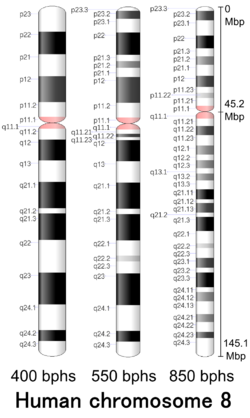
Chromosome 8
| Chromosome 8 | |
|---|---|
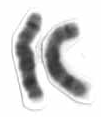 Human chromosome 8 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 8 pair in human male karyogram. | |
| Features | |
| Length (bp) | 146,259,331 (CHM13)[1] |
| No. of genes | 646 (CCDS)[2] |
| Type | Autosome |
| Centromere position | Submetacentric[3] (45.2 Mbp[4]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 8 |
| Entrez | Chromosome 8 |
| NCBI | Chromosome 8 |
| UCSC | Chromosome 8 |
| Full DNA sequences | |
| RefSeq | NC_000008 (FASTA) |
| GenBank | CM000670 (FASTA) |
Chromosome 8 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 8 spans about 146 million base pairs (the building material of DNA) and represents between 4.5 and 5.0% of the total DNA in cells.[5]
About 8% of its genes are involved in brain development and function, and about 16% are involved in cancer. A unique feature of 8p is a region of about 15 megabases that appears to have a high mutation rate. This region shows a significant divergence between human and chimpanzee, suggesting that its high mutation rates have contributed to the evolution of the human brain.[5]
Genes
Number of genes
The following are some of the gene count estimates of human chromosome 8. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[6]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 646 | — | — | [2] | 2016-09-08 |
| HGNC | 656 | 242 | 539 | [7] | 2017-05-12 |
| Ensembl | 670 | 1,052 | 613 | [8] | 2017-03-29 |
| UniProt | 703 | — | — | [9] | 2018-02-28 |
| NCBI | 719 | 848 | 682 | [10][11][12] | 2017-05-19 |
Gene list
The following is a partial list of genes on human chromosome 8. For complete list, see the link in the infobox on the right.
- ADHFE1 encoding protein Alcohol dehydrogenase, iron containing 1
- AEG1 : Astrocyte Elevated Gene (linked to hepatocellular carcinoma and neuroblastoma)
- ANK1: ankyrin 1, erythrocytic
- Arc/Arg3.1
- ASAH1: N-acylsphingosine amidohydrolase (acid ceramidase) 1
- ASPH: encoding enzyme Aspartyl/asparaginyl beta-hydroxylase
- AZIN1: encoding protein Antizyme inhibitor 1
- BRF2: transcription factor IIIB 50 kDa subunit
- C8orf32/WDYHV1: encoding enzyme Protein N-terminal glutamine amidohydrolase
- C8orf33: chromosome 8, open reading frame 33
- C8orf34: chromosome 8, open reading frame 34
- C8orf4: encoding protein Uncharacterized protein C8orf4
- C8orf48 encoding protein C8orf48
- C8orf58: chromosome 8 open reading frame 58
- C8orfk29: encoding protein TMEM249
- CCAT1: colon cancer associated transcript 1
- CCDC166: encoding protein Coiled-coil domain containing 166
- CCDC25: coiled-coil domain containing protein 25
- CHD7: chromodomain helicase DNA binding protein 7
- CHMP4C: Charged multivesicular body protein 4c
- CHRAC1 encoding protein Chromatin accessibility complex 1
- CHRNA2: cholinergic receptor, nicotinic, alpha 2 (neuronal)
- CLN8: ceroid-lipofuscinosis, neuronal 8
- CNGB3: cyclic nucleotide gated channel beta 3
- COH1: vacuolar protein sorting 13B
- CRISPLD1 encoding protein Cysteine rich secretory protein LCCL domain containing 1
- CSGALNACT1: Chondroitin sulfate N-acetylgalactosaminyltransferase 1
- CTHRC1 encoding protein Collagen triple helix repeat containing 1
- CYP11B1: cytochrome P450, family 11, subfamily B, polypeptide 1
- CYP11B2: cytochrome P450, family 11, subfamily B, polypeptide 2
- DCSTAMP: dendrocyte expressed seven transmembrane protein
- DPYS: dihydropyrimidinase
- EBAG9: Estrogen receptor binding site associated antigen 9
- ENTPD4: encoding protein Ectonucleoside triphosphate diphosphohydrolase 4
- EPPK1: epiplakin
- ERICH5 encoding protein Glutamate rich 5
- ESCO2: establishment of sister chromatid cohesion N-acetyltransferase 2
- EXT1: exostosin glycosyltransferase 1
- EXTL3: exostosin-like glycosyltransferase 3
- EYA1: EYA transcriptional coactivator and phosphatase 1
- FABP9: fatty acid binding protein 9, testis
- FAM167A: family with sequence similarity 167, member A
- FAM203B: family with sequence similarity 203, member B
- FAM83A: family with sequence similarity 83, member A
- FAM83H: family with sequence similarity 83, member H
- FAM84B: encoding protein Family with sequence similarity 84 member b
- FBXO16 encoding protein F-box protein 16
- FDFT1 encoding protein Farnesyl-diphosphate farnesyltransferase 1
- FGFR1: fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome)
- FGL1: Fibrinogen-like protein 1
- GDAP1: ganglioside-induced differentiation-associated protein 1
- GDF6: growth differentiation factor 6
- GPIHBP1: GPI-anchored high density lipoprotein binding protein 1
- GRINA: encoding protein Glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding)
- GSDMC encoding protein Gasdermin C
- GULOP pseudogene: responsible for human inability to produce Vitamin C
- HGSNAT: heparan-alpha-glucosaminide N-acetyltransferase
- HMBOX1: encoding protein Homeobox containing 1, also known as homeobox telomere-binding protein 1 (HOT1)
- HRSP12 encoding enzyme Ribonuclease UK114
- INTS8: integrator complex subunit 8
- INTS9: integrator complex subunit 9
- KCNQ3: potassium channel, voltage gated KQT-like subfamily Q, member 3.
- KIAA0196: KIAA0196
- KIF13B encoding protein Kinesin family member 13B
- LACTB2: lactamase, beta 2
- LAPTM4B: lysosomal-associated transmembrane protein 4B
- LOC642658: encoding protein Basic helix-loop-helix transcription factor scleraxis
- LPL: lipoprotein lipase
- LSM1: U6 snRNA-associated Sm-like protein LSm1
- MAK16: MAK16 homolog
- MCPH1: microcephaly, primary autosomal recessive 1
- MIR486-1: encoding MicroRNA 486-1
- MIR548V: encoding MicroRNA 548v
- MIR5680: encoding MicroRNA 5680
- MIR6850 encoding protein MicroRNA 6850
- MRPL13 encoding protein Mitochondrial ribosomal protein L13
- MYBL1 encoding protein MYB proto-oncogene like 1
- NBN: nibrin
- NDRG1: N-myc downstream regulated gene 1
- NEF3: neurofilament 3 (150kDa medium)
- NEFL: neurofilament, light polypeptide 68kDa
- ODF1: outer dense fiber protein 1
- OTUD6B: OTU domain containing 6B
- PDP1: pyruvate dehydrogenase phosphatase catalytic subunit 1
- PKIA: cAMP-dependent protein kinase inhibitor alpha
- PLEC: plectin
- PNMA2: paraneoplastic antigen Ma2
- PREX2: phosphatidylinositol-3,4,5-trisphosphate dependent Rac exchange factor 2
- PROSC: proline synthetase co-transcribe bacterial homolog protein
- PURG encoding protein Purine-rich element binding protein G
- PVT1: Pvt1 oncogene
- RECQL4: RecQ protein-like 4
- RNF5P1: ring finger protein 5 pseudogene 1
- RRS1: ribosome biogenesis regulator homolog
- RUNX1T1: runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
- SFTPC: surfactant protein C
- SLC20A2: Sodium-dependent phosphate transporter 2
- SLURP1: secreted LY6/PLAUR domain containing 1
- SNAI2: snail homolog 2 (Drosophila)
- SPAG11B: sperm-associated antigen 11B
- STAU2: staufen double-stranded RNA binding protein 2
- SYBU: Syntabulin
- TG: thyroglobulin
- THAP1: THAP domain containing, apoptosis associated protein 1
- TMEM67: encoding protein Meckelin
- TNFRSF11B: tumor necrosis factor receptor superfamily, member 11b
- TONSL: encoding protein Tonsoku-like, DNA repair protein
- TPA: tissue plasminogen activator
- TRMT12: tRNA methyltransferase 12 homolog
- TRPS1: trichorhinophalangeal syndrome I
- TSPYL5 (gene): encoding protein TSPY like 5
- TTI2 encoding protein TELO2 interacting protein 2
- VCPIP1: valosin containing protein/p47 complex interacting protein 1
- VXN: encoding vexin
- VMAT1: vesicular monoamine transporter protein
- VPS13B: vacuolar protein sorting 13 homolog B (yeast)
- VPS37A: vacuolar protein sorting 37 homolog A
- WRN: Werner syndrome
- YTHDF3: YTH N6-methyladenosine RNA binding protein 3
- ZFP41: encoding protein ZFP41 zinc finger protein
- ZHX2: zinc fingers and homeoboxes protein 2
- ZMAT4: zinc finger matrin-type 4
- ZNF16: zinc finger protein 16
- ZNF395: encoding protein Zinc finger protein 395
- ZNF517 encoding protein Zinc finger protein 517
- ZNF696 encoding protein Zinc finger protein 696
- ZNF703: zinc finger protein 703
- ZNF706: zinc finger protein 706
- ZNF707: encoding protein Zinc finger protein 707
Diseases and disorders
The following diseases and disorders are some of those related to genes on chromosome 8:
- 8p23.1 duplication syndrome
- Burkitt lymphoma
- Charcot–Marie–Tooth disease
- COACH syndrome
- Cleft lip and cleft palate
- Cohen syndrome
- Congenital hypothyroidism
- Fahr's syndrome
- Hereditary multiple exostoses
- Lipoprotein lipase deficiency, familial
- Myelodysplastic syndrome
- Pfeiffer syndrome
- Primary microcephaly
- Rothmund–Thomson syndrome
- Schizophrenia, associated with 8p21-22 locus[13][14][15]
- Waardenburg syndrome
- Werner syndrome
- Pingelapese blindness
- Langer–Giedion syndrome
- Roberts syndrome
- Hepatocellular carcinoma
- Sanfilippo syndrome
Cytogenetic band
| Chr. | Arm[20] | Band[21] | ISCN start[22] |
ISCN stop[22] |
Basepair start |
Basepair stop |
Stain[23] | Density |
|---|---|---|---|---|---|---|---|---|
| 8 | p | 23.3 | 0 | 115 | 1 | 2,300,000 | gneg | |
| 8 | p | 23.2 | 115 | 331 | 2,300,001 | 6,300,000 | gpos | 75 |
| 8 | p | 23.1 | 331 | 690 | 6,300,001 | 12,800,000 | gneg | |
| 8 | p | 22 | 690 | 992 | 12,800,001 | 19,200,000 | gpos | 100 |
| 8 | p | 21.3 | 992 | 1179 | 19,200,001 | 23,500,000 | gneg | |
| 8 | p | 21.2 | 1179 | 1380 | 23,500,001 | 27,500,000 | gpos | 50 |
| 8 | p | 21.1 | 1380 | 1639 | 27,500,001 | 29,000,000 | gneg | |
| 8 | p | 12 | 1639 | 1897 | 29,000,001 | 36,700,000 | gpos | 75 |
| 8 | p | 11.23 | 1897 | 2041 | 36,700,001 | 38,500,000 | gneg | |
| 8 | p | 11.22 | 2041 | 2156 | 38,500,001 | 39,900,000 | gpos | 25 |
| 8 | p | 11.21 | 2156 | 2343 | 39,900,001 | 43,200,000 | gneg | |
| 8 | p | 11.1 | 2343 | 2472 | 43,200,001 | 45,200,000 | acen | |
| 8 | q | 11.1 | 2472 | 2645 | 45,200,001 | 47,200,000 | acen | |
| 8 | q | 11.21 | 2645 | 2817 | 47,200,001 | 51,300,000 | gneg | |
| 8 | q | 11.22 | 2817 | 3033 | 51,300,001 | 51,700,000 | gpos | 75 |
| 8 | q | 11.23 | 3033 | 3277 | 51,700,001 | 54,600,000 | gneg | |
| 8 | q | 12.1 | 3277 | 3493 | 54,600,001 | 60,600,000 | gpos | 50 |
| 8 | q | 12.2 | 3493 | 3622 | 60,600,001 | 61,300,000 | gneg | |
| 8 | q | 12.3 | 3622 | 3809 | 61,300,001 | 65,100,000 | gpos | 50 |
| 8 | q | 13.1 | 3809 | 3938 | 65,100,001 | 67,100,000 | gneg | |
| 8 | q | 13.2 | 3938 | 4096 | 67,100,001 | 69,600,000 | gpos | 50 |
| 8 | q | 13.3 | 4096 | 4312 | 69,600,001 | 72,000,000 | gneg | |
| 8 | q | 21.11 | 4312 | 4545 | 72,000,001 | 74,600,000 | gpos | 100 |
| 8 | q | 21.12 | 4545 | 4628 | 74,600,001 | 74,700,000 | gneg | |
| 8 | q | 21.13 | 4628 | 4858 | 74,700,001 | 83,500,000 | gpos | 75 |
| 8 | q | 21.2 | 4858 | 4959 | 83,500,001 | 85,900,000 | gneg | |
| 8 | q | 21.3 | 4959 | 5289 | 85,900,001 | 92,300,000 | gpos | 100 |
| 8 | q | 22.1 | 5289 | 5577 | 92,300,001 | 97,900,000 | gneg | |
| 8 | q | 22.2 | 5577 | 5692 | 97,900,001 | 100,500,000 | gpos | 25 |
| 8 | q | 22.3 | 5692 | 5922 | 100,500,001 | 105,100,000 | gneg | |
| 8 | q | 23.1 | 5922 | 6152 | 105,100,001 | 109,500,000 | gpos | 75 |
| 8 | q | 23.2 | 6152 | 6267 | 109,500,001 | 111,100,000 | gneg | |
| 8 | q | 23.3 | 6267 | 6611 | 111,100,001 | 116,700,000 | gpos | 100 |
| 8 | q | 24.11 | 6611 | 6726 | 116,700,001 | 118,300,000 | gneg | |
| 8 | q | 24.12 | 6726 | 6942 | 118,300,001 | 121,500,000 | gpos | 50 |
| 8 | q | 24.13 | 6942 | 7244 | 121,500,001 | 126,300,000 | gneg | |
| 8 | q | 24.21 | 7244 | 7431 | 126,300,001 | 130,400,000 | gpos | 50 |
| 8 | q | 24.22 | 7431 | 7661 | 130,400,001 | 135,400,000 | gneg | |
| 8 | q | 24.23 | 7661 | 7804 | 135,400,001 | 138,900,000 | gpos | 75 |
| 8 | q | 24.3 | 7804 | 8250 | 138,900,001 | 145,138,636 | gneg |
References
- ^ "Homo sapiens isolate CHM13 chromosome 8". National Library of Medicine /accessdate=2023-01-13. 4 April 2022.
- ^ a b "Search results - 8[CHR] AND "Homo sapiens"[Organism] AND ("has ccds"[Properties] AND alive[prop]) - Gene". NCBI. CCDS Release 20 for Homo sapiens. 2016-09-08. Retrieved 2017-05-28.
- ^ Tom Strachan; Andrew Read (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN 978-1-136-84407-2.
- ^ a b c Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
- ^ a b Tabarés-Seisdedos R, Rubenstein JL; Rubenstein (2009). "Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer". Mol Psychiatry. 14 (6): 563–89. doi:10.1038/mp.2009.2. PMID 19204725. S2CID 11118479.
- ^ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human genes". Genome Biol. 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMC 2898077. PMID 20441615.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "Statistics & Downloads for chromosome 8". HUGO Gene Nomenclature Committee. 2017-05-12. Retrieved 2017-05-19.
- ^ "Chromosome 8: Chromosome summary - Homo sapiens". Ensembl Release 88. 2017-03-29. Archived from the original on 2019-12-03. Retrieved 2017-05-19.
- ^ "Human chromosome 8: entries, gene names and cross-references to MIM". UniProt. 2018-02-28. Retrieved 2018-03-16.
- ^ "Search results - 8[CHR] AND "Homo sapiens"[Organism] AND ("genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ^ "Search results - 8[CHR] AND "Homo sapiens"[Organism] AND ( ("genetype miscrna"[Properties] OR "genetype ncrna"[Properties] OR "genetype rrna"[Properties] OR "genetype trna"[Properties] OR "genetype scrna"[Properties] OR "genetype snrna"[Properties] OR "genetype snorna"[Properties]) NOT "genetype protein coding"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ^ "Search results - 8[CHR] AND "Homo sapiens"[Organism] AND ("genetype pseudo"[Properties] AND alive[prop]) - Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
- ^ Blouin JL, Dombroski BA, Nath SK, et al. (September 1998). "Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21". Nat. Genet. 20 (1): 70–3. doi:10.1038/1734. PMID 9731535. S2CID 52804924.
- ^ Gurling HM, Kalsi G, Brynjolfson J, et al. (March 2001). "Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23". Am. J. Hum. Genet. 68 (3): 661–73. doi:10.1086/318788. PMC 1274479. PMID 11179014.
- ^ Suarez BK, Duan J, Sanders AR, et al. (February 2006). "Genomewide linkage scan of 409 European-ancestry and African American families with schizophrenia: suggestive evidence of linkage at 8p23.3-p21.2 and 11p13.1-q14.1 in the combined sample". Am. J. Hum. Genet. 78 (2): 315–33. doi:10.1086/500272. PMC 1380238. PMID 16400611.
- ^ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (400 bphs, Assembly GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
- ^ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (550 bphs, Assembly GRCh38.p3). Last update 2015-08-11. Retrieved 2017-04-26.
- ^ International Standing Committee on Human Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN 978-3-318-02253-7.
- ^ Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. (2012). "Estimation of band level resolutions of human chromosome images". 2012 Ninth International Conference on Computer Science and Software Engineering (JCSSE). pp. 276–282. doi:10.1109/JCSSE.2012.6261965. ISBN 978-1-4673-1921-8. S2CID 16666470.
- ^ "p": Short arm; "q": Long arm.
- ^ For cytogenetic banding nomenclature, see article locus.
- ^ a b These values (ISCN start/stop) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
- ^ gpos: Region which is positively stained by G banding, generally AT-rich and gene poor; gneg: Region which is negatively stained by G banding, generally CG-rich and gene rich; acen Centromere. var: Variable region; stalk: Stalk.
- Gilbert F (2001). "Chromosome 8". Genet Test. 5 (4): 345–54. doi:10.1089/109065701753617516. PMID 11960583.
- Nusbaum C; et al. (2006). "DNA sequence and analysis of human chromosome 8". Nature. 439 (7074): 331–5. Bibcode:2006Natur.439..331N. doi:10.1038/nature04406. PMID 16421571.
External links
- National Institutes of Health. "Chromosome 8". Genetics Home Reference. Archived from the original on October 14, 2004. Retrieved 2017-05-06.
- "Chromosome 8". Human Genome Project Information Archive 1990–2003. Retrieved 2017-05-06.