Herpesvirales: Difference between revisions
→Taxonomy: expansion |
Citation bot (talk | contribs) Add: doi-access, authors 1-1. Removed parameters. Some additions/deletions were parameter name changes. | Use this bot. Report bugs. | #UCB_CommandLine |
||
| (35 intermediate revisions by 17 users not shown) | |||
| Line 1: | Line 1: | ||
{{Short description|Order of viruses}} |
|||
{{Virusbox |
{{Virusbox |
||
| image |
| image = Herpesvirales virion diagram.jpg |
||
| image_caption |
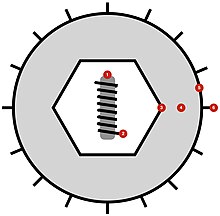
| image_caption = Simplified diagram of ''Herpesvirales'' virion structure in cross-section. (1) nucleoprotein, (2) DNA, (3) capsid, (4) tegument, (5) envelope, (6) glycoprotein. Click to enlarge. |
||
| image2 |
| image2 = Herpesviridae EM PHIL 2171 lores.jpg |
||
| image2_caption |
| image2_caption = [[Electron micrograph]] of various viruses from the ''[[Orthoherpesviridae]]'' family including ''[[Human alphaherpesvirus 3]]'' ([[Chickenpox]]), ''[[Human alphaherpesvirus 1]]'', and ''[[Human alphaherpesvirus 2]]'' |
||
| |
| taxon = Herpesvirales |
||
| taxon = Herpesvirales |
|||
| subdivision_ranks = Families |
| subdivision_ranks = Families |
||
| subdivision |
| subdivision = |
||
* ''[[Alloherpesviridae]]'' |
* ''[[Alloherpesviridae]]'' |
||
* ''[[ |
* ''[[Orthoherpesviridae]]'' |
||
* ''[[Malacoherpesviridae]]'' |
* ''[[Malacoherpesviridae]]'' |
||
And see text |
And see text |
||
}} |
}} |
||
The '''''Herpesvirales''''' is an order of dsDNA [[virus]]es (Baltimore group I) with animal hosts, characterised by a common morphology consisting of an [[icosahedron|icosahedral]] [[capsid]] enclosed in a glycoprotein-containing lipid envelope. Common infections in humans caused by members of this order include [[cold sores]], [[genital herpes]], [[chickenpox]], [[shingles]], and [[glandular fever]]. |
The '''''Herpesvirales''''' is an order of dsDNA [[virus]]es (Baltimore group I) with animal hosts, characterised by a common morphology consisting of an [[icosahedron|icosahedral]] [[capsid]] enclosed in a glycoprotein-containing lipid envelope. Common infections in humans caused by members of this order include [[cold sores]], [[genital herpes]], [[chickenpox]], [[shingles]], and [[glandular fever]]. ''Herpesvirales'' is the sole order in the class '''''Herviviricetes''''', which is the sole class in the phylum '''''Peploviricota'''''. |
||
== Virology == |
== Virology == |
||
=== Morphology === |
=== Morphology === |
||
All members of the order have a virion structure that consists of a DNA core surrounded by an icosahedral capsid composed of 12 pentavalent and 150 hexavalent [[capsomere]]s (T = 16). The capsid has a diameter of ~110 nanometers (nm) and is embedded in a proteinaceous matrix called the tegument, which in its turn is enclosed by a glycoprotein-containing lipid envelope with a diameter of about 200 nm. The DNA genome is linear and double stranded, with sizes in the range 125–290 kbp.<ref name=":0">{{Cite journal| |
All members of the order have a virion structure that consists of a DNA core surrounded by an icosahedral capsid composed of 12 pentavalent and 150 hexavalent [[capsomere]]s (T = 16). The capsid has a diameter of ~110 nanometers (nm) and is embedded in a proteinaceous matrix called the [[Viral tegument|tegument]], which in its turn is enclosed by a glycoprotein-containing lipid envelope with a diameter of about 200 nm. The DNA genome is linear and double stranded, with sizes in the range 125–290 kbp.<ref name=":0">{{Cite journal|last1=Davison|first1=Andrew J.|last2=Eberle|first2=Richard|last3=Ehlers|first3=Bernhard|last4=Hayward|first4=Gary S.|last5=McGeoch|first5=Duncan J.|last6=Minson|first6=Anthony C.|last7=Pellett|first7=Philip E.|last8=Roizman|first8=Bernard|last9=Studdert|first9=Michael J.|date=2009|title=The order Herpesvirales|journal=Archives of Virology|volume=154|issue=1|pages=171–177|doi=10.1007/s00705-008-0278-4|issn=1432-8798|pmc=3552636|pmid=19066710}}</ref> The genome contains terminal and internal reiterated sequences, with their number and disposition varying depending on the different subclades. |
||
=== Hosts === |
=== Hosts === |
||
[[File:Herpes(PHIL 1573 lores).jpg|thumb|A [[Herpes labialis|cold sore]]]] |
[[File:Herpes(PHIL 1573 lores).jpg|thumb|A [[Herpes labialis|cold sore]]]] |
||
[[File:Duck plague victim.JPG|thumb|Herpesviruses may be highly virulent in some species. This dying mallard infected with ''[[Anatid alphaherpesvirus 1]]'' is emitting a bloody nasal discharge on the ice.]] |
[[File:Duck plague victim.JPG|thumb|Herpesviruses may be highly virulent in some species. This dying mallard infected with ''[[Anatid alphaherpesvirus 1]]'' is emitting a bloody nasal discharge on the ice.]] |
||
All species in this order have animal hosts. The ''Malacoherpesviridae'' infect molluscs (abalone and oysters), the ''Alloherpesviridae'' infect [[anamniote]]s (frogs and fish), and the ''Herpesviridae'' infect [[amniote]]s (reptiles, birds, and mammals). Within the family ''Herpesviridae'', genera ''[[Iltovirus]]'' and ''[[Mardivirus]]'', and genus ''[[Scutavirus]]'' of the subfamily ''Alphaherpesvirinae'' infect birds and reptiles, respectively. All other genera in the ''Herpesviridae'' infect mammals only.<ref>{{Cite book|url=https://archive.org/details/originevolutionv00domi_525|url-access=limited|title=Origin and Evolution of Viruses|last1=McGeoch|first1=Duncan J.|last2=Davison|first2=Andrew J.|last3=Dolan|first3=Aidan|last4=Gatherer|first4=Derek|last5=Sevilla-Reyes|first5=Edgar E.|date=2008|publisher=Academic Press|isbn=9780123741530|editor-last=DOMINGO|editor-first=ESTEBAN|location=London|pages=[https://archive.org/details/originevolutionv00domi_525/page/n444 447]–475|editor-last2=PARRISH|editor-first2=COLIN R.|editor-last3=HOLLAND|editor-first3=JOHN J.|edition=Second}}</ref> |
|||
== Taxonomy == |
== Taxonomy == |
||
As of |
As of 2020, the order is made up of three families, three subfamilies, 23 genera, and 130 species. The taxonomy is shown from family, to subfamily, to genus below.<ref>{{cite web |title=Virus Taxonomy: 2020 Release |url=https://ictv.global/taxonomy |publisher=International Committee on Taxonomy of Viruses (ICTV) |date=March 2021 |access-date=10 May 2021}}</ref> |
||
* ''Alloherpesviridae'' |
* ''[[Alloherpesviridae]]'' |
||
::* ''[[Batrachovirus]]'' |
::* ''[[Batrachovirus]]'' |
||
::* ''[[Cyprinivirus]]'' |
::* ''[[Cyprinivirus]]'' |
||
::* ''[[Ictalurivirus]]'' |
::* ''[[Ictalurivirus]]'' |
||
::* ''[[Salmonivirus]]'' |
::* ''[[Salmonivirus]]'' |
||
* '' |
* ''[[Orthoherpesviridae]]'' |
||
:* ''[[Alphaherpesvirinae]] |
:* ''[[Alphaherpesvirinae]]'' |
||
::* ''[[Iltovirus]]'' |
::* ''[[Iltovirus]]'' |
||
::* ''[[Mardivirus]]'' |
::* ''[[Mardivirus]]'' |
||
| Line 44: | Line 44: | ||
::* ''[[Muromegalovirus]]'' |
::* ''[[Muromegalovirus]]'' |
||
::* ''[[Proboscivirus]]'' |
::* ''[[Proboscivirus]]'' |
||
::* ''[[Quwivirus]]'' |
|||
::* ''[[Roseolovirus]]'' |
::* ''[[Roseolovirus]]'' |
||
:* ''[[Gammaherpesvirinae]]'' |
:* ''[[Gammaherpesvirinae]]'' |
||
::* ''[[Bossavirus]]'' |
|||
::* ''[[Lymphocryptovirus]]'' |
::* ''[[Lymphocryptovirus]]'' |
||
::* ''[[Macavirus]]'' |
::* ''[[Macavirus]]'' |
||
::* ''[[Manticavirus]]'' |
|||
::* ''[[Patagivirus]]'' |
|||
::* ''[[Percavirus]]'' |
::* ''[[Percavirus]]'' |
||
::* ''[[Rhadinovirus]]'' |
::* ''[[Rhadinovirus]]'' |
||
* ''Malacoherpesviridae'' |
* ''[[Malacoherpesviridae]]'' |
||
::* ''[[Aurivirus]]'' |
::* ''[[Aurivirus]]'' |
||
::* ''[[Ostreavirus]]'' |
::* ''[[Ostreavirus]]'' |
||
=== History === |
=== History === |
||
The [[herpesvirus]] was first isolated from the [[blue wildebeest]] in 1960 by veterinary scientist [[Walter Plowright]].<ref>{{cite book|last=O.A.|first=Ryder|title=One Medicine: A Tribute to Kurt Benirschke, Director Center for Reproduction of Endangered Species Zoological Society of San Diego and Professor of Pathology and Reproductive Medicine University of California San Diego from his Students and Colleagues |year=1984 |publisher=Springer |location=Berlin, Heidelberg |isbn=978-3-642-61749-2 |pages=296–308 |author2=Byrd, M.L.}}</ref> The genus ''Herpesvirus'' was established in 1971 in the first report of the [[International Committee on Taxonomy of Viruses]] (ICTV) |
The [[herpesvirus]] was first isolated from the [[blue wildebeest]] in 1960 by veterinary scientist [[Walter Plowright]].<ref>{{cite book|last=O.A.|first=Ryder|title=One Medicine: A Tribute to Kurt Benirschke, Director Center for Reproduction of Endangered Species Zoological Society of San Diego and Professor of Pathology and Reproductive Medicine University of California San Diego from his Students and Colleagues |year=1984 |publisher=Springer |location=Berlin, Heidelberg |isbn=978-3-642-61749-2 |pages=296–308 |author2=Byrd, M.L.}}</ref> The genus ''Herpesvirus'' was established in 1971 in the first report of the [[International Committee on Taxonomy of Viruses]] (ICTV). This genus consisted of 23 viruses and 4 groups of viruses. In the second ICTV report in 1976 this genus was elevated to family level - the ''Herpetoviridae''. Because of possible confusion with viruses derived from [[reptile]]s this name was changed in the third report in 1979 to ''Herpesviridae''. In this report the family ''Herpesviridae'' was divided into 3 subfamilies (''Alphaherpesvirinae'', ''Betaherpesvirinae'' and ''Gammaherpesvirinae'') and 5 unnamed genera: 21 viruses were listed. In 2009 the family ''Herpesviridae'' was elevated to the order ''Herpesvirales''. This elevation was necessitated by the discovery that the herpesviruses of [[fish]] and [[mollusc]]s were only distantly related to those of [[bird]]s and [[mammal]]s.<ref>{{Cite book|title=Origin and Evolution of Viruses|last=Duncan J. McGeoch, Andrew J. Davison, Aidan Dolan, Derek Gatherer, Edgar E. Sevilla-Reyes|publisher=Academic Press|year=2008|pages=Chapter 20: 447–475|edition=Second}}</ref> Two new families were created - the family ''Alloherpesviridae'' which incorporates bony fish and frog viruses and the family ''Malacoherpesviridae'' which contains those of molluscs. |
||
=== Phylogenetics === |
=== Phylogenetics === |
||
The only protein with widespread conservation amongst all members of the order, albeit only at the amino-acid level, is the ATPase subunit of the DNA terminase;<ref name=":0" /> the latter is involved in the packaging of the DNA during virion assembly.<ref>{{Cite journal| |
The only protein with widespread conservation amongst all members of the order, albeit only at the amino-acid level, is the ATPase subunit of the DNA terminase;<ref name=":0" /> the latter is involved in the packaging of the DNA during virion assembly.<ref>{{Cite journal|last1=Selvarajan Sigamani|first1=Sundaresan|last2=Zhao|first2=Haiyan|last3=Kamau|first3=Yvonne N.|last4=Baines|first4=Joel D.|last5=Tang|first5=Liang|date=June 2013|title=The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses|journal=Journal of Virology|volume=87|issue=12|pages=7140–7148|doi=10.1128/JVI.00311-13|issn=1098-5514|pmc=3676077|pmid=23596306}}</ref> |
||
Phylogenies constructed with the conserved regions of the ATPase subunit of the DNA terminase suggest that ''Alloherpesviridae'' is the basal [[clade]] of the order, and that ''Herpesviridae'' and ''Malacoherpesviridae'' are sister clades.<ref>{{Cite journal| |
Phylogenies constructed with the conserved regions of the ATPase subunit of the DNA terminase suggest that ''Alloherpesviridae'' is the basal [[clade]] of the order, and that ''Herpesviridae'' and ''Malacoherpesviridae'' are sister clades.<ref>{{Cite journal|last1=Rakus|first1=Krzysztof|last2=Ouyang|first2=Ping|last3=Boutier|first3=Maxime|last4=Ronsmans|first4=Maygane|last5=Reschner|first5=Anca|last6=Vancsok|first6=Catherine|last7=Jazowiecka-Rakus|first7=Joanna|last8=Vanderplasschen|first8=Alain|date=2013|title=Cyprinid herpesvirus 3: an interesting virus for applied and fundamental research|journal=Veterinary Research|volume=44|issue=1|pages=85|doi=10.1186/1297-9716-44-85|issn=0928-4249|pmc=3850573|pmid=24073814 |doi-access=free }}</ref> Given the phylogenetic distances between vertebrates and molluscs, this suggests that herpesviruses were initially fish viruses and that they have evolved with their hosts to infect other vertebrates.{{citation needed|date=July 2017}} |
||
=== Species nomenclature === |
=== Species nomenclature === |
||
The ''Herpesvirales'' naming system originated in 1973 and has been elaborated considerably since. All herpesviruses described since this system was adopted have been named in accordance with it. The recommended naming system specifies that each species name consists of three parts: a first word, a second word, and finally a number. |
The ''Herpesvirales'' naming system originated in 1973 and has been elaborated considerably since. All herpesviruses described since this system was adopted have been named in accordance with it. The recommended naming system specifies that each species name consists of three parts: a first word, a second word, and finally a number. |
||
The first word should be derived from the taxon (family or subfamily) to which its primary natural host belongs. The subfamily name is used for viruses from members of the family ''[[Bovidae]]'' or from [[primate]]s (the virus name ending in –ine, e.g. bovine), and the host family name for other viruses (ending in –id, e.g. equid). Human herpesviruses have been treated as an exception (''human'' rather than ''hominid''). Following the host-derived term, species in the family ''Herpesviridae'', which are divided into subfamilies ''Alphaherpesvirinae'', ''Betaherpesvirinae'', and ''Gammaherpesvirinae'', will have the word ''alphaherpesvirus'', ''betaherpesvirus'', or ''gammaherpesvirus'' added, respectively. Species in the ''Herpesviridae'' that have not been assigned to a subfamily, and species in ''Alloherpesviridae'' and ''Malacoherpesviridae'' will have ''herpesvirus'' following the host-derived term instead. Finally an Arabic number (''1'', ''2'', ''3'', and so |
The first word should be derived from the taxon (family or subfamily) to which its primary natural host belongs. The subfamily name is used for viruses from members of the family ''[[Bovidae]]'' or from [[primate]]s (the virus name ending in –ine, e.g. bovine), and the host family name for other viruses (ending in –id, e.g. equid). Human herpesviruses have been treated as an exception (''human'' rather than ''hominid''). Following the host-derived term, species in the family ''Herpesviridae'', which are divided into subfamilies ''Alphaherpesvirinae'', ''Betaherpesvirinae'', and ''Gammaherpesvirinae'', will have the word ''alphaherpesvirus'', ''betaherpesvirus'', or ''gammaherpesvirus'' added, respectively. Species in the ''Herpesviridae'' that have not been assigned to a subfamily, and species in ''Alloherpesviridae'' and ''Malacoherpesviridae'' will have ''herpesvirus'' following the host-derived term instead. Finally an Arabic number (''1'', ''2'', ''3'', and so forth) is appended. Numbers are assigned in order of naming and bear no implied meaning about the taxonomic or biological properties of the virus. |
||
An example of such a name is ''[[Canid alphaherpesvirus 1]]''. From this name it can be ascertained |
An example of such a name is ''[[Canid alphaherpesvirus 1]]''. From this name it can be ascertained that the virus' primary natural host is a canid (i.e. a member of the family [[Canidae]]; dogs etc.), that it is a member of the family ''Herpesviridae'' and subfamily ''Alphaherpesvirinae'', and that it is the first herpesvirus named for which canids serve as primary natural hosts. |
||
A number of virus names (e.g. [[Epstein–Barr virus]], also known as ''Human gammaherpesvirus 4'') are so widely used that it may be impractical to attempt to insist on their replacement. This has led to a dual nomenclature in the literature for some herpesviruses. |
A number of virus names (e.g. [[Epstein–Barr virus]], also known as ''Human gammaherpesvirus 4'') are so widely used that it may be impractical to attempt to insist on their replacement. This has led to a dual nomenclature in the literature for some herpesviruses. |
||
== |
==References== |
||
{{Reflist}} |
{{Reflist}} |
||
| Line 77: | Line 81: | ||
{{Commons category}} |
{{Commons category}} |
||
{{Wikispecies}} |
{{Wikispecies}} |
||
* [http://talk.ictvonline.org/media/p/174/download.aspx ICTV Accepted Taxonomic Proposals from the ''Herpesviridae'' Study Group (2003–2005)] |
* [http://talk.ictvonline.org/media/p/174/download.aspx ICTV Accepted Taxonomic Proposals from the ''Herpesviridae'' Study Group (2003–2005)] {{Webarchive|url=https://web.archive.org/web/20200615193030/https://talk.ictvonline.org/cfs-file/__key/telligent-evolution-components-attachments/13-55-00-00-00-00-01-74/2005.020_2D00_72.04.Herpes.pdf?forcedownload=true |date=2020-06-15 }} |
||
{{Herpesvirales}} |
{{Herpesvirales}} |
||
{{Baltimore classification}} |
{{Baltimore classification}} |
||
{{Taxonbar|from=Q6542}} |
{{Taxonbar|from=Q6542}} |
||
{{Authority control}} |
|||
[[Category:Herpesvirales]] |
[[Category:Herpesvirales| ]] |
||
[[Category:Virus orders]] |
[[Category:Virus orders]] |
||
Latest revision as of 19:39, 3 December 2023
| Herpesvirales | |
|---|---|
| |
| Simplified diagram of Herpesvirales virion structure in cross-section. (1) nucleoprotein, (2) DNA, (3) capsid, (4) tegument, (5) envelope, (6) glycoprotein. Click to enlarge. | |

| |
| Electron micrograph of various viruses from the Orthoherpesviridae family including Human alphaherpesvirus 3 (Chickenpox), Human alphaherpesvirus 1, and Human alphaherpesvirus 2 | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Peploviricota |
| Class: | Herviviricetes |
| Order: | Herpesvirales |
| Families | |
|
And see text | |
The Herpesvirales is an order of dsDNA viruses (Baltimore group I) with animal hosts, characterised by a common morphology consisting of an icosahedral capsid enclosed in a glycoprotein-containing lipid envelope. Common infections in humans caused by members of this order include cold sores, genital herpes, chickenpox, shingles, and glandular fever. Herpesvirales is the sole order in the class Herviviricetes, which is the sole class in the phylum Peploviricota.
Virology
[edit]Morphology
[edit]All members of the order have a virion structure that consists of a DNA core surrounded by an icosahedral capsid composed of 12 pentavalent and 150 hexavalent capsomeres (T = 16). The capsid has a diameter of ~110 nanometers (nm) and is embedded in a proteinaceous matrix called the tegument, which in its turn is enclosed by a glycoprotein-containing lipid envelope with a diameter of about 200 nm. The DNA genome is linear and double stranded, with sizes in the range 125–290 kbp.[1] The genome contains terminal and internal reiterated sequences, with their number and disposition varying depending on the different subclades.
Hosts
[edit]

All species in this order have animal hosts. The Malacoherpesviridae infect molluscs (abalone and oysters), the Alloherpesviridae infect anamniotes (frogs and fish), and the Herpesviridae infect amniotes (reptiles, birds, and mammals). Within the family Herpesviridae, genera Iltovirus and Mardivirus, and genus Scutavirus of the subfamily Alphaherpesvirinae infect birds and reptiles, respectively. All other genera in the Herpesviridae infect mammals only.[2]
Taxonomy
[edit]As of 2020, the order is made up of three families, three subfamilies, 23 genera, and 130 species. The taxonomy is shown from family, to subfamily, to genus below.[3]
History
[edit]The herpesvirus was first isolated from the blue wildebeest in 1960 by veterinary scientist Walter Plowright.[4] The genus Herpesvirus was established in 1971 in the first report of the International Committee on Taxonomy of Viruses (ICTV). This genus consisted of 23 viruses and 4 groups of viruses. In the second ICTV report in 1976 this genus was elevated to family level - the Herpetoviridae. Because of possible confusion with viruses derived from reptiles this name was changed in the third report in 1979 to Herpesviridae. In this report the family Herpesviridae was divided into 3 subfamilies (Alphaherpesvirinae, Betaherpesvirinae and Gammaherpesvirinae) and 5 unnamed genera: 21 viruses were listed. In 2009 the family Herpesviridae was elevated to the order Herpesvirales. This elevation was necessitated by the discovery that the herpesviruses of fish and molluscs were only distantly related to those of birds and mammals.[5] Two new families were created - the family Alloherpesviridae which incorporates bony fish and frog viruses and the family Malacoherpesviridae which contains those of molluscs.
Phylogenetics
[edit]The only protein with widespread conservation amongst all members of the order, albeit only at the amino-acid level, is the ATPase subunit of the DNA terminase;[1] the latter is involved in the packaging of the DNA during virion assembly.[6]
Phylogenies constructed with the conserved regions of the ATPase subunit of the DNA terminase suggest that Alloherpesviridae is the basal clade of the order, and that Herpesviridae and Malacoherpesviridae are sister clades.[7] Given the phylogenetic distances between vertebrates and molluscs, this suggests that herpesviruses were initially fish viruses and that they have evolved with their hosts to infect other vertebrates.[citation needed]
Species nomenclature
[edit]The Herpesvirales naming system originated in 1973 and has been elaborated considerably since. All herpesviruses described since this system was adopted have been named in accordance with it. The recommended naming system specifies that each species name consists of three parts: a first word, a second word, and finally a number.
The first word should be derived from the taxon (family or subfamily) to which its primary natural host belongs. The subfamily name is used for viruses from members of the family Bovidae or from primates (the virus name ending in –ine, e.g. bovine), and the host family name for other viruses (ending in –id, e.g. equid). Human herpesviruses have been treated as an exception (human rather than hominid). Following the host-derived term, species in the family Herpesviridae, which are divided into subfamilies Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae, will have the word alphaherpesvirus, betaherpesvirus, or gammaherpesvirus added, respectively. Species in the Herpesviridae that have not been assigned to a subfamily, and species in Alloherpesviridae and Malacoherpesviridae will have herpesvirus following the host-derived term instead. Finally an Arabic number (1, 2, 3, and so forth) is appended. Numbers are assigned in order of naming and bear no implied meaning about the taxonomic or biological properties of the virus.
An example of such a name is Canid alphaherpesvirus 1. From this name it can be ascertained that the virus' primary natural host is a canid (i.e. a member of the family Canidae; dogs etc.), that it is a member of the family Herpesviridae and subfamily Alphaherpesvirinae, and that it is the first herpesvirus named for which canids serve as primary natural hosts.
A number of virus names (e.g. Epstein–Barr virus, also known as Human gammaherpesvirus 4) are so widely used that it may be impractical to attempt to insist on their replacement. This has led to a dual nomenclature in the literature for some herpesviruses.
References
[edit]- ^ a b Davison, Andrew J.; Eberle, Richard; Ehlers, Bernhard; Hayward, Gary S.; McGeoch, Duncan J.; Minson, Anthony C.; Pellett, Philip E.; Roizman, Bernard; Studdert, Michael J. (2009). "The order Herpesvirales". Archives of Virology. 154 (1): 171–177. doi:10.1007/s00705-008-0278-4. ISSN 1432-8798. PMC 3552636. PMID 19066710.
- ^ McGeoch, Duncan J.; Davison, Andrew J.; Dolan, Aidan; Gatherer, Derek; Sevilla-Reyes, Edgar E. (2008). DOMINGO, ESTEBAN; PARRISH, COLIN R.; HOLLAND, JOHN J. (eds.). Origin and Evolution of Viruses (Second ed.). London: Academic Press. pp. 447–475. ISBN 9780123741530.
- ^ "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 10 May 2021.
- ^ O.A., Ryder; Byrd, M.L. (1984). One Medicine: A Tribute to Kurt Benirschke, Director Center for Reproduction of Endangered Species Zoological Society of San Diego and Professor of Pathology and Reproductive Medicine University of California San Diego from his Students and Colleagues. Berlin, Heidelberg: Springer. pp. 296–308. ISBN 978-3-642-61749-2.
- ^ Duncan J. McGeoch, Andrew J. Davison, Aidan Dolan, Derek Gatherer, Edgar E. Sevilla-Reyes (2008). Origin and Evolution of Viruses (Second ed.). Academic Press. pp. Chapter 20: 447–475.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Selvarajan Sigamani, Sundaresan; Zhao, Haiyan; Kamau, Yvonne N.; Baines, Joel D.; Tang, Liang (June 2013). "The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses". Journal of Virology. 87 (12): 7140–7148. doi:10.1128/JVI.00311-13. ISSN 1098-5514. PMC 3676077. PMID 23596306.
- ^ Rakus, Krzysztof; Ouyang, Ping; Boutier, Maxime; Ronsmans, Maygane; Reschner, Anca; Vancsok, Catherine; Jazowiecka-Rakus, Joanna; Vanderplasschen, Alain (2013). "Cyprinid herpesvirus 3: an interesting virus for applied and fundamental research". Veterinary Research. 44 (1): 85. doi:10.1186/1297-9716-44-85. ISSN 0928-4249. PMC 3850573. PMID 24073814.
