Catalase: Difference between revisions
ClueBot NG (talk | contribs) m Reverting possible vandalism by 71.184.197.178 to version by Boghog. False positive? Report it. Thanks, ClueBot NG. (724554) (Bot) |
|||
| Line 29: | Line 29: | ||
== History == |
== History == |
||
| ⚫ | fudges a substance in 1818 when [[Louis Jacques Thénard]], who discovered H<sub>2</sub>O<sub>2</sub> ([[hydrogen peroxide]]), suggested that its breakdown is caused by a substance. In 1900, [[Oscar Loew]] was the first to give it the name catalase, and found its presence in many plants and animals.<ref name="pmid17751716">{{cite journal | author = Loew O | title = A New Enzyme of General Occurrence in Organisms | journal = Science | volume = 11 | issue = 279 | pages = 701–702 | year = 1900 | month = May | pmid = 17751716 | doi = 10.1126/science.11.279.701 | url = }}</ref> In 1937 catalase from beef liver was crystallised by [[James B. Sumner]]<ref name="pmid17776781">{{cite journal | author = Sumner JB, Dounce AL | title = Crystalline catalase | journal = Science | volume = 85 | issue = 2206 | pages = 366–367 | year = 1937 | month = April | pmid = 17776781 | doi = 10.1126/science.85.2206.366 | url = }}</ref> and the molecular weight was worked out in 1938.<ref name="pmid17831682">{{cite journal | author = Sumner JB, Gralén N | title = The molecular weight of crystalline catalase | journal = Science | volume = 87 | issue = 2256 | pages = 284–284 | year = 1938 | month = March | pmid = 17831682 | doi = 10.1126/science.87.2256.284 | url = }}</ref> |
||
| ⚫ | |||
In 1969, the [[amino acid]] sequence of [[bovine]] catalase was worked out.<ref name="pmid4892021">{{cite journal | author = Schroeder WA, Shelton JR, Shelton JB, Robberson B, Apell G | title = The amino acid sequence of bovine liver catalase: a preliminary report | journal = Arch. Biochem. Biophys. | volume = 131 | issue = 2 | pages = 653–655 | year = 1969 | month = May | pmid = 4892021 | doi = 10.1016/0003-9861(69)90441-X | url = }}</ref> Then in 1981, the 3D structure of the protein was revealed.<ref name="pmid7328661">{{cite journal | author = Murthy MR, Reid TJ, Sicignano A, Tanaka N, Rossmann MG | title = Structure of beef liver catalase | journal = J. Mol. Biol. | volume = 152 | issue = 2 | pages = 465–499 | year = 1981 | month = October | pmid = 7328661 | doi = 10.1016/0022-2836(81)90254-0 | url = }}</ref> |
In 1969, the [[amino acid]] sequence of [[bovine]] catalase was worked out.<ref name="pmid4892021">{{cite journal | author = Schroeder WA, Shelton JR, Shelton JB, Robberson B, Apell G | title = The amino acid sequence of bovine liver catalase: a preliminary report | journal = Arch. Biochem. Biophys. | volume = 131 | issue = 2 | pages = 653–655 | year = 1969 | month = May | pmid = 4892021 | doi = 10.1016/0003-9861(69)90441-X | url = }}</ref> Then in 1981, the 3D structure of the protein was revealed.<ref name="pmid7328661">{{cite journal | author = Murthy MR, Reid TJ, Sicignano A, Tanaka N, Rossmann MG | title = Structure of beef liver catalase | journal = J. Mol. Biol. | volume = 152 | issue = 2 | pages = 465–499 | year = 1981 | month = October | pmid = 7328661 | doi = 10.1016/0022-2836(81)90254-0 | url = }}</ref> |
||
Revision as of 15:23, 14 November 2011
| Catalase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | Catalase | ||||||||||
| Pfam | PF00199 | ||||||||||
| InterPro | IPR011614 | ||||||||||
| PROSITE | PDOC00395 | ||||||||||
| SCOP2 | 7cat / SCOPe / SUPFAM | ||||||||||
| |||||||||||
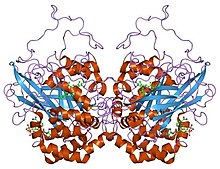
Template:PBB Catalase is a common enzyme found in nearly all living organisms that are exposed to oxygen, where it catalyzes the decomposition of hydrogen peroxide to water and oxygen.[1] Catalase has one of the highest turnover numbers of all enzymes; one catalase molecule can convert 40 million molecules of hydrogen peroxide to water and oxygen each second.[2]
Catalase is a tetramer of four polypeptide chains, each over 500 amino acids long.[3] It contains four porphyrin heme (iron) groups that allow the enzyme to react with the hydrogen peroxide. The optimum pH for human catalase is approximately 7,[4] and has a fairly broad maximum (the rate of reaction does not change appreciably at pHs between 6.8 and 7.5).[5] The pH optimum for other catalases varies between 4 and 11 depending on the species.[6] The optimum temperature also varies by species.[7]
History
fudges a substance in 1818 when Louis Jacques Thénard, who discovered H2O2 (hydrogen peroxide), suggested that its breakdown is caused by a substance. In 1900, Oscar Loew was the first to give it the name catalase, and found its presence in many plants and animals.[8] In 1937 catalase from beef liver was crystallised by James B. Sumner[9] and the molecular weight was worked out in 1938.[10]
In 1969, the amino acid sequence of bovine catalase was worked out.[11] Then in 1981, the 3D structure of the protein was revealed.[12]
Action
The reaction of catalase in the decomposition of hydrogen peroxide is:
- 2 H2O2 → 2 H2O + O2
The presence to catalse in a microbial or tissue sample can be tested by adding a volume of enzyme solution and observing the reaction. The formation of bubbles (oxygen) indicates a positive result. This is possible since catalase has a very high specific activity, which produces a detectable response.
Molecular mechanism
While the complete mechanism of catalase is not currently known,[13] the reaction is believed to occur in two stages:
- H2O2 + Fe(III)-E → H2O + O=Fe(IV)-E(.+)
- H2O2 + O=Fe(IV)-E(.+) → H2O + Fe(III)-E + O2[13]
- Here Fe()-E represents the iron center of the heme group attached to the enzyme. Fe(IV)-E(.+) is a mesomeric form of Fe(V)-E, meaning that iron is not completely oxidized to +V but receives some "supporting electron" from the heme ligand. This heme has to be drawn then as radical cation (.+).
As hydrogen peroxide enters the active site, it interacts with the amino acids Asn147 (asparagine at position 147) and His74, causing a proton (hydrogen ion) to transfer between the oxygen atoms. The free oxygen atom coordinates, freeing the newly-formed water molecule and Fe(IV)=O. Fe(IV)=O reacts with a second hydrogen peroxide molecule to reform Fe(III)-E and produce water and oxygen.[13] The reactivity of the iron center may be improved by the presence of the phenolate ligand of Tyr357 in the fifth iron ligand, which can assist in the oxidation of the Fe(III) to Fe(IV). The efficiency of the reaction may also be improved by the interactions of His74 and Asn147 with reaction intermediates.[13] In general, the rate of the reaction can be determined by the Michaelis-Menten equation.[14]
Catalase can also catalyze the oxidation, by hydrogen peroxide, of various metabolites and toxins, including formaldehyde, formic acid, phenols, acetaldehyde and alcohols. In does so according to the following reaction:
- H2O2 + H2R → 2H2O + R
The exact mechanism of this reaction is not known.
Any heavy metal ion (such as copper cations in copper(II) sulfate) can act as a noncompetitive inhibitor of catalase. Also, the poison cyanide is a competitive inhibitor of catalase, strongly binding to the heme of catalase and stopping the enzyme's action.
Three-dimensional protein structures of the peroxidated catalase intermediates are available at the Protein Data Bank. This enzyme is commonly used in laboratories as a tool for learning the effect of enzymes upon reaction rates.
Cellular role
Hydrogen peroxide is a harmful by-product of many normal metabolic processes: to prevent damage, it must be quickly converted into other, less dangerous substances. To this end, catalase is frequently used by cells to rapidly catalyze the decomposition of hydrogen peroxide into less reactive gaseous oxygen and water molecules.[15]
The true biological significance of catalase is not always straightforward to assess: Mice genetically engineered to lack catalase are phenotypically normal, indicating that this enzyme is dispensable in animals under some conditions.[16] A Catalase deficiency may increase the likelihood of developing Type II Diabetes.[17][18] Some human beings have very low levels of catalase (acatalasia), yet show few ill effects. It is likely that the predominant scavengers of H2O2 in normal mammalian cells are peroxiredoxins rather than catalase.[citation needed]
Human catalase works at an optimum temperature of 37 °C,[5] which is approximately the temperature of the human body. In contrast, catalase isolated from the hyperthermophile archaea Pyrobaculum calidifontis has a temperature optimum of 90 °C.[19]
Catalase is usually located in a cellular organelle called the peroxisome.[20] Peroxisomes in plant cells are involved in photorespiration (the use of oxygen and production of carbon dioxide) and symbiotic nitrogen fixation (the breaking apart of diatomic nitrogen (N2) to reactive nitrogen atoms). Hydrogen peroxide is used as a potent antimicrobial agent when cells are infected with a pathogen. Pathogens that are catalase-positive, such as Mycobacterium tuberculosis, Legionella pneumophila, and Campylobacter jejuni, make catalase in order to deactivate the peroxide radicals, thus allowing them to survive unharmed within the host.[21]
Catalase contributes to ethanol metabolism in the body after ingestion of alcohol, however it only breaks down a small fraction of the alcohol in the body.[22]
Distribution among organisms
All known animals use catalase in every organ, with particularly high concentrations occurring in the liver. One unique use of catalase occurs in bombardier beetle. The beetle has two sets of chemicals ordinarily stored separately in its paired glands. The larger of the pair, the storage chamber or reservoir, contains hydroquinones and hydrogen peroxide, whereas the smaller of the pair, the reaction chamber, contains catalases and peroxidases. To activate the noxious spray, the beetle mixes the contents of the two compartments, causing oxygen to be liberated from hydrogen peroxide. The oxygen oxidizes the hydroquinones and also acts as the propellant.[23] The oxidation reaction is very exothermic (ΔH = −202.8 kJ/mol) which rapidly heats the mixture to the boiling point.[24]
Catalase is also universal among plants, and many fungi are also high producers of the enzyme.[25]
Very few aerobic microorganisms that do not use catalase are known. Catalase has also been observed in some anaerobic microorganisms, such as Methanosarcina barkeri.[26]
Applications

Catalase is used in the food industry for removing hydrogen peroxide from milk prior to cheese production.[27] Another use is in food wrappers where it prevents food from oxidizing.[28] Catalase is also used in the textile industry, removing hydrogen peroxide from fabrics to make sure the material is peroxide-free.[29]
A minor use is in contact lens hygiene - a few lens-cleaning products disinfect the lens using a hydrogen peroxide solution; a solution containing catalase is then used to decompose the hydrogen peroxide before the lens is used again.[30] Recently, catalase has also begun to be used in the aesthetics industry. Several mask treatments combine the enzyme with hydrogen peroxide on the face with the intent of increasing cellular oxygenation in the upper layers of the epidermis.
Catalase test

The catalase test is also one of the main three tests used by microbiologists to identify species of bacteria. The presence of catalase enzyme in the test isolate is detected using hydrogen peroxide. If the bacteria possess catalase (i.e., are catalase-positive), when a small amount of bacterial isolate is added to hydrogen peroxide, bubbles of oxygen are observed.
In microbiology, the catalase test is used to differentiate between bacterial species in the lab. The test is done by placing a drop of hydrogen peroxide on a microscope slide. Using an applicator stick, a scientist touches the colony and then smears a sample into the hydrogen peroxide drop.
- If bubbles or froth forms, the organism is said to be catalase-positive. Staphylococci[31] and Micrococci[32] are catalase-positive. Other catalase positive organisms include Listeria, Corynebacterium diphtheriae, Burkholderia cepacia, Nocardia, the Family Enterobacteriaceae (Citrobacter, E.Coli, Enterobacter, Klebsiella, Shigella, Yersinia, Proteus, Salmonella, Serratia, Pseudomonas), Mycobacterium tuberculosis, Aspergillus, and Cryptococcus.
- If not, the organism is catalase-negative. Streptococci[33] and Enterococci are catalase-negative.
While the catalase test alone cannot identify a particular organism, combined with other tests such as antibiotic resistance, it can aid diagnosis. The presence of catalase in bacterial cells depends on both the growth condition and the medium used to grow the cells.
Grey hair
According to recent scientific studies, low levels of catalase may play a role in the greying process of human hair. Hydrogen peroxide is naturally produced by the body and catalase breaks it down. If there is a dip in catalase levels, hydrogen peroxide cannot be broken down. This causes the hydrogen peroxide to bleach the hair from the inside out. This finding may someday be incorporated into anti-greying treatments for aging hair.[34][35][36]
Role in disease
The peroxisomal disorder acatalasia is due to a deficiency in the function of catalase. Genetic polymorphisms in catalase and its altered expression and activity are associated with oxidative DNA damage and subsequently the individual’s risk of cancer susceptibility.[37]
Interactions
Catalase has been shown to interact with ABL2[38] and Abl gene.[38]
See also
References
- ^ Chelikani P, Fita I, Loewen PC (2004). "Diversity of structures and properties among catalases". Cell. Mol. Life Sci. 61 (2): 192–208. doi:10.1007/s00018-003-3206-5. PMID 14745498.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Goodsell DS (2004-09-01). "Catalase". Molecule of the Month. RCSB Protein Data Bank. Retrieved 2007-02-11.
- ^ Boon EM, Downs A, Marcey D. "Catalase: H2O2: H2O2 Oxidoreductase". Catalase Structural Tutorial Text. Retrieved 2007-02-11.
{{cite web}}: CS1 maint: multiple names: authors list (link) - ^ Maehly A, Chance B (1954). "The assay of catalases and peroxidases". Methods Biochem Anal. Methods of Biochemical Analysis. 1: 357–424. doi:10.1002/9780470110171.ch14. ISBN 9780470110171. PMID 13193536.
- ^ a b Aebi H (1984). Aebi, Hugo (ed.). "Catalase in vitro". Meth. Enzymol. Methods in Enzymology. 105: 121–126. doi:10.1016/S0076-6879(84)05016-3. ISBN 012182005X. PMID 6727660.
- ^ "EC 1.11.1.6 - catalase". BRENDA: The Comprehensive Enzyme Information System. Department of Bioinformatics and Biochemistry, Technische Universität Braunschweig. Retrieved 2009-05-26.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Toner K, Sojka G, Ellis R. "A Quantitative Enzyme Study; CATALASE". bucknell.edu. Archived from the original on 2000-06-12. Retrieved 2007-02-11.
{{cite web}}: CS1 maint: multiple names: authors list (link) - ^ Loew O (1900). "A New Enzyme of General Occurrence in Organisms". Science. 11 (279): 701–702. doi:10.1126/science.11.279.701. PMID 17751716.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Sumner JB, Dounce AL (1937). "Crystalline catalase". Science. 85 (2206): 366–367. doi:10.1126/science.85.2206.366. PMID 17776781.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Sumner JB, Gralén N (1938). "The molecular weight of crystalline catalase". Science. 87 (2256): 284–284. doi:10.1126/science.87.2256.284. PMID 17831682.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Schroeder WA, Shelton JR, Shelton JB, Robberson B, Apell G (1969). "The amino acid sequence of bovine liver catalase: a preliminary report". Arch. Biochem. Biophys. 131 (2): 653–655. doi:10.1016/0003-9861(69)90441-X. PMID 4892021.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Murthy MR, Reid TJ, Sicignano A, Tanaka N, Rossmann MG (1981). "Structure of beef liver catalase". J. Mol. Biol. 152 (2): 465–499. doi:10.1016/0022-2836(81)90254-0. PMID 7328661.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c d Boon EM, Downs A, Marcey D. "Proposed Mechanism of Catalase". Catalase: H2O2: H2O2 Oxidoreductase: Catalase Structural Tutorial. Retrieved 2007-02-11.
{{cite web}}: CS1 maint: multiple names: authors list (link) - ^ Maass E (1998-07-19). "How does the concentration of hydrogen peroxide affect the reaction". MadSci Network. Retrieved 009-03-02.
{{cite web}}: Check date values in:|accessdate=(help); Cite has empty unknown parameter:|coauthors=(help) - ^ Gaetani G, Ferraris A, Rolfo M, Mangerini R, Arena S, Kirkman H (1996). "Predominant role of catalase in the disposal of hydrogen peroxide within human erythrocytes". Blood. 87 (4): 1595–9. PMID 8608252.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Ho YS, Xiong Y, Ma W, Spector A, Ho D (2004). "Mice Lacking Catalase Develop Normally but Show Differential Sensitivity to Oxidant Tissue Injury". J Biol Chem. 279 (31): 32804–32812. doi:10.1074/jbc.M404800200. PMID 15178682.
{{cite journal}}: CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ László Góth, Ágota Lenkey, William N. Bigler (2001). "Blood Catalase Deficiency and Diabetes in Hungary". Diabetes Care. 24 (10): 1839–1840. doi:10.2337/diacare.24.10.1839. PMID 11574451.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ László Góth (2008). "Catalase Deficiency and Type 2 Diabetes". Diabetes Care. 24 (10): e93–e93. doi:10.2337/dc08-1607. PMID 19033415.
- ^ Amo T, Atomi H, Imanaka T (2002). "Unique presence of a manganese catalase in a hyperthermophilic archaeon, Pyrobaculum calidifontis VA1". J. Bacteriol. 184 (12): 3305–3312. doi:10.1128/JB.184.12.3305-3312.2002. PMC 135111. PMID 12029047.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "Peroxisomes". Molecular Biology of the Cell (4th ed.). New York: Garland Science. ISBN 0-8153-3218-1.
{{cite book}}: External link in|chapterurl=|chapterurl=ignored (|chapter-url=suggested) (help)CS1 maint: multiple names: authors list (link) - ^ Srinivasa Rao PS, Yamada Y, Leung KY (2003). "A major catalase (KatB) that is required for resistance to H2O2 and phagocyte-mediated killing in Edwardsiella tarda". Microbiology (Reading, Engl.). 149 (Pt 9): 2635–2644. doi:10.1099/mic.0.26478-0. PMID 12949187.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ http://pubs.niaaa.nih.gov/publications/AA72/AA72.htm
- ^ Eisner T, Aneshansley DJ (1999). "Spray aiming in the bombardier beetle: photographic evidence". Proc. Natl. Acad. Sci. U.S.A. 96 (17): 9705–9709. doi:10.1073/pnas.96.17.9705. PMC 22274. PMID 10449758.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Beheshti N, McIntosh AC (2006). Int. Journal of Design & Nature. 1 (1): 1–9 http://www.heveliusforum.org/Artykuly/Biomimetics.pdf.
{{cite journal}}: Missing or empty|title=(help) - ^ Isobe K, Inoue N, Takamatsu Y, Kamada K, Wakao N (2006). "Production of catalase by fungi growing at low pH and high temperature". J. Biosci. Bioeng. 101 (1): 73–76. doi:10.1263/jbb.101.73. PMID 16503295.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Brioukhanov AL, Netrusov AI, Eggen RI (2006). "The catalase and superoxide dismutase genes are transcriptionally up-regulated upon oxidative stress in the strictly anaerobic archaeon Methanosarcina barkeri". Microbiology (Reading, Engl.). 152 (Pt 6): 1671–1677. doi:10.1099/mic.0.28542-0. PMID 16735730.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "Catalase". Worthington Enzyme Manual. Worthington Biochemical Corporation. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Hengge A (1999-03-16). "Re: how is catalase used in industry?". General Biology. MadSci Network. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ "textile industry". Case study 228. International Cleaner Production Information Clearinghouse. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ US patent 5521091, Cook JN, Worsley JL, "Compositions and method for destroying hydrogen peroxide on contact lens", issued 1996-05-28
- ^ Rollins DM (2000-08-01). "Bacterial Pathogen List". BSCI 424 Pathogenic Microbiology. University of Maryland. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Johnson M. "Catalase Production". Biochemical Tests. Mesa Community College. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Fox A. "Streptococcus pneumoniae and Staphylococci". University of South Carolina. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ "Why Hair Turns Gray Is No Longer A Gray Area: Our Hair Bleaches Itself As We Grow Older". Science News. ScienceDaily. 2009-02-24. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Hitti M (2009-02-25). "Why Hair Goes Gray". Health News. WebMD. Retrieved 2009-03-01.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - ^ Wood JM, Decker H, Hartmann H, Chavan B, Rokos H, Spencer JD, Hasse S, Thornton MJ, Shalbaf M, Paus R, Schallreuter KU (2009). "Senile hair graying: H2O2-mediated oxidative stress affects human hair color by blunting methionine sulfoxide repair". FASEB J. 23 (7): 2065–2075. doi:10.1096/fj.08-125435. PMID 19237503.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Khan MA, Tania M, Zhang D, Chen H (2010). "Antioxidant enzymes and cancer". Chin J Cancer Res. 22 (2): 87–92. doi:10.1007/s11670-010-0087-7.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Cao, Cheng (2003). "Catalase activity is regulated by c-Abl and Arg in the oxidative stress response". J. Biol. Chem. 278 (32). United States: 29667–29675. doi:10.1074/jbc.M301292200. ISSN 0021-9258. PMID 12777400.
{{cite journal}}: Cite has empty unknown parameters:|laydate=,|laysummary=, and|laysource=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link)
External links
- "GenAge entry for CAT (Homo sapiens)". Human Ageing Genomic Resources. Retrieved 2009-03-05.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "Catalase". MadSci FAQ. madsci.org. Retrieved 2009-03-05.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "Catalase and oxidase test video". Regnvm Prokaryotae. Retrieved 2009-03-05.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "EC 1.11.1.6 - catalase". Brenda: The Comprehensive Enzyme Information System. Retrieved 2009-03-05.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "PeroxiBase - The peroxidase database". Swiss Institute of Bioinformatics. Retrieved 2009-03-05.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "Catalase Procedure". MicrobeID.com. Retrieved 2009-04-22.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
