Baculoviridae
This section may be too technical for most readers to understand. (August 2009) |
| Baculoviridae | |
|---|---|
| Virus classification | |
| Group: | Group I (dsDNA)
|
| Family: | Baculoviridae
|
| Genera | |
|
Alphabaculovirus | |
Baculoviridae is a family of viruses. Arthropods, lepidoptera, hymenoptera, diptera, and decapoda serve as natural hosts. There are currently 49 species in this family, divided among 4 genera.[1][2]
Baculoviruses are known to infect invertebrates, with over 600 host species having been described. Immature (larval) forms of moth species are the most common hosts, but these viruses have also been found infecting sawflies, mosquitoes, and shrimp. Although baculoviruses are capable of entering mammalian cells in culture [3] they are not known to be capable of replication in mammalian or other vertebrate animal cells.
Starting in the 1940s they were used and studied widely as biopesticides in crop fields. Baculoviruses contain circular double-stranded genome ranging from 80–180 kbp.
Historical influence
The earliest records of baculoviruses can be found in the literature from as early as the sixteenth century in reports of “wilting disease” infecting silk-producing larva. Starting in the 1940s they were used and studied widely as biopesticides in crop fields. Since the 1990s they have been utilized for producing complex eukaryotic proteins in insect cell cultures (see Sf21, High Five cells). These recombinant proteins have been used in research and as vaccines in both human and veterinary medical treatments (for example, the most widely used vaccine for prevention of H5N1 avian influenza in chickens was produced in a baculovirus expression vector). More recently it has been found that baculoviruses can transduce mammalian cells with a suitable promoter.[4] These medical and potential medical uses have accelerated the number of publications on baculoviruses since 1995.
Baculovirus life cycle

The baculovirus life cycle involves two distinct forms of virus. Occlusion derived virus (ODV) is present in a protein matrix (polyhedrin or granulin) and is responsible for the primary infection of the host while the budded virus (BV) is released from the infected host cells later during the secondary infection.
Baculoviruses have very species-specific tropisms among the invertebrates with over 600 host species having been described. Immature (larval) forms of moth species are the most common hosts, but these viruses have also been found infecting sawflies, mosquitoes, and shrimp.
Typically, the initial infection occurs when a susceptible host insect feeds on plants that are contaminated with the occluded form of the virus. The protein matrix dissolves in the alkaline environment of the host midgut (stomach), releasing ODV that then fuse to the columnar epithelial cell membrane of the host intestine and are taken into the cell in endosomes. Nucleocapsids escape from the endosomes and are transported to nucleus. This step is possibly mediated by actin filaments. Viral transcription and replication occur in the cell nucleus and new BV particles are budded out from the basolateral side to spread the infection systemically. During budding, BV acquires a loosely fitting host cell membrane with expressed and displayed viral glycoproteins.
Baculovirus infection can be divided to three distinct phases:
- early (0–6 h post-infection),
- late (6–24 h p.i.), and
- very late phase (18–24 to 72 h p.i.).
While BV is produced in the late phase, the ODV form is produced in the very late phase acquiring the envelope from host cell nucleus and embedded in the matrix of occlusion body protein. These occlusion bodies are released when cells lyse to further spread baculovirus infection to next host. The extensive lysis of cells frequently causes the host insect to literally disintegrate, thus the reason for the historic name "wilting disease." the complete ODV-polyhedrin particles are resistant to heat and light inactivation, whereas the naked BV virion is more sensitive to environment.
Structure of the virion
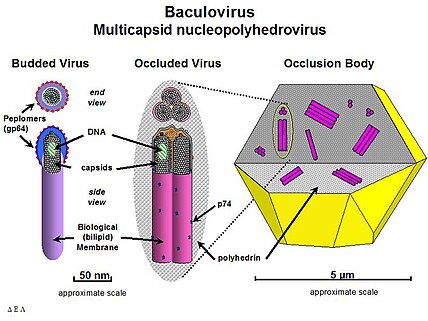
The most studied baculovirus is Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV). The virus was originally isolated from the alfalfa looper (a lepidopteran) and contains a 134-kbp genome with 154 open reading frames (ORF). The major capsid protein VP39 together with some minor proteins forms the nucleocapsid (21 nm x 260 nm) that encloses the DNA with p6.9 protein.
BV acquires its envelope from the cell membrane and requires a glycoprotein, gp64, to be able to spread systemic infection. This protein forms structures called peplomers on one end of the budded virus particle but is not found on ODV (although several other proteins are only associated with the ODV form). Some differences also exist in the lipid composition of the viral envelope of the two forms. While the BV envelope consists of phosphatidylserine, ODV contains phosphatidylcholine and phosphatidylethanolamine.
Major envelope glycoprotein gp64
During periods of evolution, the baculoviral envelope glycoproteins have undergone changes. Ld130, also known as baculovirus F-protein from Lymantria dispar (LdMNPV) is suggested to be an ancestral envelope fusion protein which has been replaced by non-orthologous gene replacement with gp64 in AcMNPV, Bombyx mori (BmNPV) and Orgyia pseudotsugata (OpMNPV) while they still retain the ld130 gene.
Gp64 is a homotrimeric membrane glycoprotein which is polarly present on the rod-shaped virion. It consists of 512 amino acids (aa) with four glycosylation sites at asparagine residues and has a N-terminal signal sequence (20 aa), oligomerization and fusion domain and a hydrophobic transmembrane domain near the C-terminus (7 aa).
It is produced in both early and late phases of the infection cycle with a maximal rate of synthesis occurring in 24–26 h p.i.. Trimerization with intermolecular cysteine-bonds seems to be a crucial step for protein transport to cell surface, since only 33% of synthesized protein reaches cell surface as monomeric gp64 is degraded within the cells.
Gp64 is essential for efficient budding of the virion and for the cell-to-cell transmission during the infection cycle as well as viral entry i.e. causing viral trophism and endosome mediated uptake to the cell. The major function of the gp64 envelope protein is to cause the pH-mediated envelope fusion to the endosome. Although gp64 has variety of essential functions, it has been reported that gp64-null baculoviruses can be substituted with other viral glycoproteins such as Ld130, G-protein of Vesicular stomatitis virus. These substitutions will result in functional virons.
Applications
Baculovirus expression in insect cells represents a robust method for producing recombinant glycoproteins.[5][6] Baculovirus-produced proteins are currently under study as therapeutic cancer vaccines with several immunologic advantages over proteins derived from mammalian sources.[7]
Biosafety
Baculoviruses are incapable of infecting mammals and plants.[8] They have a restricted range of hosts that they can infect that is typically restricted to a limited number of closely related insect species. Because baculoviruses are not harmful to humans, they are considered a safe option for use in research applications.
Taxonomy
This family has been divided into four genera: Alphabaculovirus (lepidopteran-specific nucleopolyhedroviruses), Betabaculovirus (lepidopteran-specific Granuloviruses), Gammabaculovirus (hymenopteran-specific nucleopolyhedroviruses) and Deltabaculovirus (dipteran-specific nucleopolyhedroviruses).[9]
Group: dsDNA
- Family: Baculoviridae
- Genus: Alphabaculovirus
- Adoxophyes honmai nucleopolyhedrovirus
- Agrotis ipsilon multiple nucleopolyhedrovirus
- Agrotis segetum nucleopolyhedrovirus
- Antheraea pernyi nucleopolyhedrovirus
- Anticarsia gemmatalis multiple nucleopolyhedrovirus
- Autographa californica multiple nucleopolyhedrovirus
- Bombyx mori nucleopolyhedrovirus
- Buzura suppressaria nucleopolyhedrovirus
- Choristoneura fumiferana DEF multiple nucleopolyhedrovirus
- Choristoneura fumiferana multiple nucleopolyhedrovirus
- Choristoneura rosaceana nucleopolyhedrovirus
- Chrysodeixis chalcites nucleopolyhedrovirus
- Clanis bilineata nucleopolyhedrovirus
- Ectropis obliqua nucleopolyhedrovirus
- Epiphyas postvittana nucleopolyhedrovirus
- Euproctis pseudoconspersa nucleopolyhedrovirus
- Helicoverpa armigera nucleopolyhedrovirus
- Hyphantria cunea nucleopolyhedrovirus
- Leucania separata nucleopolyhedrovirus
- Lymantria dispar multiple nucleopolyhedrovirus
- Mamestra brassicae multiple nucleopolyhedrovirus
- Mamestra configurata nucleopolyhedrovirus A
- Mamestra configurata nucleopolyhedrovirus B
- Maruca vitrata nucleopolyhedrovirus
- Orgyia pseudotsugata multiple nucleopolyhedrovirus
- Spodoptera exigua multiple nucleopolyhedrovirus
- Spodoptera frugiperda multiple nucleopolyhedrovirus
- Spodoptera littoralis nucleopolyhedrovirus
- Spodoptera litura nucleopolyhedrovirus
- Thysanoplusia orichalcea nucleopolyhedrovirus
- Trichoplusia ni single nucleopolyhedrovirus
- Wiseana signata nucleopolyhedrovirus
- Genus: Betabaculovirus
- Adoxophyes orana granulovirus
- Artogeia rapae granulovirus
- Choristoneura fumiferana granulovirus
- Cryptophlebia leucotreta granulovirus
- Cydia pomonella granulovirus
- Harrisina brillians granulovirus
- Helicoverpa armigera granulovirus
- Lacanobia oleracea granulovirus
- Phthorimaea operculella granulovirus
- Plodia interpunctella granulovirus
- Plutella xylostella granulovirus
- Pseudalatia unipuncta granulovirus
- Trichoplusia ni granulovirus
- Xestia c-nigrum granulovirus
Evolution
Baculoviruses are thought to have evolved from the Nudivirus family of viruses 310 million years ago.[10]
See also
References
This article includes a list of general references, but it lacks sufficient corresponding inline citations. (February 2008) |
- ^ "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ^ a b ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ^ Hofmann, C.; Sandig, V.; Jennings, G.; Rudolph, M.; Schlag, P.; Strauss, M. (1995). "Efficient Gene Transfer into Human Hepatocytes by Baculovirus Vectors". Proceedings of the National Academy of Sciences. 92 (22): 10099. doi:10.1073/pnas.92.22.10099.
- ^ Lackner, A; Genta, K; Koppensteiner, H; Herbacek, I; Holzmann, K; Spiegl-Kreinecker, S; Berger, W; Grusch, M (2008). "A bicistronic baculovirus vector for transient and stable protein expression in mammalian cells". Analytical Biochemistry. 380 (1): 146–8. doi:10.1016/j.ab.2008.05.020. PMID 18541133.
- ^ Altmann, Friedrich; Staudacher, E; Wilson, IB; März, L (1999). "Insect cells as hosts for the expression of recombinant glycoproteins". Glycoconjugate Journal. 16 (2): 109–23. doi:10.1023/A:1026488408951. PMID 10612411.
- ^ Kost, T; Condreay, JP (1999). "Recombinant baculoviruses as expression vectors for insect and mammalian cells". Current Opinion in Biotechnology. 10 (5): 428–33. doi:10.1016/S0958-1669(99)00005-1. PMID 10508635.
- ^ Betting, David J.; Mu, Xi Y.; Kafi, Kamran; McDonnel, Desmond; Rosas, Francisco; Gold, Daniel P.; Timmerman, John M. (2009). "Enhanced immune stimulation by a therapeutic lymphoma tumor antigen vaccine produced in insect cells involves mannose receptor targeting to antigen presenting cells". Vaccine. 27 (2): 250–9. doi:10.1016/j.vaccine.2008.10.055. PMC 2683685. PMID 19000731.
- ^ Ignoffo CM. (1975) Baculoviruses for Insect Pest Control: Safety Considerations, Summers M, Engler R, Falcon LA, Vail PV (eds.) American Society for Microbiology, Washington DC, p52
- ^ Jehle, JA; Blissard, GW; Bonning, BC; Cory, JS; Herniou, EA; Rohrmann, GF; Theilmann, DA; Thiem, SM; Vlak, JM; et al. (2006). "On the classification and nomenclature of baculoviruses: a proposal for revision". Arch Virol. 151 (7): 1257–1266. doi:10.1007/s00705-006-0763-6. PMID 16648963.
- ^ Theze, J.; Bezier, A.; Periquet, G.; Drezen, J.-M.; Herniou, E. A. (2011). "Paleozoic origin of insect large dsDNA viruses". Proceedings of the National Academy of Sciences. 108 (38): 15931–5. doi:10.1073/pnas.1105580108. PMC 3179036. PMID 21911395.
Further reading
- Federici, Brian A.; Granados, Robert R. (1986). The Biology of baculoviruses. Boca Raton: CRC Press. ISBN 0-8493-5988-0.
{{cite book}}: CS1 maint: multiple names: authors list (link) - Miller, Lois (1997). The baculoviruses. New York: Plenum Press. ISBN 0-306-45641-9.
External links
- Viralzone: Baculoviridae
- ICTV
- Index of Viruses - Baculoviridae (2006). In: ICTVdB - The Universal Virus Database, version 4. Büchen-Osmond, C (Ed), Columbia University, New York, USA. http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_index.htm
- http://www.ncbi.nlm.nih.gov/books/NBK49500/
- http://ir.library.oregonstate.edu/dspace/handle/1957/9989

