Determinations of Y Haplogroup F*]</ref> |
It is found in two of the north [[Portuguese people|Portuguese]] populations (0.5%) and it may have possibly occurred as a result of admixture since Portugal had significant contacts with India about 500 years ago.<ref>T. Whit Athey 2005, [http://www.jogg.info/12/Pitfalls.pdf Pitfalls in Determinations of Y Haplogroup F*]</ref><ref>ISSOG 2010, [http://www.isogg.org/tree/ISOGG_HapgrpF.html Y-DNA Haplogroup F and its Subclades - 2010]</ref> It is wide spread and is found at between 7-41% among [[Kurdish people]].<ref>Nasidze, Iván et al 2005, [http://www.eva.mpg.de/genetics/pdf/Kurds.pdf MtDNA and Y-chromosome Variation in Kurdish Groups]</ref> The [[Ailao Mountains]] of [[Yunnan Province]] in southwestern [[China]] appear to be the only regions where such lineages, which are grouped for convenience as Haplogroup F[[* (haplogroup)|*]], comprise a significant portion of the Y-chromosome diversity of the modern populations; F* Y-chromosomes have been found to be particularly common among the [[Kucong]] or [[Lahu people|Yellow Lahu]], a group of hunter-gatherers who live in the Ailao Mountains of Yunnan.<ref>{{cite journal |doi=10.1353/hub.2006.0041 |author=Black ML, Wise CA, Wang W, Bittles AH |title=Combining genetics and population history in the study of ethnic diversity in the People's Republic of China |journal=Hum. Biol. |volume=78 |issue=3 |pages=277–93 |year=2006 |month=June |pmid=17216801 }}</ref> Also have been found in the [[Malay Archipelago]] and in 8% of sampled [[Korea]]n males,<ref name="Kayser">Kayser et al 2002, [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC379223/ Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea.]</ref> but other researches have not found it in Korea.<ref>[http://dienekes.blogspot.com/2009/01/y-chromosomes-and-mtdna-of-koreans.html Y chromosomes and mtDNA of Koreans.] Dienekes' Anthropology Blog.</ref> |
||
==Subclades== |
==Subclades== |
||
Revision as of 07:42, 1 October 2010
| Haplogroup F | |
|---|---|
 | |
| Possible time of origin | 48,000 years BP(38,700-55,700);[1] 50,300±6500, Hammer and Zegura 2002 |
| Possible place of origin | South Asia or Southwest Asia |
| Ancestor | CF |
| Descendants | F*, F1, F2, F3, F4, F5, G, H, IJK |
| Defining mutations | P14, M89, M213, P133, P134, P135, P136, P138, P139, P140, P141, P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187 |
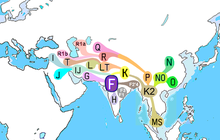
In human genetics, haplogroup F or FT is an enormous Y-chromosome haplogroup spanning all the continents. This haplogroup and its subclades contain more than 90% of the world's existing male population.
Haplogroup F is an ancestral haplogroup to Y-chromosome haplogroups G (M201), H (M69), IJ (M429), and K (M9) along with K's descendant haplogroups (L, M, N, O, Q, R, S, and T).
Origins
This supercluster contains lineages that are not typically found in sub-Saharan Africa, suggesting that his ancestral C-F chromosome may have been carried out of Africa very early in the modern human diaspora, and F may have appeared 48,000(38,700-55,700) years ago.[1]
According to the phylogeographic distribution of haplotypes observed among South Asian populations defined by social and linguistic criteria, the most plausibly arose of haplogroup F might be in India and its potentially paraphyletic group F* might share a common demographic history with H, C5, R2 and L1.[2] The presence of several subclusters of F and K that are largely restricted to the Indian subcontinent is consistent with the scenario that the coastal (southern route) migration(s) from Africa carried the ancestral Eurasian lineages first to the coast of Indian subcontinent, or that some of them originated there.[3]
Other sources mention that this ancient haplogroup may have first appeared in India, North Africa, the Levant, or the Arabian Peninsula as much as 50,000 years ago (50,300±6500).[4] It is sometimes believed to represent a "second-wave" of expansion out of Africa. However, the location of this lineage's first expansion and rise to dominance appears to have been in South Asia or somewhere close to it within extended Middle East; all of Haplogroup F's descendant haplogroups also show a pattern of radiation from South Asia (haplogroups H, F* and K) or the Middle East (haplogroups G and IJ).
Several lineages derived from Haplogroup F appear to have migrated into Africa from a homeland in Southwest Asia sometime during prehistory. Y-chromosome haplogroups associated with this hypothetical "Back to Africa" migration include J, R1b, and T.
Distribution
F is the most important macro-haplogroup outside of Africa, except for Tibet, Kazakhstan, Mongolia, Japan, Polynesia, and communities of indigenous Australians and indigenous Siberians, while also including many men within those regions.
Paragroup F*
Besides the major clades G, H and IJK, other patrilines derived from Haplogroup F-M89 can still be detected at a very low frequency among many populations of the southern fringe of Eurasia and Oceania. Haplogroup F-M89 is a “default” haplogroup potentially comprising several lineages.[5] Originally it was believed all M89+ men would be found to belong to F descendant haplogroups G-T, but an increasing number of men have continued to test positive only for the mutation that defines F.[6]
The F* haplogroup is paraphyletic and often rare, except in India, which codistributes with H.[7] India has a frequency of 12.5%, being the most frequent haplogroup in tribal groups after that of H: among tribal groups is at 18.1% with a moderate distribution among caste groups (approx. 9.6%),[5] so the gene flow between castes was low but it is important in panchamas and sudras.[8] So it is important in Tamil Nadu and Andhra Pradesh,[2] in Koya tribes is at 27% and in Sinhalese people 10%.[3]
It is found in two of the north Portuguese populations (0.5%) and it may have possibly occurred as a result of admixture since Portugal had significant contacts with India about 500 years ago.[9][10] It is wide spread and is found at between 7-41% among Kurdish people.[11] The Ailao Mountains of Yunnan Province in southwestern China appear to be the only regions where such lineages, which are grouped for convenience as Haplogroup F*, comprise a significant portion of the Y-chromosome diversity of the modern populations; F* Y-chromosomes have been found to be particularly common among the Kucong or Yellow Lahu, a group of hunter-gatherers who live in the Ailao Mountains of Yunnan.[12] Also have been found in the Malay Archipelago and in 8% of sampled Korean males,[13] but other researches have not found it in Korea.[14]
Subclades

This phylogenetic tree of haplogroup subclades is based on the YCC 2009 tree[1] and subsequent published research.
- CF
- F (P14, M89, M213, P133, P134, P135, P136, P138, P139, P140, P141, P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187)
- F* Specially in India[7] and rare through Asia.
- F1 (P91, P104) - Found in Sri Lanka.[1]
- F2 (M427, M428) - In Lahu people (China).[2]
- F3 (P96, M282) - In South Iran and South India.[15] Also in Armenia[16] and rare in Netherlands.[1]
- F4 (P254) - In Sri Lanka.[1]
- F5 (M481) - Found in southeastern India.[17]
- G (M201) - Mainly in Caucasus and Near East.
- H (M201) - Typical for the Indian subcontinent.
- IJK (L15/S137, L16/S138)
- F (P14, M89, M213, P133, P134, P135, P136, P138, P139, P140, P141, P142, P145, P146, P148, P149, P151, P157, P158, P159, P160, P161, P163, P166, P187)
References
- ^ a b c d e f Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF (2008). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research. 18 (5): 830–8. doi:10.1101/gr.7172008. PMC 2336805. PMID 18385274.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c Sengupta, Sanghamitra et al 2005-2006, Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists American Journal of Human Genetics, 2006 February; 78(2): 202–221. Published online 2005 December 16.
- ^ a b Kivisild T et al 2003, The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations
- ^ Hammer and Zegura 2002, The Human Y Chromosome Haplogroup Tree: Nomenclature and Phylogeography of Its Major Divisions
- ^ a b Cordaux, Richard et al 2004, Independent Origins of Indian Caste and Tribal Paternal Lineages
- ^ The Haplogroup F Y-DNA Project - Family Project Website
- ^ a b Chiaroni J. et al 2009, Y chromosome diversity, human expansion, drift, and cultural evolution
- ^ Zerjal Tatiana et al 2007, Y-chromosomal insights into the genetic impact of the caste system in India
- ^ T. Whit Athey 2005, Pitfalls in Determinations of Y Haplogroup F*
- ^ ISSOG 2010, Y-DNA Haplogroup F and its Subclades - 2010
- ^ Nasidze, Iván et al 2005, MtDNA and Y-chromosome Variation in Kurdish Groups
- ^ Black ML, Wise CA, Wang W, Bittles AH (2006). "Combining genetics and population history in the study of ethnic diversity in the People's Republic of China". Hum. Biol. 78 (3): 277–93. doi:10.1353/hub.2006.0041. PMID 17216801.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Kayser et al 2002, Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea.
- ^ Y chromosomes and mtDNA of Koreans. Dienekes' Anthropology Blog.
- ^ Regueiro M et al 2006 Iran: tricontinental nexus for Y-chromosome driven migration
- ^ Armenian DNA Project Family Tree DNA - Genealogy by Genetics, Ltd. World Headquarters, 2010
- ^ Fornarino, Simona et al 2009 Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation
