Nucleolus
This article may be too technical for most readers to understand. (October 2012) |

| Cell biology | |
|---|---|
| Animal cell diagram | |
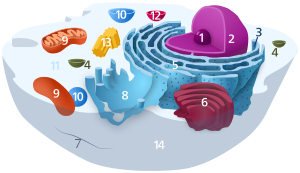 Components of a typical animal cell:
|
The nucleolus (/njuːˈkliːələs/ or /ˌnjuːkliˈoʊləs/, plural nucleoli /njuːˈkliːəˌlaɪ/ or /ˌnjuːkliˈoʊlaɪ/[1][2]) is the largest structure in the nucleus of eukaryotic cells, where it primarily serves as the site of ribosome synthesis and assembly. Nucleoli also have other important functions like assembly of signal recognition particles and playing a role in the cell's response to stress.[3] Nucleoli are made of proteins and RNA and form around specific chromosomal regions. Malfunction of nucleoli can be the cause of several human diseases.[citation needed]
History
Until 1964 little was known about the function of the nucleolus, even though it could be easily seen through microscopy and was recognized as part of the nucleus. During that year, a study of nucleoli by John Gurdon and Donald Brown in the African clawed frog Xenopus laevis generated increasing interest in the function and detailed structure of the nucleolus. They found that 25% of the frog eggs had no nucleolus and that such eggs were not capable of life. Half of the eggs had one nucleolus and 25% had two. They concluded that the nucleolus had a function necessary for life. In 1966 Max L. Birnstiel and Hugh Wallace showed via hybridization experiments that genes within nucleoli code for ribosomal RNA.
Structure
Three major components of the nucleolus are recognized: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC).[4] The DFC consists of newly transcribed rRNA bound to ribosomal proteins, while the GC contains RNA bound to ribosomal proteins that are being assembled into immature ribosomes.
However, it has been proposed that this particular organization is only observed in higher eukaryotes and that it evolved from a bipartite organization with the transition from anamniotes to amniotes. Reflecting the substantial increase in the DNA intergenic region, an original fibrillar component would have separated into the FC and the DFC.[5]
Another structure identified within many nucleoli (particularly in plants) is a clear area in the center of the structure referred to as a nucleolar vacuole.[6] Nucleoli of various plant species have been shown to have very high concentrations of iron[7] in contrast to human and animal cell nucleoli.
The nucleolus ultrastructure can be visualized through an electron microscope, while the organization and dynamics can be studied through fluorescent protein tagging and fluorescent recovery after photobleaching (FRAP). Antibodies against the PAF49 protein can also be used as a marker for the nucleolus in immunofluorescence experiments.[8]
Function and ribosome assembly

Nucleoli are formed around specific genetic loci called nucleolar organizing regions (NORs), first described by Barbara McClintock. Because of this non-random organization, the nucleolus is defined as a "genetically determined element."[9] An NOR is composed of tandem repeats of rDNA, which can be found on several different chromosomes. The human genome, for example, contains more than 200 clustered copies of rDNA on five different chromosomes (13, 14, 15, 21, 22). In a typical eukaryote rDNA consists of a promoter, internal and external transcribed spacers (ITS/ETS), rRNA coding sequences (18S, 5.8S, 28S) and an intergenic spacer.[10]
In ribosome biogenesis, two of the three eukaryotic RNA polymerases (pol I and III) are required, and these function in a coordinated manner. In an initial stage, the rRNA genes are transcribed as a single unit within the nucleolus by RNA polymerase I. In order for this transcription to occur, several pol I-associated factors and DNA-specific trans-acting factors are required. In yeast, the most important are: UAF (upstream activating factor), TBP (TATA-box binding protein), and CF (core factor), which bind promoter elements and form the preinitiation complex (PIC), which is in turn recognized by RNA pol. In humans, a similar PIC is assembled with SL1, the promoter selectivity factor (composed of TBP and TBP-associated factors, or TAFs), transcription initiation factors, and UBF (upstream binding factor). RNA polymerase I transcribes most rRNA transcripts (28S, 18S, and 5.8S) but the 5S rRNA subunit (component of the 60S ribosomal subunit) is transcribed by RNA polymerase III.[11]
Transcription of rRNA yields a long precursor molecule (45S pre-rRNA) which still contains the ITS and ETS. Further processing is needed to generate the 18S RNA, 5.8S and 28S RNA molecules. In eukaryotes, the RNA-modifying enzymes are brought to their respective recognition sites by interaction with guide RNAs, which bind these specific sequences. These guide RNAs belong to the class of small nucleolar RNAs (snoRNAs) which are complexed with proteins and exist as small-nucleolar-ribonucleoproteins (snoRNPs). Once the rRNA subunits are processed, they are ready to be assembled into larger ribosomal subunits. However, an additional rRNA molecule, the 5S rRNA, is also necessary. In yeast, the 5S rDNA sequence is localized in the intergenic spacer and is transcribed in the nucleolus by RNA pol.
In higher eukaryotes and plants, the situation is more complex, for the 5S DNA sequence lies outside the NOR and is transcribed by RNA pol III in the nucleoplasm, after which it finds its way into the nucleolus to participate in the ribosome assembly. This assembly not only involves the rRNA, but ribosomal proteins as well. The genes encoding these r-proteins are transcribed by pol II in the nucleoplasm by a "conventional" pathway of protein synthesis (transcription, pre-mRNA processing, nuclear export of mature mRNA and translation on cytoplasmic ribosomes). The mature r-proteins are then "imported" back into the nucleus and finally the nucleolus. Association and maturation of rRNA and r-proteins result in the formation of the 40S (small) and 60S (large) subunits of the complete ribosome. These are exported through the nuclear pore complexes to the cytoplasm, where they remain free or become associated with the endoplasmic reticulum, forming rough endoplasmic reticulum (RER).[10][12]
A continuous chain between the nucleoplasm and the inner parts of the nucleolus exists through a network of nucleolar channels. In this way, macromolecules with a molecular weight up to 2000 kDa are easily distributed throughout the nucleolus. [citation needed]
Sequestration of proteins
In addition to its role in ribosomal biogenesis, the nucleolus is known to capture and immobilize proteins, a process known as nucleolar detention. Proteins that are detained in the nucleolus are unable to diffuse and to interact with their binding partners. Targets of this post-translational regulatory mechanism include VHL, PML, MDM2, POLD1, RelA, HAND1 and hTERT, among many others. It is now known that long noncoding RNAs originating from intergenic regions of the nucleolus are responsible for this phenomenon.[13]
References
- ^ "nucleolus". Merriam-Webster.com Dictionary. Merriam-Webster.
- ^ "nucleolus - definition of nucleolus in English from the Oxford dictionary". OxfordDictionaries.com. Retrieved 20 January 2016.
- ^ Olsen, Mark OJ (December 2010). "Nucleolus: Structure and Function". eLS. doi:10.1002/9780470015902.a0005975.pub2. Retrieved 17 October 2014.
- ^ Koberna K, Malínský J, Pliss A, et al. (2002). "Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of "Christmas trees" in situ". J. Cell Biol. 157 (5): 743–8. doi:10.1083/jcb.200202007. PMC 2173423. PMID 12034768.
- ^ Thiry M, Lafontaine DL (2005). "Birth of a nucleolus: the evolution of nucleolar compartments". Trends Cell Biol. 15 (4): 194–9. doi:10.1016/j.tcb.2005.02.007. PMID 15817375. as PDF
- ^ Beven AF, Lee R, Razaz M, Leader DJ, Brown JW, Shaw PJ (1 June 1996). "The organization of ribosomal RNA processing correlates with the distribution of nucleolar snRNAs". J. Cell. Sci. 109 (6): 1241–51. PMID 8799814.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Roschzttardtz H, Grillet L, Isaure MP, et al. (2011). "Plant cell nucleolus as a hot spot for iron". J. Biol. Chem. 286 (32): 27863–6. doi:10.1074/jbc.C111.269720. PMC 3151030. PMID 21719700.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ PAF49 antibody | GeneTex Inc. Genetex.com. Retrieved on 2013-03-03.
- ^ Raška I, Shaw PJ, Cmarko D (2006). "New insights into nucleolar architecture and activity". Int. Rev. Cytol. International Review of Cytology. 255: 177–235. doi:10.1016/S0074-7696(06)55004-1. ISBN 9780123735997. PMID 17178467.
{{cite journal}}: CS1 maint: multiple names: authors list (link) as PDF - ^ a b Alberts, Bruce; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2002). Molecular Biology of the Cell (4th ed.). New York: Garland Science. pp. 331–3. ISBN 0-8153-3218-1.
- ^ Champe, Pamela C.; Harvey, Richard A.; Ferrier, Denise R. (2005). Lippincott's Illustrated Reviews: Biochemistry. Lippincott Williams & Wilkins. ISBN 978-0-7817-2265-0.
- ^ Cooper, Geoffrey M.; Hausman, Robert E. (2007). The Cell: A Molecular Approach (4th ed.). Sinauer Associates. pp. 371–9. ISBN 0878932208.
- ^ Audas TE, Jacob MD, Lee S (2012). "Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA". Mol Cell. 45 (2): 147–57. doi:10.1016/j.molcel.2011.12.012. PMID 22284675.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
External links
- Cooper, Geoffrey M. (2000). "The Nucleolus". The Cell: A Molecular Approach (2nd ed.). Sunderland MA: Sinauer Associates. ISBN 0-87893-106-6.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help) - Nucleolus under electron microscope II at uni-mainz.de
- Nuclear Protein Database – search under compartment
- Cell+Nucleolus at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- Histology image: 20104loa – Histology Learning System at Boston University
