Chloroplast: Difference between revisions
m Disambiguated: daltons → dalton (unit) |
Tic20 |
||
| Line 29: | Line 29: | ||
<div style="float: right; clear: right; width: 500px; height: 50px; " /> |
<div style="float: right; clear: right; width: 500px; height: 50px; " /> |
||
Somewhere around a billion years ago,<ref name=CurrentBiology-Endosymbiosis /> a free-living [[cyanobacterium]] entered an early [[eukaryotic]] cell, either as food or an internal [[parasite]],<ref name=Biology-Campbell&Reece /> and managed to escape the [[phagocytic vacuole]] it was contained in.<ref name="Springer—The Chloroplast—Chloroplast shapes" /> The two innermost [[cell membrane|lipid-bilayer membranes]] that surround all chloroplasts<ref>{{cite web|title=Chloroplasts|url=http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/C/Chloroplasts.html|work=Kimball's Biology Pages|accessdate=27 December 2012}}</ref> correspond to the outer and inner [[cell membrane|membranes]] of the ancestral cyanobacterium's [[gram negative]] cell wall,<ref>{{cite journal |author=Joyard J Block MA, Douce R|title=Molecular aspects of plastid envelope biochemistry |journal=Eur J Biochem. |volume=199 |pages=489–509 |year=1991 |pmid=1868841 |doi=10.1111/j.1432-1033.1991.tb16148.x |issue=3}}</ref><ref>{{cite web|title=Chloroplast|url=http://www.daviddarling.info/encyclopedia/C/chloroplasts.html|work=Encyclopedia of Science|accessdate=27 December 2012}}</ref><ref name=AJB-historyofplastids>{{cite journal|last=Keeling|first=Patrick J|title=Diversity and evolutionary history of plastids and their hosts|journal=American Journal of Botany|year=2004|month=October|volume=91|issue=10|doi=10.3732/ajb.91.10.1481|url=http://www.amjbot.org/content/91/10/1481.full|accessdate=27 January 2013|pages=1481–93|pmid=21652304}}</ref> and not the [[phagosomal]] membrane from the host, which was probably lost.<ref name=AJB-historyofplastids /> |
Somewhere around a billion years ago,<ref name=CurrentBiology-Endosymbiosis /> a free-living [[cyanobacterium]] entered an early [[eukaryotic]] cell, either as food or an internal [[parasite]],<ref name=Biology-Campbell&Reece /> and managed to escape the [[phagocytic vacuole]] it was contained in.<ref name="Springer—The Chloroplast—Chloroplast shapes" /> The two innermost [[cell membrane|lipid-bilayer membranes]] that surround all chloroplasts<ref>{{cite web|title=Chloroplasts|url=http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/C/Chloroplasts.html|work=Kimball's Biology Pages|accessdate=27 December 2012}}</ref> correspond to the outer and inner [[cell membrane|membranes]] of the ancestral cyanobacterium's [[gram negative]] cell wall,<ref>{{cite journal |author = Joyard J Block MA, Douce R|title = Molecular aspects of plastid envelope biochemistry |journal = Eur J Biochem. |volume = 199 |pages = 489–509 |year = 1991 |pmid = 1868841 |doi = 10.1111/j.1432-1033.1991.tb16148.x |issue = 3}}</ref><ref>{{cite web|title=Chloroplast|url=http://www.daviddarling.info/encyclopedia/C/chloroplasts.html|work=Encyclopedia of Science|accessdate=27 December 2012}}</ref><ref name=AJB-historyofplastids>{{cite journal|last=Keeling|first=Patrick J|title=Diversity and evolutionary history of plastids and their hosts|journal=American Journal of Botany|year=2004|month=October|volume=91|issue=10|doi=10.3732/ajb.91.10.1481|url=http://www.amjbot.org/content/91/10/1481.full|accessdate=27 January 2013|pages=1481–93|pmid=21652304}}</ref> and not the [[phagosomal]] membrane from the host, which was probably lost.<ref name=AJB-historyofplastids /> |
||
The new cellular resident quickly became an advantage, providing food for the eukaryotic host, which allowed it to live within it.<ref name=Biology-Campbell&Reece /> Over time, the cyanobacterium was assimilated, and many of its genes were lost or transferred to the [[nuclear genome|nucleus]] of the host.<ref name=BMCBiology-EvolvinganOrganelle /> Some of its proteins were then synthesized in the cytoplasm of the host cell, and imported back into the chloroplast.<ref name=BMCBiology-EvolvinganOrganelle /><ref name=PlantPhysiology-ChloroplastOrigin /> |
The new cellular resident quickly became an advantage, providing food for the eukaryotic host, which allowed it to live within it.<ref name=Biology-Campbell&Reece /> Over time, the cyanobacterium was assimilated, and many of its genes were lost or transferred to the [[nuclear genome|nucleus]] of the host.<ref name=BMCBiology-EvolvinganOrganelle /> Some of its proteins were then synthesized in the cytoplasm of the host cell, and imported back into the chloroplast.<ref name=BMCBiology-EvolvinganOrganelle /><ref name=PlantPhysiology-ChloroplastOrigin /> |
||
This event is called ''[[endosymbiosis]]'', or "cell living inside another cell". The cell living inside the other cell is called the ''endosymbiont''; the endosymbiont is found inside the ''host cell''.<ref name=Biology-Campbell&Reece /> |
This event is called ''[[endosymbiosis]]'', or "cell living inside another cell". The cell living inside the other cell is called the ''endosymbiont''; the endosymbiont is found inside the ''host cell''.<ref name=Biology-Campbell&Reece /> |
||
Chloroplasts are believed to have arisen after [[mitochondria]], since all [[eukaryotes]] contain mitochondria, but not all have chloroplasts.<ref name=Biology-Campbell&Reece>{{cite book|title=Biology 8th Edition Campbell & Reece|year=2009|publisher=Benjamin Cummings (Pearson)|page=516}}</ref><ref>{{cite journal|author=Archibald, J.M.|year=2009|title=The Puzzle of Plastid Evolution|journal=Current biology|volume=19|issue=2|pages=R81–R88|url=http://linkinghub.elsevier.com/retrieve/pii/S0960982208014851|doi=10.1016/j.cub.2008.11.067|pmid=19174147}}</ref> This is called ''[[serial endosymbiosis]]''—an early eukaryote engulfing the [[mitochondrion]] ancestor, and some descendants of it then engulfing the chloroplast ancestor, creating a cell with both chloroplasts and mitochondria.<ref name=Biology-Campbell&Reece /> |
Chloroplasts are believed to have arisen after [[mitochondria]], since all [[eukaryotes]] contain mitochondria, but not all have chloroplasts.<ref name=Biology-Campbell&Reece>{{cite book|title=Biology 8th Edition Campbell & Reece|year=2009|publisher=Benjamin Cummings (Pearson)|page=516}}</ref><ref>{{cite journal|author = Archibald, J.M.|year = 2009|title = The Puzzle of Plastid Evolution|journal = Current biology|volume = 19|issue = 2|pages = R81–R88|url = http://linkinghub.elsevier.com/retrieve/pii/S0960982208014851|doi = 10.1016/j.cub.2008.11.067|pmid = 19174147}}</ref> This is called ''[[serial endosymbiosis]]''—an early eukaryote engulfing the [[mitochondrion]] ancestor, and some descendants of it then engulfing the chloroplast ancestor, creating a cell with both chloroplasts and mitochondria.<ref name=Biology-Campbell&Reece /> |
||
Whether or not chloroplasts came from a single endosymbiotic event, or many independent engulfments across various eukaryotic lineages has been long debated, but it is now generally held that all organisms with chloroplasts either share [[monophyly|a single ancestor]] or obtained their chloroplast from organisms that share a common ancestor that took in a [[cyanobacterium]] 600–1600 million years ago.<ref name=CurrentBiology-Endosymbiosis>{{cite journal|last=McFadden|first=Geoffrey I|coauthors=van Dooren, Giel G|title=Evolution: Red Algal Genome Affirms a Common Origin of All Plastids|journal=Current Biology|date=NaN undefined NaN|volume=14|issue=13|pages=R514–R516|doi=10.1016/j.cub.2004.06.041|url=http://www.cell.com/current-biology/retrieve/pii/S0960982204004464|accessdate=7 April 2013|pmid=15242632}}</ref> |
Whether or not chloroplasts came from a single endosymbiotic event, or many independent engulfments across various eukaryotic lineages has been long debated, but it is now generally held that all organisms with chloroplasts either share [[monophyly|a single ancestor]] or obtained their chloroplast from organisms that share a common ancestor that took in a [[cyanobacterium]] 600–1600 million years ago.<ref name=CurrentBiology-Endosymbiosis>{{cite journal|last=McFadden|first=Geoffrey I|coauthors=van Dooren, Giel G|title=Evolution: Red Algal Genome Affirms a Common Origin of All Plastids|journal=Current Biology|date=NaN undefined NaN|volume=14|issue=13|pages=R514–R516|doi=10.1016/j.cub.2004.06.041|url=http://www.cell.com/current-biology/retrieve/pii/S0960982204004464|accessdate=7 April 2013|pmid=15242632}}</ref> |
||
| Line 299: | Line 299: | ||
===Chromatophores=== |
===Chromatophores=== |
||
While most chloroplasts originate from that first set of endosymbiotic events, ''[[Paulinella]] chromatophora'' is an exception, which has acquired a photosynthetic cyanobacterial endosymbiont more recently. It is not closely related to chloroplasts of other eukaryotes.<ref>{{cite journal | title = Diversity and evolutionary history of plastids and their hosts | author = Patrick J. Keeling | url = http://www.amjbot.org/cgi/content/full/91/10/z1481 | journal = American Journal of Botany | year = 2004 | volume = 91 | pages = 1481–1493 | doi = 10.3732/ajb.91.10.1481 | issue = 10 | pmid = 21652304 }}{{dead link|date=July 2010}}</ref> Being in the early stages of endosymbiosis, Paulinella chromatophora can offer some insights into how chloroplasts evolved.<ref name="BMCBiology-EvolvinganOrganelle">{{cite journal|last=Nakayama|first=Takuro|coauthors=John M Archibald|title=Evolving a photosynthetic organelle|journal=BMC Biology|year=2012|month=April|url=http://www.biomedcentral.com/1741-7007/10/35|accessdate=27 January 2013|doi=10.1186/1741-7007-10-35|volume=10|page=35|pmid=22531210|pmc=3337241}}</ref><ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer>{{cite journal|last=Nowack|first=EC|coauthors=Vogel H, Groth M, Grossman AR, Melkonian M, Glöckner G.|title=Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora|journal=Molecular Biology Evol.|year=2011|month=January|doi=10.1093/molbev/msq209|volume=28|pages=407–22|pmid=20702568|issue=1}}</ref> ''Paulinella'' cells contain one or two sausage shaped blue-green photosynthesizing structures called [[chromatophores]],<ref name=BMCBiology-EvolvinganOrganelle /><ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer /> descended from the cyanobacterium ''[[Synechococcus]]''. Chromatophores cannot survive outside their host.<ref name=BMCBiology-EvolvinganOrganelle /> Chromatophore DNA is about a million [[base pairs]] long, containing around 850 protein encoding [[genes]]—far less than the three million base pair ''Synechococcus'' genome,<ref name=BMCBiology-EvolvinganOrganelle /> but much larger than the ~150,000 base pair genome of the more assimilated chloroplast.<ref name=BioscienceExplained /><ref name=PNAS-RatesofctDNAevolution /><ref name=AmericanJournalofBotany /> Chromatophores have transferred much less of their DNA to the nucleus of their host. About 0.3–0.8% of the nuclear DNA in ''Paulinella'' is from the chromatophore, compared with 11–14% from the chloroplast in plants.<ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer /> |
While most chloroplasts originate from that first set of endosymbiotic events, ''[[Paulinella]] chromatophora'' is an exception, which has acquired a photosynthetic cyanobacterial endosymbiont more recently. It is not closely related to chloroplasts of other eukaryotes.<ref>{{cite journal | title = Diversity and evolutionary history of plastids and their hosts | author = Patrick J. Keeling | url = http://www.amjbot.org/cgi/content/full/91/10/z1481 | journal = American Journal of Botany | year = 2004 | volume = 91 | pages = 1481–1493 | doi = 10.3732/ajb.91.10.1481 | issue = 10 | pmid = 21652304 }}{{dead link|date=July 2010}}</ref> Being in the early stages of endosymbiosis, Paulinella chromatophora can offer some insights into how chloroplasts evolved.<ref name="BMCBiology-EvolvinganOrganelle">{{cite journal|last = Nakayama|first = Takuro|coauthors = John M Archibald|title = Evolving a photosynthetic organelle|journal = BMC Biology|year = 2012|month = April|url = http://www.biomedcentral.com/1741-7007/10/35|accessdate = 27 January 2013|doi = 10.1186/1741-7007-10-35|volume = 10|page = 35|pmid = 22531210|pmc = 3337241}}</ref><ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer>{{cite journal|last=Nowack|first=EC|coauthors=Vogel H, Groth M, Grossman AR, Melkonian M, Glöckner G.|title=Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora|journal=Molecular Biology Evol.|year=2011|month=January|doi=10.1093/molbev/msq209|volume=28|pages=407–22|pmid=20702568|issue=1}}</ref> ''Paulinella'' cells contain one or two sausage shaped blue-green photosynthesizing structures called [[chromatophores]],<ref name=BMCBiology-EvolvinganOrganelle /><ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer /> descended from the cyanobacterium ''[[Synechococcus]]''. Chromatophores cannot survive outside their host.<ref name=BMCBiology-EvolvinganOrganelle /> Chromatophore DNA is about a million [[base pairs]] long, containing around 850 protein encoding [[genes]]—far less than the three million base pair ''Synechococcus'' genome,<ref name=BMCBiology-EvolvinganOrganelle /> but much larger than the ~150,000 base pair genome of the more assimilated chloroplast.<ref name=BioscienceExplained /><ref name=PNAS-RatesofctDNAevolution /><ref name=AmericanJournalofBotany /> Chromatophores have transferred much less of their DNA to the nucleus of their host. About 0.3–0.8% of the nuclear DNA in ''Paulinella'' is from the chromatophore, compared with 11–14% from the chloroplast in plants.<ref name=MolecularBiologyEvol-EndosymbioticGeneTransfer /> |
||
===Kleptoplastidy=== |
===Kleptoplastidy=== |
||
| Line 329: | Line 329: | ||
====Linear structure==== |
====Linear structure==== |
||
Chloroplast DNAs have long been thought to have a [[circular genome|circular structure]], but some evidence suggests that chloroplast DNA more commonly takes a [[linear]] shape.<ref name="The Plant Cell—Linear chloroplast DNA">{{cite journal|last=Bendich|first=Arnold J|title=Circular Chloroplast Chromosomes: The Grand Illusion|journal=The Plant Cell|year=2004|month=July|volume=16|issue=7|pages=1661–1666|doi=10.1105/tpc.160771|url=http://www.plantcell.org/content/16/7/1661.full|accessdate=12 June 2013}}</ref> Over 95% of the chloroplast DNA in [[corn]] chloroplasts has been observed to be in branched linear form rather than individual circles.<ref name=Springer-TheChloroplast-18 /> |
Chloroplast DNAs have long been thought to have a [[circular genome|circular structure]], but some evidence suggests that chloroplast DNA more commonly takes a [[linear]] shape.<ref name="The Plant Cell—Linear chloroplast DNA">{{cite journal|last = Bendich|first = Arnold J|title = Circular Chloroplast Chromosomes: The Grand Illusion|journal = The Plant Cell|year = 2004|month = July|volume = 16|issue = 7|pages = 1661–1666|doi = 10.1105/tpc.160771|url = http://www.plantcell.org/content/16/7/1661.full|accessdate = 12 June 2013}}</ref> Over 95% of the chloroplast DNA in [[corn]] chloroplasts has been observed to be in branched linear form rather than individual circles.<ref name=Springer-TheChloroplast-18 /> |
||
====Nucleoids==== |
====Nucleoids==== |
||
| Line 341: | Line 341: | ||
===Gene content and protein synthesis=== |
===Gene content and protein synthesis=== |
||
The chloroplast genome most commonly includes around 100 genes<ref name="PlantPhysiology-ChloroplastOrigin">{{cite journal|last=McFadden|first=Geoffrey I|title=Chloroplast Origin and Integration|journal=Plant Physiology|year=2001|month=January|url=http://www.plantphysiology.org/content/125/1/50.full|accessdate=27 January 2013|doi=10.1104/pp.125.1.50|volume=125|pages=50–3|pmid=11154294|issue=1|pmc=1539323}}</ref><ref name=PNAS-RatesofctDNAevolution>{{cite journal|last=Clegg|first=MT|coauthors=B S Gaut, G H Learn, Jr, and B R Morton|title=Rates and patterns of chloroplast DNA evolution|journal=Proceedings of the National Academy of Sciences|year=1994|pmc=44285|volume=91|issue=15|pages=6795–6801|doi=10.1073/pnas.91.15.6795|pmid=8041699}}</ref> which code for a variety of things, mostly to do with the [[protein synthesis|protein pipeline]] and [[photosynthesis]]. As in [[prokaryotes]], genes in chloroplast DNA are organized into [[operons]].<ref name=PlantPhysiology-ChloroplastOrigin /> |
The chloroplast genome most commonly includes around 100 genes<ref name="PlantPhysiology-ChloroplastOrigin">{{cite journal|last = McFadden|first = Geoffrey I|title = Chloroplast Origin and Integration|journal = Plant Physiology|year = 2001|month = January|url = http://www.plantphysiology.org/content/125/1/50.full|accessdate = 27 January 2013|doi = 10.1104/pp.125.1.50|volume = 125|pages = 50–3|pmid = 11154294|issue = 1|pmc = 1539323}}</ref><ref name=PNAS-RatesofctDNAevolution>{{cite journal|last=Clegg|first=MT|coauthors=B S Gaut, G H Learn, Jr, and B R Morton|title=Rates and patterns of chloroplast DNA evolution|journal=Proceedings of the National Academy of Sciences|year=1994|pmc=44285|volume=91|issue=15|pages=6795–6801|doi=10.1073/pnas.91.15.6795|pmid=8041699}}</ref> which code for a variety of things, mostly to do with the [[protein synthesis|protein pipeline]] and [[photosynthesis]]. As in [[prokaryotes]], genes in chloroplast DNA are organized into [[operons]].<ref name=PlantPhysiology-ChloroplastOrigin /> |
||
Among land plants, the contents of the chloroplast genome are fairly similar<ref name=AmericanJournalofBotany />—they code for four [[ribosomal RNAs]], 30–31 [[tRNAs]], 21 [[ribosomal proteins]], and four [[RNA polymerase]] subunits,<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /><ref>{{cite journal|last=Wakasugi|first=T|coauthors=M. Sugita, T. Tsudzuki and M. Sugiura|title=Updated Gene Map of Tobacco Chloroplast DNA|journal=Plant Molecular Biology Reporter|year=1998|url=http://link.springer.com/article/10.1023%2FA%3A1007564209282?LI=true|accessdate=2 January 2013|doi=10.1023/A:1007564209282|volume=16|issue=3|page=231}}</ref> involved in protein synthesis. For photosynthesis, the chloroplast DNA includes genes for 28 [[thylakoid]] proteins and the large [[Rubisco]] subunit.<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /> In addition, its genes encode eleven subunits of a protein complex which mediates [[redox]] reactions to recycle electrons,<ref>{{cite journal |author=Krause K |title=From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants |journal=Curr. Genet. |volume=54 |issue=3 |pages=111–21 |year=2008 |month=September |pmid=18696071 |doi=10.1007/s00294-008-0208-8}}</ref> which is similar to the [[NADH dehydrogenase (ubiquinone)|NADH dehydrogenase]] found in mitochondria.<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /><ref>{{cite journal|last=Peng|first=Lianwei|coauthors=Yoichiro Fukao, Masayuki Fujiwara, and Toshiharu Shikanaia|title=Multistep Assembly of Chloroplast NADH Dehydrogenase-Like Subcomplex A Requires Several Nucleus-Encoded Proteins, Including CRR41 and CRR42, in Arabidopsis|journal=Plant Cell|date=24|year=2012|month=January|doi=10.1105/tpc.111.090597|pmc=3289569|volume=24|issue=1|pages=202–214|pmid=22274627}}</ref> |
Among land plants, the contents of the chloroplast genome are fairly similar<ref name=AmericanJournalofBotany />—they code for four [[ribosomal RNAs]], 30–31 [[tRNAs]], 21 [[ribosomal proteins]], and four [[RNA polymerase]] subunits,<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /><ref>{{cite journal|last=Wakasugi|first=T|coauthors=M. Sugita, T. Tsudzuki and M. Sugiura|title=Updated Gene Map of Tobacco Chloroplast DNA|journal=Plant Molecular Biology Reporter|year=1998|url=http://link.springer.com/article/10.1023%2FA%3A1007564209282?LI=true|accessdate=2 January 2013|doi=10.1023/A:1007564209282|volume=16|issue=3|page=231}}</ref> involved in protein synthesis. For photosynthesis, the chloroplast DNA includes genes for 28 [[thylakoid]] proteins and the large [[Rubisco]] subunit.<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /> In addition, its genes encode eleven subunits of a protein complex which mediates [[redox]] reactions to recycle electrons,<ref>{{cite journal |author = Krause K |title = From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants |journal = Curr. Genet. |volume = 54 |issue = 3 |pages = 111–21 |year = 2008 |month = September |pmid = 18696071 |doi = 10.1007/s00294-008-0208-8}}</ref> which is similar to the [[NADH dehydrogenase (ubiquinone)|NADH dehydrogenase]] found in mitochondria.<ref name=ASM-ChloroplastRibosomesandProteinSynthesis /><ref>{{cite journal|last=Peng|first=Lianwei|coauthors=Yoichiro Fukao, Masayuki Fujiwara, and Toshiharu Shikanaia|title=Multistep Assembly of Chloroplast NADH Dehydrogenase-Like Subcomplex A Requires Several Nucleus-Encoded Proteins, Including CRR41 and CRR42, in Arabidopsis|journal=Plant Cell|date=24|year=2012|month=January|doi=10.1105/tpc.111.090597|pmc=3289569|volume=24|issue=1|pages=202–214|pmid=22274627}}</ref> |
||
====Chloroplast genome reduction and gene transfer==== |
====Chloroplast genome reduction and gene transfer==== |
||
Over time, many parts of the chloroplast genome were transferred to the [[nuclear genome]] of the host,<ref name=BioscienceExplained /><ref name=PNAS-RatesofctDNAevolution /><ref>{{cite journal |author=Huang CY, Ayliffe MA, Timmis JN |title=Direct measurement of the transfer rate of chloroplast DNA into the nucleus |journal=Nature |volume=422 |issue=6927 |pages=72–6 |year=2003 |month=March |pmid=12594458 |doi=10.1038/nature01435 }}</ref> a process called ''[[endosymbiotic gene transfer]]''. |
Over time, many parts of the chloroplast genome were transferred to the [[nuclear genome]] of the host,<ref name=BioscienceExplained /><ref name=PNAS-RatesofctDNAevolution /><ref>{{cite journal |author = Huang CY, Ayliffe MA, Timmis JN |title = Direct measurement of the transfer rate of chloroplast DNA into the nucleus |journal = Nature |volume = 422 |issue = 6927 |pages = 72–6 |year = 2003 |month = March |pmid = 12594458 |doi = 10.1038/nature01435 }}</ref> a process called ''[[endosymbiotic gene transfer]]''. |
||
As a result, the chloroplast genome is heavily [[genome reduction|reduced]] compared to that of free-living cyanobacteria. Chloroplasts may contain 60–100 genes whereas cyanobacteria often have more than 1500 genes in their genome.<ref name="PNAS—Cyanobacterial genes in Arabidopsis">{{cite journal |author=Martin W, Rujan T, Richly E |title=Evolutionary analysis of ''Arabidopsis'', cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=99 |issue=19 |pages=12246–51 |year=2002 |month=September |pmid=12218172 |pmc=129430 |doi=10.1073/pnas.182432999 |url=http://www.pnas.org/cgi/pmidlookup?view=long&pmid=12218172|archiveurl = http://www.webcitation.org/5uKjjsvvn |archivedate = 2010-11-18|deadurl=no}}</ref> |
As a result, the chloroplast genome is heavily [[genome reduction|reduced]] compared to that of free-living cyanobacteria. Chloroplasts may contain 60–100 genes whereas cyanobacteria often have more than 1500 genes in their genome.<ref name="PNAS—Cyanobacterial genes in Arabidopsis">{{cite journal |author=Martin W, Rujan T, Richly E |title=Evolutionary analysis of ''Arabidopsis'', cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=99 |issue=19 |pages=12246–51 |year=2002 |month=September |pmid=12218172 |pmc=129430 |doi=10.1073/pnas.182432999 |url=http://www.pnas.org/cgi/pmidlookup?view=long&pmid=12218172|archiveurl = http://www.webcitation.org/5uKjjsvvn |archivedate = 2010-11-18|deadurl=no}}</ref> |
||
| Line 365: | Line 365: | ||
{{See also|Translation (biology)|label 1=Translation}} |
{{See also|Translation (biology)|label 1=Translation}} |
||
The movement of so many chloroplast genes to the nucleus means that lots of chloroplast [[proteins]] that were supposed to be [[Translation (biology)|translated]] in the chloroplast are now synthesized in the cytoplasm. This means that these proteins must be directed back to the chloroplast, and imported through at least two chloroplast membranes.<ref name="Nature—Protein import">{{cite journal|last=Soll|first=Jürgen|coauthors=Schleiff, Enrico|title=Plant cell biology: Protein import into chloroplasts|journal=Nature Reviews Molecular Cell Biology|year=2004|month=March|volume=5|issue=3|pages=198–208|doi=10.1038/nrm1333|accessdate=12 June 2013}}</ref> |
The movement of so many chloroplast genes to the nucleus means that lots of chloroplast [[proteins]] that were supposed to be [[Translation (biology)|translated]] in the chloroplast are now synthesized in the cytoplasm. This means that these proteins must be directed back to the chloroplast, and imported through at least two chloroplast membranes.<ref name="Nature—Protein import">{{cite journal|last = Soll|first = Jürgen|coauthors = Schleiff, Enrico|title = Plant cell biology: Protein import into chloroplasts|journal = Nature Reviews Molecular Cell Biology|year = 2004|month = March|volume = 5|issue = 3|pages = 198–208|doi = 10.1038/nrm1333|accessdate = 12 June 2013}}</ref> |
||
Curiously, around half of the protein products of transferred genes aren't even targeted back to the chloroplast. Many became [[exaptations]], taking on new functions like participating in [[cell division]], [[protein routing]], and even [[disease resistance]]. A few chloroplast genes found new homes in the [[mitochondrial genome]]—most became nonfunctional [[pseudogenes]], though a few [[tRNA]] genes still work in the [[mitochondrion]].<ref name="PNAS—Cyanobacterial genes in Arabidopsis" /> Some transferred chloroplast DNA protein products get directed to the [[secretory pathway]]<ref name="PNAS—Cyanobacterial genes in Arabidopsis" /> (though it should be noted that many [[secondary plastids]] are bounded by an outermost membrane derived from the host's [[cell membrane]], and therefore [[topologically]] outside of the cell, because to reach the chloroplast from the [[cytosol]], you have to cross the [[cell membrane]], just like if you were headed for the [[extracellular space]]. In those cases, chloroplast-targeted proteins do initially travel along the secretory pathway).<ref name="RSTB—Endosymbiosis of plastids" /> |
Curiously, around half of the protein products of transferred genes aren't even targeted back to the chloroplast. Many became [[exaptations]], taking on new functions like participating in [[cell division]], [[protein routing]], and even [[disease resistance]]. A few chloroplast genes found new homes in the [[mitochondrial genome]]—most became nonfunctional [[pseudogenes]], though a few [[tRNA]] genes still work in the [[mitochondrion]].<ref name="PNAS—Cyanobacterial genes in Arabidopsis" /> Some transferred chloroplast DNA protein products get directed to the [[secretory pathway]]<ref name="PNAS—Cyanobacterial genes in Arabidopsis" /> (though it should be noted that many [[secondary plastids]] are bounded by an outermost membrane derived from the host's [[cell membrane]], and therefore [[topologically]] outside of the cell, because to reach the chloroplast from the [[cytosol]], you have to cross the [[cell membrane]], just like if you were headed for the [[extracellular space]]. In those cases, chloroplast-targeted proteins do initially travel along the secretory pathway).<ref name="RSTB—Endosymbiosis of plastids" /> |
||
| Line 385: | Line 385: | ||
====Phosphorylation, chaperones, and transport==== |
====Phosphorylation, chaperones, and transport==== |
||
After a chloroplast [[polypeptide]] is synthesized on a [[ribosome]] in the [[cytosol]], [[Adenosine triphosphate|ATP]] energy can be used to [[phosphorylate]], or add a [[phosphate group]] to many (but not all) of them in their transit sequences.<ref name="Nature—Protein import" /> [[Serine]] and [[threonine]] (both very common in chloroplast transit sequences—making up 20–30% of the sequence)<ref name="JBC—Transit sequence phosphorylation" /> are often the [[amino acids]] that accept the [[phosphate group]].<ref name="JBC—Transit sequence phosphorylation" /><ref name="The Plant Cell—14-3-3 proteins">{{cite journal|last=May|first=Timo|title=14-3-3 Proteins Form a Guidance Complex with Chloroplast Precursor Proteins in Plants|journal=The Plant Cell|year=2000|month=January|volume=12|issue=1|pages=53–64|doi=10.1105/tpc.12.1.53|accessdate=18 June 2013}}</ref> The [[protein kinase|enzyme]] that carries out the phosphorylation is [[enzyme specificity|specific]] for chloroplast polypeptides, and ignores ones meant for [[mitochondria]] or [[peroxisomes]].<ref name="JBC—Transit sequence phosphorylation">{{cite journal|last=Soll|first=Jr.|title=Phosphorylation of the Transit Sequence of Chloroplast Precursor Proteins|journal=Journal of Biological Chemistry|date=15|year=1996|month=March|volume=271|issue=11|pages=6545–6554|doi=10.1074/jbc.271.11.6545}}</ref> |
After a chloroplast [[polypeptide]] is synthesized on a [[ribosome]] in the [[cytosol]], [[Adenosine triphosphate|ATP]] energy can be used to [[phosphorylate]], or add a [[phosphate group]] to many (but not all) of them in their transit sequences.<ref name="Nature—Protein import" /> [[Serine]] and [[threonine]] (both very common in chloroplast transit sequences—making up 20–30% of the sequence)<ref name="JBC—Transit sequence phosphorylation" /> are often the [[amino acids]] that accept the [[phosphate group]].<ref name="JBC—Transit sequence phosphorylation" /><ref name="The Plant Cell—14-3-3 proteins">{{cite journal|last = May|first = Timo|title = 14-3-3 Proteins Form a Guidance Complex with Chloroplast Precursor Proteins in Plants|journal = The Plant Cell|year = 2000|month = January|volume = 12|issue = 1|pages = 53–64|doi = 10.1105/tpc.12.1.53|accessdate = 18 June 2013}}</ref> The [[protein kinase|enzyme]] that carries out the phosphorylation is [[enzyme specificity|specific]] for chloroplast polypeptides, and ignores ones meant for [[mitochondria]] or [[peroxisomes]].<ref name="JBC—Transit sequence phosphorylation">{{cite journal|last = Soll|first = Jr.|title = Phosphorylation of the Transit Sequence of Chloroplast Precursor Proteins|journal = Journal of Biological Chemistry|date = 15|year = 1996|month = March|volume = 271|issue = 11|pages = 6545–6554|doi = 10.1074/jbc.271.11.6545}}</ref> |
||
Phosphorylation changes the polypeptide's shape,<ref name="JBC—Transit sequence phosphorylation" /> making it easier for [[14-3-3 proteins]] to attach to the polypeptide.<ref name="Nature—Protein import" /><ref name="BBA—Tic-toc" /> In plants, [[14-3-3 proteins]] only bind to chloroplast preproteins.<ref name="The Plant Cell—14-3-3 proteins" /> It is also bound by the [[heat shock protein|'''h'''eat '''s'''hock '''p'''rotein]] [[Hsp70]] that keeps the polypeptide from [[protein folding|folding]] prematurely.<ref name="Nature—Protein import" /> This is important because it prevents chloroplast proteins from assuming their active form and carrying out their chloroplast functions in the wrong place—the [[cytosol]].<ref name="The Plant Cell—14-3-3 proteins" /><ref name="BBA—Tic-toc">{{cite journal|last=Jarvis|first=Paul|coauthors=Soll, Jürgen|title=Toc, Tic, and chloroplast protein import|journal=Biochimica et Biophysica Acta (BBA) — Molecular Cell Research|date=1 December 2001|volume=1541|issue=1-2|pages=64–79|doi=10.1016/S0167-4889(01)00147-1}}</ref> At the same time, they have to keep just enough shape so that they can be recognized and imported into the chloroplast.<ref name="The Plant Cell—14-3-3 proteins" /> |
Phosphorylation changes the polypeptide's shape,<ref name="JBC—Transit sequence phosphorylation" /> making it easier for [[14-3-3 proteins]] to attach to the polypeptide.<ref name="Nature—Protein import" /><ref name="BBA—Tic-toc" /> In plants, [[14-3-3 proteins]] only bind to chloroplast preproteins.<ref name="The Plant Cell—14-3-3 proteins" /> It is also bound by the [[heat shock protein|'''h'''eat '''s'''hock '''p'''rotein]] [[Hsp70]] that keeps the polypeptide from [[protein folding|folding]] prematurely.<ref name="Nature—Protein import" /> This is important because it prevents chloroplast proteins from assuming their active form and carrying out their chloroplast functions in the wrong place—the [[cytosol]].<ref name="The Plant Cell—14-3-3 proteins" /><ref name="BBA—Tic-toc">{{cite journal|last = Jarvis|first = Paul|coauthors = Soll, Jürgen|title = Toc, Tic, and chloroplast protein import|journal = Biochimica et Biophysica Acta (BBA) — Molecular Cell Research|date = 1 December 2001|volume = 1541|issue = 1-2|pages = 64–79|doi = 10.1016/S0167-4889(01)00147-1}}</ref> At the same time, they have to keep just enough shape so that they can be recognized and imported into the chloroplast.<ref name="The Plant Cell—14-3-3 proteins" /> |
||
The heat shock protein and the 14-3-3 proteins together form a cytosolic guidance complex that makes it easier for the chloroplast polypeptide to get imported into the chloroplast.<ref name="Nature—Protein import" /> |
The heat shock protein and the 14-3-3 proteins together form a cytosolic guidance complex that makes it easier for the chloroplast polypeptide to get imported into the chloroplast.<ref name="Nature—Protein import" /> |
||
| Line 438: | Line 438: | ||
The [[TIC translocon]], or '''''t'''ranslocon on the '''i'''nner '''c'''hloroplast membrane [[translocon]]''<!--Yes, that's what the source says--><ref name="Nature—Protein import" /> is another protein complex that imports proteins across the [[inner chloroplast envelope]]. Chloroplast polypeptide chains probably often travel through the two complexes at the same time, but the TIC complex can also retrieve preproteins lost in the [[chloroplast intermembrane space|intermembrane space]].<ref name="Nature—Protein import" /> |
The [[TIC translocon]], or '''''t'''ranslocon on the '''i'''nner '''c'''hloroplast membrane [[translocon]]''<!--Yes, that's what the source says--><ref name="Nature—Protein import" /> is another protein complex that imports proteins across the [[inner chloroplast envelope]]. Chloroplast polypeptide chains probably often travel through the two complexes at the same time, but the TIC complex can also retrieve preproteins lost in the [[chloroplast intermembrane space|intermembrane space]].<ref name="Nature—Protein import" /> |
||
Like the [[Toc translocon|TOC translocon]], the TIC translocon has a large core [[Protein complex|complex]] surrounded by some loosely associated peripheral proteins like [[Tic110]], [[Tic40]], and [[Tic21]].<ref name="Science—TIC">{{cite journal|doi=10.1126/science.1229262|title=Uncovering the Protein Translocon at the Chloroplast Inner Envelope Membrane|author=Shingo Kikuchi et al|journal=Science|volume=339|issue=571|pages=571–573|year="2013|month=February}}</ref> |
Like the [[Toc translocon|TOC translocon]], the TIC translocon has a large core [[Protein complex|complex]] surrounded by some loosely associated peripheral proteins like [[Tic110]], [[Tic40]], and [[Tic21]].<ref name="Science—TIC">{{cite journal|doi = 10.1126/science.1229262|title = Uncovering the Protein Translocon at the Chloroplast Inner Envelope Membrane|author = Shingo Kikuchi et al|journal = Science|volume = 339|issue = 571|pages = 571–573|year = "2013|month = February}}</ref> |
||
The core complex weighs about one million [[dalton (unit)|dalton]]s and contains [[Tic214]], [[Tic100]], [[Tic56]], and [[Tic20 I]].<ref name="Science—TIC" /> |
The core complex weighs about one million [[dalton (unit)|dalton]]s and contains [[Tic214]], [[Tic100]], [[Tic56]], and [[Tic20 I]].<ref name="Science—TIC" /> |
||
{{clear}} |
|||
=====Tic20===== |
|||
[[Tic20]] is an [[Integral membrane protein|integral]] protein thought to have four [[Transmembrane domain|transmembrane]] [[α-helices]].<ref name="Nature—Protein import" /> It's found in the 1 million [[Dalton (unit)|dalton]] TIC complex.<ref name="Science—TIC" /> Because it's similar to [[Bacterial protein|bacterial]] [[amino acid]] transporters and the [[mitochondrial]] import protein [[Tim17]]<ref name="Nature—Protein import" /> ('''''[[Translocase of the inner membrane|t]]'''''<nowiki/>''[[Translocase of the inner membrane|ranslocase]] on the '''[[Inner mitochondrial membrane|i]]'''[[Inner mitochondrial membrane|nner '''m'''itochondrial '''m'''embrane]]''),<ref>{{Cite book|isbn = 9783540214892|title = Mitochondrial Function and Biogenesis|url = http://books.google.com/books?id=ie5ZmOYHpVEC&pg=PA59|publisher = Springer|page = 59|year = 2004|authors = Sean P Curran and Carla M Koehler}}</ref> it's been proposed to be part of the TIC import channel.<ref name="Nature—Protein import" /> There is no [[In vitro|''in vitro'']] evidence for this though.<ref name="Nature—Protein import" /> In [[Arabidopsis thaliana|''Arabidopsis thaliana'']], it is known that for about every five [[Toc75]] proteins in the outer chloroplast membrane, there are two [[Tic20 I]] proteins (the main [[Isoform|form]] of Tic20 in [[Arabidopsis thaliana|''Arabidopsis'']]) in the inner chloroplast membrane.<ref name="Science—TIC" />{{clear}} |
|||
<!-- |
<!-- |
||
| Line 474: | Line 476: | ||
The outer chloroplast membrane is a semi-porous membrane that small molecules and [[ions]] can easily diffuse across.<ref>{{cite journal|last=Koike|first=H|coauthors=Yoshio, M; Kashino, Y; Satoh, K|title=Polypeptide composition of envelopes of spinach chloroplasts: two major proteins occupy 90% of outer envelope membranes|journal=Plant & cell physiology|date=1998 May|volume=39|issue=5|pages=526–32|pmid=9664716|url=http://pcp.oxfordjournals.org/content/39/5/526.full.pdf|accessdate=18 May 2013|doi=10.1093/oxfordjournals.pcp.a029400}}</ref> However, it's not permeable to larger [[proteins]], so chloroplast [[polypeptides]] being synthesized in the cell [[cytoplasm]] must be transported across the outer chloroplast membrane by the [[TOC complex]], or ''[[translocon|'''t'''ranslocon]] on the '''o'''uter '''c'''hloroplast'' membrane.<ref name="Nature—Chloroplast protein import" /> |
The outer chloroplast membrane is a semi-porous membrane that small molecules and [[ions]] can easily diffuse across.<ref>{{cite journal|last=Koike|first=H|coauthors=Yoshio, M; Kashino, Y; Satoh, K|title=Polypeptide composition of envelopes of spinach chloroplasts: two major proteins occupy 90% of outer envelope membranes|journal=Plant & cell physiology|date=1998 May|volume=39|issue=5|pages=526–32|pmid=9664716|url=http://pcp.oxfordjournals.org/content/39/5/526.full.pdf|accessdate=18 May 2013|doi=10.1093/oxfordjournals.pcp.a029400}}</ref> However, it's not permeable to larger [[proteins]], so chloroplast [[polypeptides]] being synthesized in the cell [[cytoplasm]] must be transported across the outer chloroplast membrane by the [[TOC complex]], or ''[[translocon|'''t'''ranslocon]] on the '''o'''uter '''c'''hloroplast'' membrane.<ref name="Nature—Chloroplast protein import" /> |
||
The chloroplast membranes sometimes protrude out into the cytoplasm, forming a [[stromule]], or [[stroma (fluid)|'''strom'''a]]-containing tub'''ule'''. Stromules are very rare in chloroplasts, and are much more common in other [[plastids]] like [[chromoplasts]] and [[amyloplasts]] in petals and roots, respectively.<ref name="Köhler2000">{{cite journal |author=Köhler RH, Hanson MR |title=Plastid tubules of higher plants are tissue-specific and developmentally regulated |journal=J. Cell. Sci. |volume=113 |issue=Pt 1 |pages=81–9 |date=1 January 2000|pmid=10591627 |url=http://jcs.biologists.org/cgi/pmidlookup?view=long&pmid=10591627 |archiveurl = http://www.webcitation.org/5uKjkRKBB |archivedate = 2010-11-18|deadurl=no}}</ref><ref name="Gray2001">{{cite journal |author=Gray JC, Sullivan JA, Hibberd JM, Hansen MR |title=Stromules: mobile protrusions and interconnections between plastids |journal=Plant Biology |volume=3 |issue=3|pages=223–33 |year=2001 |doi=10.1055/s-2001-15204}}</ref> They may exist to increase the chloroplast's [[surface area to volume ratio|surface area]] for cross-membrane transport, connect two or more chloroplasts allowing them to exchange metabolites, or both.<ref name="Wise—Plastid diversity" /> |
The chloroplast membranes sometimes protrude out into the cytoplasm, forming a [[stromule]], or [[stroma (fluid)|'''strom'''a]]-containing tub'''ule'''. Stromules are very rare in chloroplasts, and are much more common in other [[plastids]] like [[chromoplasts]] and [[amyloplasts]] in petals and roots, respectively.<ref name="Köhler2000">{{cite journal |author=Köhler RH, Hanson MR |title=Plastid tubules of higher plants are tissue-specific and developmentally regulated |journal=J. Cell. Sci. |volume=113 |issue=Pt 1 |pages=81–9 |date=1 January 2000|pmid=10591627 |url=http://jcs.biologists.org/cgi/pmidlookup?view=long&pmid=10591627 |archiveurl = http://www.webcitation.org/5uKjkRKBB |archivedate = 2010-11-18|deadurl=no}}</ref><ref name="Gray2001">{{cite journal |author = Gray JC, Sullivan JA, Hibberd JM, Hansen MR |title = Stromules: mobile protrusions and interconnections between plastids |journal = Plant Biology |volume = 3 |issue = 3|pages = 223–33 |year = 2001 |doi = 10.1055/s-2001-15204}}</ref> They may exist to increase the chloroplast's [[surface area to volume ratio|surface area]] for cross-membrane transport, connect two or more chloroplasts allowing them to exchange metabolites, or both.<ref name="Wise—Plastid diversity" /> |
||
===Intermembrane space and peptidoglycan wall=== |
===Intermembrane space and peptidoglycan wall=== |
||
| Line 492: | Line 494: | ||
====Peripheral reticulum==== |
====Peripheral reticulum==== |
||
Some chloroplasts contain a structure called the [[chloroplast peripheral reticulum]].<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> It is often found in the chloroplasts of [[C4 plant|{{C4}} plants]], though it's also been found in some {{C3}} [[angiosperms]],<ref name="Wise—Plastid diversity" /> and even some [[gymnosperms]].<ref name="NewPhytologist-GymnospermPeripheralReticulum">{{cite journal|last=Whatley|first=Jean M|title=The occurrence of a peripheral reticulum in plastids of the gymnosperm Welwitschia mirabilis|journal=New Phytologist|date=5|year=1994|month=July|volume=74|issue=2|pages=215–220|doi=10.1111/j.1469-8137.1975.tb02608.x}}</ref> The chloroplast peripheral reticulum consists of a maze of membranous tubes and vesicles continuous with the [[inner chloroplast membrane]] that extends into the internal [[stroma (fluid)|stromal]] fluid of the chloroplast. Its purpose is thought to be to increase the chloroplast's [[surface area to volume ratio|surface area]] for cross-membrane transport between its stroma and the cell [[cytoplasm]]. The small vesicles sometimes observed may serve as [[transport vesicles]] to shuttle stuff between the [[thylakoids]] and intermembrane space.<ref name=StructureandFunctionofPlastids /> |
Some chloroplasts contain a structure called the [[chloroplast peripheral reticulum]].<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> It is often found in the chloroplasts of [[C4 plant|{{C4}} plants]], though it's also been found in some {{C3}} [[angiosperms]],<ref name="Wise—Plastid diversity" /> and even some [[gymnosperms]].<ref name="NewPhytologist-GymnospermPeripheralReticulum">{{cite journal|last = Whatley|first = Jean M|title = The occurrence of a peripheral reticulum in plastids of the gymnosperm Welwitschia mirabilis|journal = New Phytologist|date = 5|year = 1994|month = July|volume = 74|issue = 2|pages = 215–220|doi = 10.1111/j.1469-8137.1975.tb02608.x}}</ref> The chloroplast peripheral reticulum consists of a maze of membranous tubes and vesicles continuous with the [[inner chloroplast membrane]] that extends into the internal [[stroma (fluid)|stromal]] fluid of the chloroplast. Its purpose is thought to be to increase the chloroplast's [[surface area to volume ratio|surface area]] for cross-membrane transport between its stroma and the cell [[cytoplasm]]. The small vesicles sometimes observed may serve as [[transport vesicles]] to shuttle stuff between the [[thylakoids]] and intermembrane space.<ref name=StructureandFunctionofPlastids /> |
||
===Stroma=== |
===Stroma=== |
||
| Line 512: | Line 514: | ||
[[Starch granules]] are very common in chloroplasts, typically taking up 15% of the organelle's volume,<ref name="Plant Physiology—Starch granules in Arabidopsis" /> though in some other plastids like [[amyloplasts]], they can be big enough to distort the shape of the organelle.<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> Starch granules are simply accumulations of starch in the stroma, and are not bounded by a membrane.<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> |
[[Starch granules]] are very common in chloroplasts, typically taking up 15% of the organelle's volume,<ref name="Plant Physiology—Starch granules in Arabidopsis" /> though in some other plastids like [[amyloplasts]], they can be big enough to distort the shape of the organelle.<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> Starch granules are simply accumulations of starch in the stroma, and are not bounded by a membrane.<ref name="Introduction to Plant Cell Development—Chloroplast structure 46" /> |
||
Starch granules appear and grow throughout the day, as the chloroplast synthesizes [[sugars]], and are consumed at night to fuel [[Cellular respiration|respiration]] and continue sugar export into the [[phloem]],<ref name="FPB—Starch breakdown">{{cite journal|last=Zeeman|first=Samuel C.|coauthors=Delatte, Thierry; Messerli, Gaëlle; Umhang, Martin; Stettler, Michaela; Mettler, Tabea; Streb, Sebastian; Reinhold, Heike; Kötting, Oliver|title=Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms|journal=Functional Plant Biology|date=1 January 2007|volume=34|issue=6|page=465|doi=10.1071/FP06313|url=http://www.pbc.ethz.ch/publications/Publi_nr_28.pdf|accessdate=19 May 2013}}</ref> though in mature chloroplasts, it's rare for a starch granule to be completely consumed or for a new granule to accumulate.<ref name="Plant Physiology—Starch granules in Arabidopsis">{{cite journal|last=Crumpton-Taylor|first=Matilda|coauthors=Crumpton-Taylor, Scott Grandison, Kenneth M.Y. Png, Andrew J. Bushby and Alison M. Smith|title=Control of Starch Granule Numbers in Arabidopsis Chloroplasts|journal=Plant Physiology|year=2011|month=December|volume=158|issue=2|pages=905–916|doi=10.1104/pp.111.186957|url=http://www.plantphysiol.org/content/158/2/905.full|accessdate=19 May 2013|pmid=22135430|pmc=3271777}}</ref> |
Starch granules appear and grow throughout the day, as the chloroplast synthesizes [[sugars]], and are consumed at night to fuel [[Cellular respiration|respiration]] and continue sugar export into the [[phloem]],<ref name="FPB—Starch breakdown">{{cite journal|last = Zeeman|first = Samuel C.|coauthors = Delatte, Thierry; Messerli, Gaëlle; Umhang, Martin; Stettler, Michaela; Mettler, Tabea; Streb, Sebastian; Reinhold, Heike; Kötting, Oliver|title = Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms|journal = Functional Plant Biology|date = 1 January 2007|volume = 34|issue = 6|page = 465|doi = 10.1071/FP06313|url = http://www.pbc.ethz.ch/publications/Publi_nr_28.pdf|accessdate = 19 May 2013}}</ref> though in mature chloroplasts, it's rare for a starch granule to be completely consumed or for a new granule to accumulate.<ref name="Plant Physiology—Starch granules in Arabidopsis">{{cite journal|last=Crumpton-Taylor|first=Matilda|coauthors=Crumpton-Taylor, Scott Grandison, Kenneth M.Y. Png, Andrew J. Bushby and Alison M. Smith|title=Control of Starch Granule Numbers in Arabidopsis Chloroplasts|journal=Plant Physiology|year=2011|month=December|volume=158|issue=2|pages=905–916|doi=10.1104/pp.111.186957|url=http://www.plantphysiol.org/content/158/2/905.full|accessdate=19 May 2013|pmid=22135430|pmc=3271777}}</ref> |
||
Starch granules vary in composition and location across different chloroplast lineages. In [[red algae]], starch granules are found in the [[cytoplasm]] rather than in the chloroplast.<ref name="Molecular Biology of Chloroplasts and Mitochondria—Starch">{{cite book|last=J D|first=Rochaix|title=The molecular biology of chloroplasts and mitochondria in Chlamydomonas|year=1998|publisher=Kluwer Acad. Publ.|location=Dordrecht [u.a.]|isbn=9780792351740|pages=550–565|url=http://books.google.com/books?id=apv1hktfq_8C&pg=PA550#v=onepage&q&f=false}}</ref> In [[C4 plant|{{C4}} plants]], [[mesophyll tissue|mesophyll]] chloroplasts, which do not synthesize sugars, lack starch granules.<ref name="Wise—Plastid diversity" /> |
Starch granules vary in composition and location across different chloroplast lineages. In [[red algae]], starch granules are found in the [[cytoplasm]] rather than in the chloroplast.<ref name="Molecular Biology of Chloroplasts and Mitochondria—Starch">{{cite book|last=J D|first=Rochaix|title=The molecular biology of chloroplasts and mitochondria in Chlamydomonas|year=1998|publisher=Kluwer Acad. Publ.|location=Dordrecht [u.a.]|isbn=9780792351740|pages=550–565|url=http://books.google.com/books?id=apv1hktfq_8C&pg=PA550#v=onepage&q&f=false}}</ref> In [[C4 plant|{{C4}} plants]], [[mesophyll tissue|mesophyll]] chloroplasts, which do not synthesize sugars, lack starch granules.<ref name="Wise—Plastid diversity" /> |
||
| Line 778: | Line 780: | ||
Chloroplasts can serve as cellular sensors. After detecting stress in a cell, which might be due to a pathogen, chloroplasts begin producing molecules like [[salicylic acid]], [[jasmonic acid]], [[nitric oxide]] and [[reactive oxygen species]] which can serve as defense-signals. As cellular signals, reactive oxygen species are unstable molecules, so they probably don't leave the chloroplast, but instead pass on their signal to an unknown second messenger molecule. All these molecules initiate [[retrograde signaling (cell biology)|retrograde signaling]]—signals from the chloroplast that regulate [[gene expression]] in the nucleus.<ref name=MPMI-AllHandsonDeck /> |
Chloroplasts can serve as cellular sensors. After detecting stress in a cell, which might be due to a pathogen, chloroplasts begin producing molecules like [[salicylic acid]], [[jasmonic acid]], [[nitric oxide]] and [[reactive oxygen species]] which can serve as defense-signals. As cellular signals, reactive oxygen species are unstable molecules, so they probably don't leave the chloroplast, but instead pass on their signal to an unknown second messenger molecule. All these molecules initiate [[retrograde signaling (cell biology)|retrograde signaling]]—signals from the chloroplast that regulate [[gene expression]] in the nucleus.<ref name=MPMI-AllHandsonDeck /> |
||
In addition to defense signaling, chloroplasts, with the help of the [[peroxisome]]s,<ref name="katsir">{{cite journal|last=Katsir|first=L.|coauthors=Chung, H. S., Koo, A. J. K., Howe, G. A.|title=Jasmonate signaling: a conserved mechanism of hormone sensing|journal=Curr Bio|year=2008|volume=11|pages=428–435|doi=10.1016/j.pbi.2008.05.004|issue=4}}</ref> help synthesize an important defense molecule, [[jasmonate]]. Chloroplasts synthesize all the [[fatty acids]] in a plant cell<ref name=MPMI-AllHandsonDeck /><ref name=PlantPhysiology-FattyAcidExport>{{cite journal|last=Schnurr|first=Judy|coauthors=Jay M. Shockey, Gert-Jan de Boer and John A. Browse|title=Fatty Acid Export from the Chloroplast. Molecular Characterization of a Major Plastidial Acyl-Coenzyme A Synthetase from ''Arabidopsis''|journal=Plant Physiology|year=2002|month=July|volume=129|issue=4|pages=1700–1709|doi=10.1104/pp.003251|url=http://www.plantphysiol.org/content/129/4/1700.full|accessdate=4 February 2013|pmid=12177483|pmc=166758}}</ref>—[[linoleic acid]], a fatty acid, is a precursor to jasmonate.<ref name=MPMI-AllHandsonDeck /> |
In addition to defense signaling, chloroplasts, with the help of the [[peroxisome]]s,<ref name="katsir">{{cite journal|last = Katsir|first = L.|coauthors = Chung, H. S., Koo, A. J. K., Howe, G. A.|title = Jasmonate signaling: a conserved mechanism of hormone sensing|journal = Curr Bio|year = 2008|volume = 11|pages = 428–435|doi = 10.1016/j.pbi.2008.05.004|issue = 4}}</ref> help synthesize an important defense molecule, [[jasmonate]]. Chloroplasts synthesize all the [[fatty acids]] in a plant cell<ref name=MPMI-AllHandsonDeck /><ref name=PlantPhysiology-FattyAcidExport>{{cite journal|last=Schnurr|first=Judy|coauthors=Jay M. Shockey, Gert-Jan de Boer and John A. Browse|title=Fatty Acid Export from the Chloroplast. Molecular Characterization of a Major Plastidial Acyl-Coenzyme A Synthetase from ''Arabidopsis''|journal=Plant Physiology|year=2002|month=July|volume=129|issue=4|pages=1700–1709|doi=10.1104/pp.003251|url=http://www.plantphysiol.org/content/129/4/1700.full|accessdate=4 February 2013|pmid=12177483|pmc=166758}}</ref>—[[linoleic acid]], a fatty acid, is a precursor to jasmonate.<ref name=MPMI-AllHandsonDeck /> |
||
===Photosynthesis=== |
===Photosynthesis=== |
||
Revision as of 15:59, 26 June 2013

the many-fruited thyme moss
Chloroplasts /ˈklɔːrəplæsts/ are organelles found in plant cells and some other eukaryotic organisms. As well as conducting photosynthesis, they carry out almost all fatty acid synthesis in plants, and are involved in a plant's immune response. A chloroplast is a type of plastid which specializes in photosynthesis. During photosynthesis, chloroplasts capture the sun's light energy, and store it in the energy storage molecules ATP and NADPH while freeing oxygen from water. They then use the ATP and NADPH to make organic molecules from carbon dioxide in a process known as the Calvin cycle.[1]
The word chloroplast (χλωροπλάστης) is derived from the Greek words chloros (χλωρός), which means green, and plastes (πλάστης), which means "the one who forms".[2]
Chloroplast lineages and evolution
Chloroplasts are one of the many different types of organelles in the plant cell. However, unlike most cellular organelles, they are considered to have originated from cyanobacteria through endosymbiosis—when a eukaryotic cell engulfed a photosynthesizing cyanobacterium which remained and became a permanent resident in the cell. Mitochondria are thought to have come from a similar event, where a ærobic prokaryote was engulfed.[3] This origin of chloroplasts was first suggested by Konstantin Mereschkowski in 1905[4] after Andreas Schimper observed that chloroplasts closely resemble cyanobacteria in 1883.[5]
Chloroplasts are similar to mitochondria in that they both originate from an endosymbiotic event, but chloroplasts are found only in plants and some protists.
Cyanobacterial ancestor
Cyanobacteria are considered the ancestors of chloroplasts. They are sometimes called blue-green algae even though they are prokaryotes. They are a diverse phylum of bacteria capable of carrying out photosynthesis, and are gram-negative, meaning they have two cell membranes. They also contain a thick peptidoglycan cell wall which is located between their two cell membranes.[6] Like chloroplasts, they have thylakoids inside of them.[7] On the thylakoid membranes are photosynthetic pigments, including chlorophyll a.[8] Phycobilins are also common cyanobacterial pigments, usually organized into hemispherical phycobilisomes attached to the outside of the thylakoid membranes. (Phycobilins are not shared with all chloroplasts though).[9][8]

Primary endosymbiosis

A eukaryote with mitochondria engulfed a cyanobacterium in an event of serial primary endosymbiosis, creating a lineage of cells with both organelles.[3] It is important to note that the cyanobacterial endosymbiont already had a double membrane—the phagosomal vacuole-derived membrane was lost.[10]
Somewhere around a billion years ago,[11] a free-living cyanobacterium entered an early eukaryotic cell, either as food or an internal parasite,[3] and managed to escape the phagocytic vacuole it was contained in.[8] The two innermost lipid-bilayer membranes that surround all chloroplasts[12] correspond to the outer and inner membranes of the ancestral cyanobacterium's gram negative cell wall,[13][14][10] and not the phagosomal membrane from the host, which was probably lost.[10] The new cellular resident quickly became an advantage, providing food for the eukaryotic host, which allowed it to live within it.[3] Over time, the cyanobacterium was assimilated, and many of its genes were lost or transferred to the nucleus of the host.[15] Some of its proteins were then synthesized in the cytoplasm of the host cell, and imported back into the chloroplast.[15][16]
This event is called endosymbiosis, or "cell living inside another cell". The cell living inside the other cell is called the endosymbiont; the endosymbiont is found inside the host cell.[3]
Chloroplasts are believed to have arisen after mitochondria, since all eukaryotes contain mitochondria, but not all have chloroplasts.[3][17] This is called serial endosymbiosis—an early eukaryote engulfing the mitochondrion ancestor, and some descendants of it then engulfing the chloroplast ancestor, creating a cell with both chloroplasts and mitochondria.[3]
Whether or not chloroplasts came from a single endosymbiotic event, or many independent engulfments across various eukaryotic lineages has been long debated, but it is now generally held that all organisms with chloroplasts either share a single ancestor or obtained their chloroplast from organisms that share a common ancestor that took in a cyanobacterium 600–1600 million years ago.[11]
These chloroplasts, which can be traced back directly to a cyanobacterial ancestor are known as primary plastids[18] ("plastid" in this context means the almost the same thing as chloroplast[3]). All primary chloroplasts belong to one of three chloroplast lineages—the glaucophyte chloroplast lineage, the rhodophyte, or red algal chloroplast lineage, or the chloroplastidan, or green chloroplast lineage.[19] The second two are the largest,[10] and the green chloroplast lineage is the one that contains the land plants.[10]
| Glaucophyta | Chloroplast lineages A primary endosymbiosis event gave rise to three main lineages of chloroplasts in the glaucophytes, chlorophyta, and rhodophyta.[19] Some of these algae were subsequently engulfed by other algae, becoming secondary (or tertiary) endosymbionts.[8][10] a The apicomplexans (malaria parasites), contain a red algal endosymbiont with a non- photosynthetic chloroplast.[20] b 2–3 chloroplast membranes[8] a c 2–4 chloroplast membranes[8] | ||||||
| Chloroplastida |
Euglenophyta | ||||||
| Chlorarachniophyta | |||||||
| Green algal dinophytes | |||||||
| Rhodophyceæ (Red algae) |
Apicomplexa a | ||||||
| Peridinin-type dinophytes b | |||||||
| Cryptophyta | |||||||
| Haptophyta | Haptophyte dinophytes c | ||||||
| Heterokontophyta | Diatom dinophytes | ||||||
| Primary endosymbiosis | Secondary endosymbiosis | Tertiary endosymbiosis | |||||
Glaucophyta
The alga Cyanophora, a glaucophyte, is thought to be one of the first organisms to contain a chloroplast.[16] The glaucophyte chloroplast group is the smallest of the three primary chloroplast lineages, being found in only thirteen species,[10] and is thought to be the one that branched off the earliest.[11][10][21] Glaucophytes have chloroplasts which retain a peptidoglycan wall between their double membranes,[18] like their cyanobacterial parent.[6] For this reason, glaucophyte chloroplasts are also known as muroplasts.[18] Glaucophyte chloroplasts also contain concentric unstacked thylakoids which surround a carboxysome, an icosahedral structure that glaucophyte chloroplasts and cyanobacteria keep their carbon fixation enzyme rubisco in. The starch they synthesize collects outside the chloroplast.[8] Like cyanobacteria, glaucophyte chloroplast thylakoids are studded with light collecting structures called phycobilisomes.[8][18] For these reasons, glaucophyte chloroplasts are considered a primitive intermediate between cyanobacteria and the more evolved chloroplasts in red algae and plants.[18]
Rhodophyceæ (red algae)
The rhodophyte, or red algal chloroplast group is another large and diverse chloroplast lineage.[10] Rhodophyte chloroplasts are also called rhodoplasts,[18] literally "red chloroplasts".[23]
Rhodoplasts have a double membrane with an intermembrane space and phycobilin pigments organized into phycobilisomes on the thylakoid membranes, preventing their thylakoids from stacking.[8] Some contain pyrenoids.[18] Rhodoplasts have chlorophyll a and phycobilins[21] for photosynthetic pigments; the phycobilin phycoerytherin is responsible for giving many red algae their distinctive red color.[22] However, since they also contain the blue-green chlorophyll a and other pigments, many are reddish to purple from the combination.[18] The red phycoerytherin pigment is an adaptation to help red algae catch more sunlight in deep water[18]—as such, some red algae that live in shallow water have less phycoerytherin in their rhodoplasts, and can appear more greenish.[22] Rhodoplasts synthesize a form of starch called floridean,[18] which collects into granules outside the rhodoplast, in the cytoplasm of the red alga.[8]
Chloroplastida (green algae and plants)
The chloroplastidan chloroplasts, or green chloroplasts, are another large, highly diverse primary chloroplast lineage. They are commonly known as the green algae and land plants.[24] They differ from glaucophyte and red algal chloroplasts in that they have lost their phycobilisomes, and contain chlorophyll b instead.[8] Most green chloroplasts are (obviously) green, though some, like some forms of Hæmatococcus pluvialis aren't, due to accessory pigments that override the chlorophylls' green colors. Chloroplastidan chloroplasts have lost the peptidoglycan wall between their double membrane, and have replaced it with an intermembrane space.[8] Some plants seem to have kept the genes for the synthesis of the peptidoglycan layer, though they've been repurposed for use in chloroplast division instead.[25]
Most of the chloroplasts depicted in this article are green chloroplasts.
Green algae and plants keep their starch inside their chloroplasts,[8][21][24] and in plants and some algae, the chloroplast thylakoids are arranged in grana stacks. Some green algal chloroplasts contain a structure called a pyrenoid,[8] which is functionally similar to the glaucophyte carboxysome in that it's where rubisco and CO2 are concentrated in the chloroplast.[26]

Helicosproidia
The helicosproidia are nonphotosynthetic parasitic green algae that are thought to contain a vestigal chloroplast.[21] Genes from a chloroplast[27] and nuclear genes indicating the presence of a chloroplast have been found in helicosporoidia.[21] even if nobody's seen the chloroplast itself.[21]
Secondary and tertiary endosymbiosis
Many other organisms obtained chloroplasts from the primary chloroplast lineages through secondary endosymbiosis—engulfing a red or green alga that contained a chloroplast. These chloroplasts are known as secondary plastids.[18]
While primary chloroplasts have a double membrane from their cyanobacterial ancestor, secondary chloroplasts have additional membranes outside of the original two, as a result of the secondary endosymbiotic event, when a nonphotosynthetic eukaryote engulfed an chloroplast-containing alga but failed to digest it—much like the cyanobacterium at the beginning of this story.[10] The engulfed alga was broken down, leaving only its chloroplast, and sometimes its cell membrane and nucleus, forming a chloroplast with three to four membranes[28]—the two cyanobacterial membranes, sometimes the eaten alga's cell membrane, and the phagosomal vacuole from the host's cell membrane.[10]


The genes in the phagocytosed eukaryote's nucleus are often transferred to the secondary host's nucleus.[10] Cryptomonads and chlorarachniophytes retain the phagocytosed eukaryote's nucleus, an object called a nucleomorph,[10] located between the second and third membranes of the chloroplast.[8][16]
All secondary chloroplasts come from green and red algae—no secondary chloroplasts from glaucophytes have been observed, probably because glaucophytes are relatively rare in nature, making them less likely to have been taken up by another eukaryote.[10]
Green algal derived chloroplasts
Green algae have been taken up by the euglenids, chlorarachniophytes, a lineage of dinoflagellates,[21] and possibly the ancestor of the chromalveolates[29] in three or four separate engulfments.[30] Many green algal derived chloroplasts contain pyrenoids, but unlike chloroplasts in their green algal ancestors, starch collects in granules outside the chloroplast.[8]
Euglenophytes
Euglenophytes are a group of common flagellated protists that contain chloroplasts derived from a green alga.[10] Euglenophyte chloroplasts have three membranes—it's thought that the membrane of the primary endosymbiont was lost, leaving the cyanobacterial membranes, and the secondary host's phagosomal membrane.[10] Euglenophyte chloroplasts have a pyrenoid and thylakoids stacked in groups of three. Starch is stored in the form of paramylon, which is contained in membrane-bound granules in the cytoplasm of the euglenophyte.[8][21]

Chlorarachniophytes
Chlorarachniophytes (/ˌklɔːrəˈrækni[invalid input: 'ɵ']ˌfaɪts/) are a rare group of organisms that also contain chloroplasts derived from green algae,[10] though their story is more complicated than that of the euglenophytes. The ancestor of chlorarachniophytes is thought to have been a chromalveolate, a eukaryote with a red algal derived chloroplast. It's then thought to have lost its first red algal chloroplast, and later engulfed a green alga, giving it its second, green algal derived chloroplast.[21]
Chlorarachniophyte chloroplasts are bounded by four membranes, except near the cell membrane, where the chloroplast membranes fuse into a double membrane.[8] Their thylakoids are arranged in loose stacks of three.[8] Chlorarachniophytes have a form of starch called chrysolaminarin, which they store in the cytoplasm,[21] often collected around the chloroplast pyrenoid, which bulges into the cytoplasm.[8]
Chlorarachniophyte chloroplasts are notable because the green alga they are derived from has not been completely broken down—its nucleus still persists as a nucleomorph[10] found between the second and third chloroplast membranes[8]—the periplastid space, which corresponds to the green alga's cytoplasm.[21]
Early chromalveolates
Recent research has suggested that the ancestor of the chromalveolates acquired a green algal prasinophyte endosymbiont. The green algal derived chloroplast was lost and replaced with a red algal derived chloroplast, but not before contributing some of its genes to the early chromalveolate's nucleus. The presence of both green algal and red algal genes in chromalveolates probably helps them thrive under fluctuating light conditions.[29]
Red algal derived chloroplasts (chromalveolate chloroplasts)
Like green algae, red algae have also been taken up in secondary endosymbiosis, though it's thought that all red algal derived chloroplasts are descended from a single red alga that was engulfed by an early chromalveolate, giving rise to the chromalveolates, some of which, like the ciliates, subsequently lost the chloroplast.[10][21][22] This is still debated though.[21][22]
Pyrenoids and stacked thylakoids are common in chromalveolate chloroplasts, and the outermost membrane of many are continuous with the rough endoplasmic reticulum and studded with ribosomes.[8][21] They have lost their phycobilisomes and exchanged them for chlorophyll c, which isn't found in primary red algal chloroplasts themselves.[8]
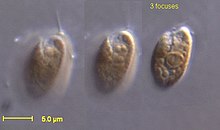
Cryptophytes
Cryptophytes, or cryptomonads are a group of algae that contain a red-algal derived chloroplast. Cryptophyte chloroplasts contain a nucleomorph that superficially resembles that of the chlorarachniophytes.[10] Cryptophyte chloroplasts have four membranes, the outermost of which is continuous with the rough endoplasmic reticulum. They synthesize ordinary starch, which is stored in granules found in the periplastid space—outside the original double membrane, in the place that corresponds to the red alga's cytoplasm. Inside cryptophyte chloroplasts is a pyrenoid and thylakoids in stacks of two.[8]
Their chloroplasts do not have phycobilisomes,[8] but they do have phycobilin pigments which they keep in their thylakoid space, rather than anchored on the outside of their thylakoid membranes.[8][10]

Haptophytes
Haptophytes are similar and closely related to cryptophytes, and are thought to be the first chromalveolates to branch off.[21] Their chloroplasts lack a nucleomorph,[8][10] their thylakoids are in stacks of three, and they synthesize chrysolaminarin sugar, which they store completely outside of the chloroplast, in the cytoplasm of the haptophyte.[8]
Heterokontophytes (stramenopiles)

The heterokontophytes, also known as the stramenopiles, are a very large and diverse group of algae that also contain red algal derived chloroplasts.[21] Heterokonts include the diatoms and the brown algae, golden algae,[22] and yellow-green algae.
Heterokont chloroplasts are very similar to haptophyte chloroplasts, containing a pyrenoid, triplet thylakoids, and with some exceptions,[8] having an epiplastid membrane connected to the endoplasmic reticulum. Like haptophytes, heterokontophytes store sugar in chrysolaminarin granules in the cytoplasm.[8] Heterokontophyte chloroplasts contain chlorophyll a and with a few exceptions[8] chlorophyll c,[10] but also have carotenoids which give them their many colors.[22]
Apicomplexans
Apicomplexans are another group of chromalveolates. Like the helicosproidia, they're parasitic, and have a nonphotosynthetic chloroplast.[21] They were once thought to be related to the helicosproidia, but it is now known that the helicosproida are green algae rather than chromalveolates.[21] The apicomplexans include Plasmodium, the malaria parasite. Many apicomplexans keep a vestigial red algal derived chloroplast[20][21] called an apicoplast, which they inherited from their ancestors. Other apicomplexans like Cryptosporidium have lost the chloroplast completely.[20] Apicomplexans store their energy in amylopectin starch granules that are located in their cytoplasm, even though they are nonphotosynthetic.[8]
Apicoplasts have lost all photosynthetic function, and contain no photosynthetic pigments or true thylakoids. They are bounded by four membranes, but the membranes are not connected to the endoplasmic reticulum.[8] The fact that apicomplexans still keep their nonphotosynthetic chloroplast around demonstrates how the chloroplast carries out important functions other than photosynthesis. Plant chloroplasts provide plant cells with many important things besides sugar, and apicoplasts are no different—they synthesize fatty acids, isopentenyl pyrophosphate, iron-sulfur clusters, and carry out part of the heme pathway.[20] This makes the apicoplast an attractive target for drugs to cure apicomplexan-related diseases.[18] The most important apicoplast function is isopentenyl pyrophosphate synthesis—in fact, apicomplexans die when something interferes with this apicoplast function, and when apicomplexans are grown in an isopentenyl pyrophosphate-rich medium, they dump the organelle.[20]
Dinophytes
The dinoflagellates are yet another very large and diverse group of protists, around half of which are (at least partially) photosynthetic.[22][31]
Most dinophyte chloroplasts are secondary red algal derived chloroplasts, like other chromalveolate chloroplasts. Many other dinophytes have lost the chloroplast (becoming the nonphotosynthetic kind of dinoflagellate), or replaced it though tertiary endosymbiosis[32]—the engulfment of another chromalveolate containing a red algal derived chloroplast. Others replaced their original chloroplast with a green algal derived one.[10][21][31]
Most dinophyte chloroplasts contain at least the photosynthetic pigments chlorophyll a, chlorophyll c2, beta-carotene, and at least one dinophyte-unique xanthophyll (peridinin, dinoxanthin, or diadinoxanthin), giving many a golden-brown color.[31] All dinophytes store starch in their cytoplasm, and most have chloroplasts with thylakoids arranged in stacks of three.[8]
Peridinin-containing dinophyte chloroplast

The most common dinophyte chloroplast is the peridinin-type chloroplast, characterized by the carotenoid pigment peridinin in their chloroplasts, along with chlorophyll a and chlorophyll c2.[10][31] Peridinin is not found in any other group of chloroplasts.[31] The peridinin chloroplast is bounded by three membranes (occasionally two),[8] having lost the red algal endosymbiont's original cell membrane.[21][10] The outermost membrane is not connected to the endoplasmic reticulum.[8][31] They contain a pyrenoid, and have triplet-stacked thylakoids. Starch is found outside the chloroplast[8] An important feature of these chloroplasts is that their chloroplast DNA is highly reduced and fragmented into many small circles. Most of the genome has migrated to the nucleus, and only critical photosynthesis-related genes remain in the chloroplast.[31]
The peridinin chloroplast is thought to be the dinophytes' "original" chloroplast,[31] which has been lost, reduced, replaced, or has company in several other dinophyte lineages.[21]
Fucoxanthin-containing dinophyte chloroplasts (haptophyte endosymbionts)

The fucoxanthin dinophyte lineages (including Karlodinium and Karenia)[21] lost their original red algal derived chloroplast, and replaced it with a new chloroplast derived from a haptophyte endosymbiont. Karlodinium and Karenia probably took up different heterokontophytes.[21] Because the haptophyte chloroplast has four membranes, tertiary endosymbiosis would be expected to create a six membraned chloroplast, adding the haptophyte's cell membrane and the dinophyte's phagosomal vacuole.[34] However, the haptophyte was heavily reduced, stripped of a few membranes and its nucleus, leaving only its chloroplast (with its original double membrane), and possibly one or two additional membranes around it.[21][34]
Fucoxanthin-containing chloroplasts are characterized by having the pigment fucoxanthin (actually 19′-hexanoyloxy-fucoxanthin and/or 19′-butanoyloxy-fucoxanthin) and no peridinin. Fucoxanthin is also found in haptophyte chloroplasts, providing evidence of ancestry.[31]

Cryptophyte derived dinophyte chloroplast
Members of the genus Dinophysis have a phycobilin-containing[34] chloroplast taken from a cryptophyte.[10] However, the cryptophyte is not an endosymbiont—only the chloroplast seems to have been taken, and the chloroplast has been stripped of its nucleomorph and outermost two membranes, leaving just a two-membraned chloroplast. Cryptophyte chloroplasts require their nucleomorph to maintain themselves, and Dinophysis species grown in cell culture alone cannot survive, so it's possible (but not confirmed) that the Dinophysis chloroplast is a kleptoplast—if so, Dinophysis chloroplasts wear out and Dinophysis species must continually engulf cryptophytes to obtain new chloroplasts to replace the old ones.[31]
Diatom derived dinophyte chloroplasts
Some dinophytes, like Kryptoperidinium and Durinskia[21] have a diatom (heterokontophyte) derived chloroplast.[10] These chloroplasts are bounded by up to five membranes,[10] (depending on whether you count the entire diatom endosymbiont as the chloroplast, or just the red algal derived chloroplast inside it). The diatom endosymbiont has been reduced relatively little—it still retains its original mitochondria,[21] and has endoplasmic reticulum, ribosomes, a nucleus, and of course, red algal derived chloroplasts—practically a complete cell,[35] all inside the host's endoplasmic reticulum lumen.[21] However the diatom endosymbiont can't store its own food—its starch is found in granules in the dinophyte host's cytoplasm instead.[35][8] The diatom endosymbiont's nucleus is present, but it probably can't be called a nucleomorph because it shows no sign of genome reduction, and might have even been expanded.[21] Diatoms have been engulfed by dinoflagellates at least three times.[21]
The diatom endosymbiont is bounded by a single membrane,[31] inside it are chloroplasts with four membranes. Like in the diatom endosymbiont's diatom ancestor, the chloroplasts have triplet thylakoids and pyrenoids.[35]
In some of these genera, the diatom endosymbiont's chloroplasts aren't the only chloroplasts in the dinophyte. The original three-membraned peridinin chloroplast is still around, converted to an eyespot.[10][21]
Prasinophyte (green algal) derived dinophyte chloroplast
Lepidodinium viride and its close relatives are dinophytes that lost their original peridinin chloroplast and replaced it with a green algal derived chloroplast (more specifically, a prasinophyte).[31][8] Lepidodinium is the only dinophyte that has a chloroplast that's not from the rhodoplast lineage. The chloroplast is surrounded by two membranes and has no nucleomorph—all the nucleomorph genes have been transferred to the dinophyte nucleus.[31] The endosymbiotic event that led to this chloroplast was serial secondary endosymbiosis rather than tertiary endosymbiosis—the endosymbiont was a green alga containing a primary chloroplast (making a secondary chloroplast).[21]
Chromatophores
While most chloroplasts originate from that first set of endosymbiotic events, Paulinella chromatophora is an exception, which has acquired a photosynthetic cyanobacterial endosymbiont more recently. It is not closely related to chloroplasts of other eukaryotes.[36] Being in the early stages of endosymbiosis, Paulinella chromatophora can offer some insights into how chloroplasts evolved.[15][37] Paulinella cells contain one or two sausage shaped blue-green photosynthesizing structures called chromatophores,[15][37] descended from the cyanobacterium Synechococcus. Chromatophores cannot survive outside their host.[15] Chromatophore DNA is about a million base pairs long, containing around 850 protein encoding genes—far less than the three million base pair Synechococcus genome,[15] but much larger than the ~150,000 base pair genome of the more assimilated chloroplast.[38][39][40] Chromatophores have transferred much less of their DNA to the nucleus of their host. About 0.3–0.8% of the nuclear DNA in Paulinella is from the chromatophore, compared with 11–14% from the chloroplast in plants.[37]
Kleptoplastidy
In some groups of mixotrophic protists, like some dinoflagellates, chloroplasts are separated from a captured alga or diatom and used temporarily. These klepto chloroplasts may only have a lifetime of a few days and are then replaced.[41]
Chloroplast DNA
Chloroplasts have their own DNA,[42] often abbreviated as ctDNA,[43] or cpDNA.[44] It is also known as the plastome. Its existence was first proved in 1962,[38] and first sequenced in 1986—when two Japanese research teams sequenced the chloroplast DNA of liverwort and tobacco.[45] Since then, hundreds of chloroplast DNAs from various species have been sequenced, but they're mostly those of land plants and green algae—glaucophytes, red algae, and other algal groups are extremely underrepresented, potentially introducing some bias in views of "typical" chloroplast DNA structure and content.[46]
Molecular structure
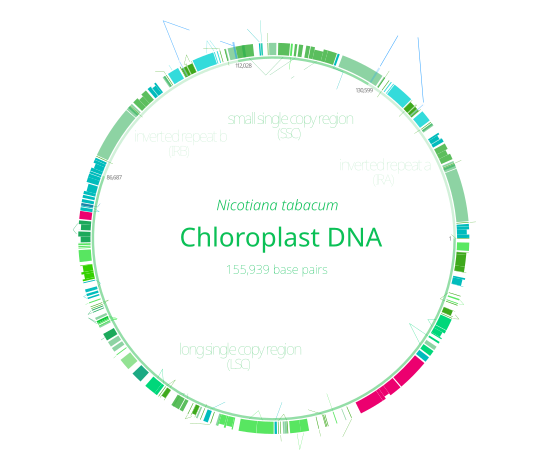
Chloroplast DNAs are circular, and are typically 120,000–170,000 base pairs long.[38][39][40] They can have a contour length of around 30–60 micrometers, and have a mass of about 80–130 million daltons.[47]
Most chloroplasts have their entire chloroplast genome combined into a single large ring, though those of dinophyte algae are a notable exception—their genome is broken up into about forty small plasmids, each 2,000–10,000 base pairs long.[46] Each minicircle contains one to three genes,[16][46] but blank plasmids, with no coding DNA, have also been found.
Inverted repeats
Many chloroplast DNAs contain two inverted repeats, which separate a long single copy section (LSC) from a short single copy section (SSC).[40]
The inverted repeats vary wildly in length, ranging from 4,000 to 25,000 base pairs long each.[46] Inverted repeats in plants tend to be at the upper end of this range, each being 20,000–25,000 base pairs long.[40][48] The inverted repeat regions usually contain three ribosomal RNA and two tRNA genes, but they can be expanded or reduced to contain as few as four or as many as over 150 genes.[46] While a given pair of inverted repeats are rarely completely identical, they are always very similar to each other, apparently resulting from concerted evolution.[46]
The inverted repeat regions are highly conserved among land plants, and accumulate few mutations.[40][48] Similar inverted repeats exist in the genomes of cyanobacteria and the other two chloroplast lineages (glaucophyta and rhodophyceæ), suggesting that they predate the chloroplast,[46] though some chloroplast DNAs like those of peas and a few red algae[46] have since lost the inverted repeats.[48][49] Others, like the red alga Porphyra flipped one of its inverted repeats (making them direct repeats).[46] It is possible that the inverted repeats help stabilize the rest of the chloroplast genome, as chloroplast DNAs which have lost some of the inverted repeat segments tend to get rearranged more.[49]
Linear structure
Chloroplast DNAs have long been thought to have a circular structure, but some evidence suggests that chloroplast DNA more commonly takes a linear shape.[50] Over 95% of the chloroplast DNA in corn chloroplasts has been observed to be in branched linear form rather than individual circles.[46]
Nucleoids
New chloroplasts may contain up to 100 copies of their DNA,[38] though the number of chloroplast DNA copies decreases to about 15–20 as the chloroplasts age.[51] They are usually packed into nucleoids which can contain several identical chloroplast DNA rings. Many nucleoids can be found in each chloroplast.[47]
Though chloroplast DNA is not associated with true histones,[3] in red algae, a histone-like chloroplast protein (HC) coded by the chloroplast DNA that tightly packs each chloroplast DNA ring into a nucleoid has been found.[52]
In primitive red algae, the chloroplast DNA nucleoids are clustered in the center of the chloroplast, while in green plants and green algae, the nucleoids are dispersed throughout the stroma.[52]
Gene content and protein synthesis
The chloroplast genome most commonly includes around 100 genes[16][39] which code for a variety of things, mostly to do with the protein pipeline and photosynthesis. As in prokaryotes, genes in chloroplast DNA are organized into operons.[16]
Among land plants, the contents of the chloroplast genome are fairly similar[40]—they code for four ribosomal RNAs, 30–31 tRNAs, 21 ribosomal proteins, and four RNA polymerase subunits,[53][54] involved in protein synthesis. For photosynthesis, the chloroplast DNA includes genes for 28 thylakoid proteins and the large Rubisco subunit.[53] In addition, its genes encode eleven subunits of a protein complex which mediates redox reactions to recycle electrons,[55] which is similar to the NADH dehydrogenase found in mitochondria.[53][56]
Chloroplast genome reduction and gene transfer
Over time, many parts of the chloroplast genome were transferred to the nuclear genome of the host,[38][39][57] a process called endosymbiotic gene transfer. As a result, the chloroplast genome is heavily reduced compared to that of free-living cyanobacteria. Chloroplasts may contain 60–100 genes whereas cyanobacteria often have more than 1500 genes in their genome.[58]
Endosymbiotic gene transfer is how we know about the lost chloroplasts in many chromalveolate lineages. Even if a chloroplast is eventually lost, the genes it donated to the former host's nucleus persist, providing evidence for the lost chloroplast's existence. For example, while diatoms (a heterokontophyte) now have a red algal derived chloroplast, the presence of many green algal genes in the diatom nucleus provide evidence that the diatom ancestor (probably the ancestor of all chromalveolates too) had a green algal derived chloroplast at some point, which was subsequently replaced by the red chloroplast.[29]
In land plants, some 11–14% of the DNA in their nuclei can be traced back to the chloroplast,[37] up to 18% in Arabidopsis, corresponding to about 4,500 protein-coding genes.[59] There have been a few recent transfers of genes from the chloroplast DNA to the nuclear genome in land plants.[39]
Of the approximately three-thousand proteins found in chloroplasts, some 95% of them are encoded by nuclear genes. Many of the chloroplast's protein complexes consist of subunits from both the chloroplast genome and the host's nuclear genome. As a result, protein synthesis must be coordinated between the chloroplast and the nucleus. The chloroplast is mostly under nuclear control, though chloroplasts can also give out signals regulating gene expression in the nucleus, called retrograde signaling.[60]
Protein synthesis
Protein synthesis within chloroplasts relies on an RNA polymerase coded by the chloroplast's own genome, which is related to RNA polymerases found in bacteria. Chloroplasts also contain a mysterious second RNA polymerase that is encoded by the plant's nuclear genome. The two RNA polymerases may recognize and bind to different kinds of promoters within the chloroplast genome.[61] The ribosomes in chloroplasts are similar to bacterial ribosomes.[53]
This section needs expansion with: Genome size differences between algae and land plants, chloroplast stuff coded by the nucleus, DNA replication, NADPH redox, special tRNA synthetases, etc.. You can help by adding to it. (January 2013) |
Protein targeting and import
The movement of so many chloroplast genes to the nucleus means that lots of chloroplast proteins that were supposed to be translated in the chloroplast are now synthesized in the cytoplasm. This means that these proteins must be directed back to the chloroplast, and imported through at least two chloroplast membranes.[62]
Curiously, around half of the protein products of transferred genes aren't even targeted back to the chloroplast. Many became exaptations, taking on new functions like participating in cell division, protein routing, and even disease resistance. A few chloroplast genes found new homes in the mitochondrial genome—most became nonfunctional pseudogenes, though a few tRNA genes still work in the mitochondrion.[58] Some transferred chloroplast DNA protein products get directed to the secretory pathway[58] (though it should be noted that many secondary plastids are bounded by an outermost membrane derived from the host's cell membrane, and therefore topologically outside of the cell, because to reach the chloroplast from the cytosol, you have to cross the cell membrane, just like if you were headed for the extracellular space. In those cases, chloroplast-targeted proteins do initially travel along the secretory pathway).[21]
Because the cell acquiring a chloroplast already had mitochondria (and peroxisomes, and a cell membrane for secretion), the new chloroplast host had to develop a unique protein targeting system to avoid having chloroplast proteins being sent to the wrong organelle, and the wrong proteins being sent to the chloroplast.[62]
Cytoplasmic translation and N-terminal transit sequences
Polypeptides, the precursors of proteins, are chains of amino acids. The two ends of a polypeptide are called the N-terminus, or amino end, and the C-terminus, or carboxyl end.[63] For many (but not all)[64] chloroplast proteins encoded by nuclear genes, cleavable transit peptides are added to the N-termini of the polypeptides, which are used to help direct the polypeptide to the chloroplast for import[62][65] (N-terminal transit peptides are also used to direct polypeptides to plant mitochondria).[66] N-terminal transit sequences are also called presequences[62] because they are located at the "front" end of a polypeptide—ribosomes synthesize polypeptides from the N-terminus to the C-terminus.[63]
Chloroplast transit peptides exhibit huge variation in length and amino acid sequence.[65] They can be from 20–150 amino acids long[62]—an unusually long length, suggesting that transit peptides are actually collections of domains with different functions.[65] Transit peptides tend to be positively charged,[62] rich in hydroxylated amino acids such as serine, threonine, and proline, and poor in acidic amino acids like aspartic acid and glutamic acid.[65] In an aqueous solution, the transit sequence forms a random coil.[62]
Not all chloroplast proteins include a N-terminal cleavable transit peptide though.[62] Some include the transit sequence within the functional part of the protein itself.[62] A few have their transit sequence appended to their C-terminus instead.[67] Most of the polypeptides that lack N-terminal targeting sequences are the ones that are sent to the outer chloroplast membrane, plus at least one sent to the inner chloroplast membrane.[62]
Phosphorylation, chaperones, and transport
After a chloroplast polypeptide is synthesized on a ribosome in the cytosol, ATP energy can be used to phosphorylate, or add a phosphate group to many (but not all) of them in their transit sequences.[62] Serine and threonine (both very common in chloroplast transit sequences—making up 20–30% of the sequence)[68] are often the amino acids that accept the phosphate group.[68][66] The enzyme that carries out the phosphorylation is specific for chloroplast polypeptides, and ignores ones meant for mitochondria or peroxisomes.[68]
Phosphorylation changes the polypeptide's shape,[68] making it easier for 14-3-3 proteins to attach to the polypeptide.[62][69] In plants, 14-3-3 proteins only bind to chloroplast preproteins.[66] It is also bound by the heat shock protein Hsp70 that keeps the polypeptide from folding prematurely.[62] This is important because it prevents chloroplast proteins from assuming their active form and carrying out their chloroplast functions in the wrong place—the cytosol.[66][69] At the same time, they have to keep just enough shape so that they can be recognized and imported into the chloroplast.[66]
The heat shock protein and the 14-3-3 proteins together form a cytosolic guidance complex that makes it easier for the chloroplast polypeptide to get imported into the chloroplast.[62]
Alternatively, if a chloroplast preprotein's transit peptide is not phosphorylated, a chloroplast preprotein can still attach to a heat shock protein or Toc159. These complexes can bind to the TOC complex on the outer chloroplast membrane using GTP energy.[62]
The translocon on the outer chloroplast membrane (TOC)
This section needs expansion with: Minor TOC proteins, Arabidopsis mutants. You can help by adding to it. (June 2013) |
The TOC complex, or translocon on the outer chloroplast membrane, is a collection of proteins that imports preproteins across the outer chloroplast envelope. Five subunits of the TOC complex have been identified—two GTP-binding proteins Toc34 and Toc159, the protein import tunnel Toc75, plus the proteins Toc64[62] and Toc12.[64]
The first three proteins form a core complex that consists of one Toc159, four to five Toc34s, and four Toc75s that form four holes in a disk 13 nanometers across. The whole core complex weighs about 500 kilodaltons. The other two proteins, Toc64 and Toc12, are associated with the core complex but are not part of it.[64]
Toc34 and 33
Toc34 is an integral protein in the outer chloroplast membrane that's anchored into it by its hydrophobic[71] C-terminal tail.[62][69] Most of the protein, however, including its large guanosine triphosphate (GTP)-binding domain projects out into the stroma.[69]
Toc34's job is to catch some chloroplast preproteins in the cytosol and hand them off to the rest of the TOC complex.[62] When GTP, an energy molecule similar to ATP attaches to Toc34, the protein becomes much more able to bind to many chloroplast preproteins in the cytosol.[62] The chloroplast preprotein's presence causes Toc34 to break GTP into guanosine diphosphate (GDP) and inorganic phosphate. This loss of GTP makes the Toc34 protein release the chloroplast preprotein, handing it off to the next TOC protein.[62] Toc34 then releases the depleted GDP molecule, probably with the help of an unknown GDP exchange factor. A domain of Toc159 might be the exchange factor that carry out the GDP removal. The Toc34 protein can then take up another molecule of GTP and begin the cycle again.[62]
Toc34 can be turned off through phosphorylation. A protein kinase drifting around on the outer chloroplast membrane can use ATP to add a phosphate group to the Toc34 protein, preventing it from being able to receive another GTP molecule, inhibiting the protein's activity. This might provide a way to regulate protein import into chloroplasts.[62][69]
Arabidopsis thaliana has two homologous proteins, AtToc33 and AtToc34 (The At stands for Arabidopsis thaliana),[62][69] which are each about 60% identical in amino acid sequence to Toc34 in peas (called psToc34).[69] AtToc33 is the most common in Arabidopsis,[69] and it's the functional analogue of Toc34 because it can be turned off by phosphorylation. AtToc34 on the other hand cannot be phosphorylated.[62][69]
Toc159
Toc159 is another GTP binding TOC subunit, like Toc34. Toc159 has three domains. At the N-terminal end is the A-domain, which is rich in acidic amino acids and takes up about half the protein length.[71][62] The A-domain is often cleaved off, leaving an 86 kilodalton fragment called Toc86.[71] In the middle is its GTP binding domain, which is very similar to the homologous GTP-binding domain in Toc34.[71][62] At the C-terminal end is the hydrophilic M-domain,[62] which anchors the protein to the outer chloroplast membrane.[71]
Toc159 probably works a lot like Toc34, recognizing proteins in the cytosol using GTP. It can be regulated through phosphorylation, but by a different protein kinase than the one that phosphorylates Toc34.[64] Its M-domain forms part of the tunnel that chloroplast preproteins travel through, and seems to provide the force that pushes preproteins through, using the energy from GTP.[62]
Toc159 is not always found as part of the TOC complex—it has also been found dissolved in the cytosol. This suggests that it might act as a shuttle that finds chloroplast preproteins in the cytosol and carries them back to the TOC complex. There isn't a lot of direct evidence for this behavior though.[62]
A family of Toc159 proteins, Toc159, Toc132, Toc120, and Toc90 have been found in Arabidopsis thaliana. They vary in the length of their A-domains, which is completely gone in Toc90. Toc132, Toc120, and Toc90 seem to have specialized functions in importing stuff like nonphotosynthetic preproteins, and can't replace Toc159.[62]
Toc75

Toc75 is the most abundant protein on the outer chloroplast envelope. It's a transmembrane tube that forms most of the TOC pore itself. Toc75 is a β-barrel channel lined by 16 β-pleated sheets.[62] The hole it forms is about 2.5 nanometers wide at the ends, and shrinks to about 1.4–1.6 nanometers in diameter at its narrowest point—wide enough to allow partially folded chloroplast preproteins to pass through.[62]
Toc75 can also bind to chloroplast preproteins, but is a lot worse at this than Toc34 or Toc159.[62]
Arabidopsis thaliana has multiple isoforms of Toc75 that are named by the chromosomal positions of the genes that code for them. AtToc75 III is the most abundant of these.[62]
The translocon on the inner chloroplast membrane (TIC)
This section needs expansion with: June 2013. You can help by adding to it. (June 2013) |
The TIC translocon, or translocon on the inner chloroplast membrane translocon[62] is another protein complex that imports proteins across the inner chloroplast envelope. Chloroplast polypeptide chains probably often travel through the two complexes at the same time, but the TIC complex can also retrieve preproteins lost in the intermembrane space.[62]
Like the TOC translocon, the TIC translocon has a large core complex surrounded by some loosely associated peripheral proteins like Tic110, Tic40, and Tic21.[72] The core complex weighs about one million daltons and contains Tic214, Tic100, Tic56, and Tic20 I.[72]
Tic20
Tic20 is an integral protein thought to have four transmembrane α-helices.[62] It's found in the 1 million dalton TIC complex.[72] Because it's similar to bacterial amino acid transporters and the mitochondrial import protein Tim17[62] (translocase on the inner mitochondrial membrane),[73] it's been proposed to be part of the TIC import channel.[62] There is no in vitro evidence for this though.[62] In Arabidopsis thaliana, it is known that for about every five Toc75 proteins in the outer chloroplast membrane, there are two Tic20 I proteins (the main form of Tic20 in Arabidopsis) in the inner chloroplast membrane.[72]Structure

In land plants, chloroplasts are generally lens-shaped, 5–8 μm in diameter and 1–3 μm thick.[74] Greater diversity in chloroplast shapes exists among the algae, which often contain a single chloroplast[8] that can be shaped like a net (e.g., Oedogonium),[75] a cup (e.g., Chlamydomonas),[76] a ribbon-like spiral around the edges of the cell (e.g., Spirogyra),[77] or slightly twisted bands at the cell edges (e.g., Sirogonium).[78] Some algae have two chloroplasts in each cell; they are star-shaped in Zygnema,[79] or may follow the shape of half the cell in order Desmidiales.[80] In some algae, the chloroplast takes up most of the cell, with pockets for the nucleus and other organelles[8] (for example some species of Chlorella have a cup-shaped chloroplast that occupies much of the cell).[81]
All chloroplasts have at least three membrane systems—the outer chloroplast membrane, the inner chloroplast membrane, and the thylakoid system. Chloroplasts that are the product of secondary endosymbiosis may have additional membranes surrounding these three.[28] Inside the outer and inner chloroplast membranes is the chloroplast stroma, a semi-gel-like fluid[18] that makes up much of a chloroplast's volume, and in which the thylakoid system floats.
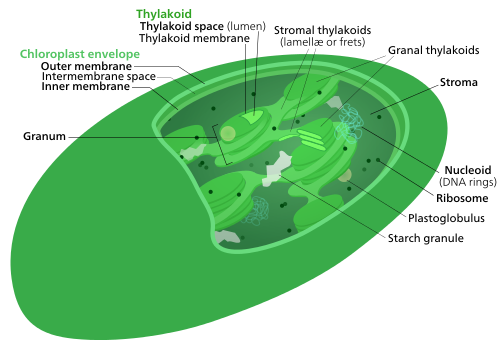
There are some common misconceptions about the outer and inner chloroplast membranes. The fact that chloroplasts are surrounded by a double membrane is often cited as evidence that they are the descendants of endosymbiotic cyanobacteria. This is often interpreted as meaning the outer chloroplast membrane is the product of the host's cell membrane infolding to form a vesicle to surround the ancestral cyanobacterium—which is not true—both chloroplast membranes are homologous to the cyanobacterium's original double membranes.[10]
The chloroplast double membrane is also often compared to the mitochondrial double membrane. This is not a valid comparison—the inner mitochondria membrane is used to run proton pumps and carry out oxidative phosphorylation across to generate ATP energy. The only chloroplast structure that can considered analogous to it is the internal thylakoid system. Even so, in terms of "in-out", the direction of chloroplast H+ ion flow is in the opposite direction compared to oxidative phosphorylation in mitochondria.[18][82] In addition, in terms of function, the inner chloroplast membrane, which regulates metabolite passage and synthesizes some materials, has no counterpart in the mitochondrion.[18]
Outer chloroplast membrane
The outer chloroplast membrane is a semi-porous membrane that small molecules and ions can easily diffuse across.[83] However, it's not permeable to larger proteins, so chloroplast polypeptides being synthesized in the cell cytoplasm must be transported across the outer chloroplast membrane by the TOC complex, or translocon on the outer chloroplast membrane.[84]
The chloroplast membranes sometimes protrude out into the cytoplasm, forming a stromule, or stroma-containing tubule. Stromules are very rare in chloroplasts, and are much more common in other plastids like chromoplasts and amyloplasts in petals and roots, respectively.[85][86] They may exist to increase the chloroplast's surface area for cross-membrane transport, connect two or more chloroplasts allowing them to exchange metabolites, or both.[18]
Intermembrane space and peptidoglycan wall

Usually, a thin intermembrane space about 10–20 nanometers thick exists between the outer and inner chloroplast membranes.[87]
Glaucophyte algal chloroplasts have a peptidoglycan layer between the chloroplast membranes. It corresponds to the peptidoglycan cell wall of their cyanobacterial ancestors, which is located between their two cell membranes. These chloroplasts are called muroplasts (from Latin "mura", meaning "wall"). Other chloroplasts have lost the cyanobacterial wall, leaving an intermembrane space between the two chloroplast envelope membranes.[18]
Inner chloroplast membrane
The inner chloroplast membrane borders the stroma and regulates passage of materials in and out of the chloroplast. After passing through the TOC complex in the outer chloroplast membrane, polypeptides must pass through the TIC complex (translocon on the inner chloroplast membrane) which is located in the inner chloroplast membrane.[84]
In addition to regulating the passage of materials, the inner chloroplast membrane is where fatty acids, lipids, and carotenoids are synthesized.[18]
Peripheral reticulum
Some chloroplasts contain a structure called the chloroplast peripheral reticulum.[87] It is often found in the chloroplasts of C4 plants, though it's also been found in some C3 angiosperms,[18] and even some gymnosperms.[88] The chloroplast peripheral reticulum consists of a maze of membranous tubes and vesicles continuous with the inner chloroplast membrane that extends into the internal stromal fluid of the chloroplast. Its purpose is thought to be to increase the chloroplast's surface area for cross-membrane transport between its stroma and the cell cytoplasm. The small vesicles sometimes observed may serve as transport vesicles to shuttle stuff between the thylakoids and intermembrane space.[89]
Stroma
The protein-rich,[18] alkaline,[82] aqueous fluid within the inner chloroplast membrane and outside of the thylakoid space is called the stroma,[18] which corresponds to the cytosol of the original cyanobacterium. Nucleoids of chloroplast DNA, chloroplast ribosomes, the thylakoid system with plastoglobuli, starch granules, and many proteins can be found floating around in it. The Calvin cycle, which fixes CO2 into sugar takes place in the stroma.
Chloroplast ribosomes
Chloroplasts have their own ribosomes, which they use to synthesize a small fraction of their proteins. Chloroplast ribosomes are about two-thirds the size of cytoplasmic ribosomes (around 17 nm vs 25 nm).[87] They take mRNAs transcribed from the chloroplast DNA and translate them into protein. While similar to bacterial ribosomes,[3] chloroplast translation is more complex than in bacteria, so chloroplast ribosomes include some chloroplast-unique features.[90]
Plastoglobuli
Plastoglobuli (singular plastoglobulus, sometimes spelled plastoglobule(s)), are spherical globules of lipids and proteins[18] about 10–15 nanometers across.[87] They are surrounded by a lipid monolayer.[91] Plastoglobuli are found in all chloroplasts,[87] but become more common when the chloroplast is under oxidative stress,[91] or when it ages and transitions into a gerontoplast.[18] They are also common in etioplasts, but decrease in number as the etioplasts mature into chloroplasts.[91]
Plastoglobuli were once thought to be free-floating in the stroma, but it is now thought that they are permanently attached either to a thylakoid or to another plastoglobulus attached to a thylakoid. Plastoglubuli contain both structural proteins and enzymes involved in lipid synthesis and metabolism. They can form when a bubble appears between the layers of the lipid bilayer of the thylakoid membrane, or bud from existing plastoglubuli—though they never detach and float off into the stroma.[91]
Starch granules
Starch granules are very common in chloroplasts, typically taking up 15% of the organelle's volume,[92] though in some other plastids like amyloplasts, they can be big enough to distort the shape of the organelle.[87] Starch granules are simply accumulations of starch in the stroma, and are not bounded by a membrane.[87]
Starch granules appear and grow throughout the day, as the chloroplast synthesizes sugars, and are consumed at night to fuel respiration and continue sugar export into the phloem,[93] though in mature chloroplasts, it's rare for a starch granule to be completely consumed or for a new granule to accumulate.[92]
Starch granules vary in composition and location across different chloroplast lineages. In red algae, starch granules are found in the cytoplasm rather than in the chloroplast.[94] In C4 plants, mesophyll chloroplasts, which do not synthesize sugars, lack starch granules.[18]
Rubisco

The chloroplast stroma contains many proteins, though the most common and important is Rubisco, which is probably also the most abundant protein on the planet.[82] Rubisco is the enzyme that fixes CO2 into sugar molecules. In C3 plants, rubisco is abundant in all chloroplasts, though in C4 plants, it's confined to the bundle sheath chloroplasts, where the Calvin cycle is carried out in C4 plants.[95]
Pyrenoids
The chloroplasts of some hornworts[96] and algae contain structures called pyrenoids. They are not found in higher plants.[97] Pyrenoids are roughly spherical and highly refractive bodies which are a site of starch accumulation in plants that contain them. They consist of an matrix opaque to electrons, surrounded by two hemispherical starch plates. The starch is accumulated as the pyrenoids mature.[98] In algae with carbon concentrating mechanisms, the enzyme rubisco is found in the pyrenoids. Starch can also accumulate around the pyrenoids when CO2 is scarce.[97] Pyrenoids can divide to form new pyrenoids, or be produced "de novo".[98][99]
Thylakoid system

Suspended within the chloroplast stroma is the thylakoid system, a highly dynamic collection of membranous sacks called thylakoids where chlorophyll is found and the light reactions of photosynthesis happen.[7] In most vascular plant chloroplasts, the thylakoids are arranged in stacks called grana,[100] though in certain C4 plant chloroplasts[95] and some algal chloroplasts, the thylakoids are free floating.[8]
Granal structure
Using a light microscope, it's just barely possible to see tiny green granules—which were named grana.[87] With electron microscopy, it became possible to see the thylakoid system in more detail, revealing it to consist of stacks of flat thylakoids which made up the grana, and long interconnecting stromal thylakoids which linked different grana.[87] In the transmission electron microscope, thylakoid membranes appear as alternating light-and-dark bands, 8.5 nanometers thick.[87]
For a long time, the three dimensional structure of the thylakoid system has been unknown or disputed. One model has the granum as a stack of thylakoids linked by helical stromal thylakoids; the other has the granum as a single folded thylakoid connected in a "hub and spoke" way to other grana by stromal thylakoids. While the thylakoid system is still commonly depicted according to the folded thylakoid model,[7] it was determined in 2011 that the stacked and helical thylakoids model is correct.[101]
In the helical thylakoid model, grana consist of a stack of flattened circular granal thylakoids that resemble pancakes. Each granum can contain anywhere from two to a hundred thylakoids,[87] though grana with 10–20 thylakoids are most common.[100] Wrapped around the grana are helicoid stromal thylakoids, also known as frets or lamellar thylakoids. The helices ascend at an angle of 20–25°, connecting to each granal thylakoid at a bridge-like slit junction. The helicoids may extend as large sheets that link multiple grana, or narrow to tube-like bridges between grana.[101] While different parts of the thylakoid system contain different membrane proteins, the thylakoid membranes are continuous and the thylakoid space they enclose form a single continuous labyrinth.[100]
Thylakoids
Thylakoids (sometimes spelled thylakoïds),[102] are small interconnected sacks which contain the membranes that the light reactions of photosynthesis take place on. The word thylakoid comes from the Greek word thylakos which means "sack".[103]
Embedded in the thylakoid membranes are important protein complexes which carry out the light reactions of photosynthesis. Photosystem II and photosystem I contain light-harvesting complexes with chlorophyll and carotenoids that absorb light energy and use it to energize electrons. Molecules in the thylakoid membrane use the energized electrons to pump hydrogen ions into the thylakoid space, decreasing the pH and turning it acidic. ATP synthase is a large protein complex that harnesses the concentration gradient of the hydrogen ions in the thylakoid space to generate ATP energy as the hydrogen ions flow back out into the stroma—much like a dam turbine.[82]
There are two types of thylakoids—granal thylakoids, which are arranged in grana, and stromal thylakoids, which are in contact with the stroma. Granal thylakoids are pancake-shaped circular disks about 300–600 nanometers in diameter. Stromal thylakoids are helicoid sheets that spiral around grana.[100] The flat tops and bottoms of granal thylakoids contain only the relatively flat photosystem II protein complex. This allows them to stack tightly, forming grana with many layers of tightly appressed membrane, called granal membrane, increasing stability and surface area for light capture.[100]
In contrast, photosystem I and ATP synthase are large protein complexes which jut out into the stroma. They can't fit in the appressed granal membranes, and so are found in the stromal thylakoid membrane—the edges of the granal thylakoid disks and the stromal thylakoids. These large protein complexes may act as spacers between the sheets of stromal thylakoids.[100]
The number of thylakoids and the total thylakoid area of a chloroplast is influenced by light exposure. Shaded chloroplasts contain larger and more grana with more thylakoid membrane area than chloroplasts exposed to bright light, which have smaller and fewer grana and less thylakoid area. Thylakoid extent can change within minutes of light exposure or removal.[89]
Pigments and chloroplast colors
Inside the photosystems embedded in chloroplast thylakoid membranes are various photosynthetic pigments, which absorb and transfer light energy. The types of pigments found are different in various groups of chloroplasts, and are responsible for a wide variety of chloroplast colorations.
Chlorophylls
Chlorophyll a is found in all chloroplasts, as well as their cyanobacterial ancestors. Chlorophyll a is a blue-green pigment[104] partially responsible for giving most cyanobacteria and chloroplasts their color. Other forms of chlorophyll exist, such as the accessory pigments chlorophyll b, chlorophyll c, chlorophyll d,[8] and chlorophyll f.
Chlorophyll b is an olive green pigment found only in the chloroplasts of plants, green algae, any secondary chloroplasts obtained through the secondary endosymbiosis of a green alga, and a few cyanobacteria.[8] It's the chlorophylls a and b together that make most plant and green algal chloroplasts green.[104]
Chlorophyll c is mainly found in secondary endosymbiotic chloroplasts that originated from a red alga, though it's not found in chloroplasts of red algae themselves. Chlorophyll c is also found in some green algae and cyanobacteria.[8]
Chlorophylls d and f are pigments found only in some cyanobacteria.[8][105]
Carotenoids
![Delesseria sanguinea, a red alga, has chloroplasts that contain red pigments like phycoerytherin that mask their blue-green chlorophyll a.[22]](http://upload.wikimedia.org/wikipedia/commons/thumb/1/19/Delesseria_sanguinea_Helgoland.JPG/300px-Delesseria_sanguinea_Helgoland.JPG)
In addition to chlorophylls, another group of yellow–orange[104] pigments called carotenoids are also found in the photosystems. There are about thirty photosynthetic carotenoids.[106] They help transfer and dissipate excess energy,[8] and their bright colors sometimes override the chlorophyll green, like during the fall, when the leaves of some land plants change color.[107] β-carotene is a bright red-orange carotenoid found in nearly all chloroplasts, like chlorophyll a.[8] Xanthophylls, especially the orange-red zeaxanthin, are also common.[106] Many other forms of carotenoids exist that are only found in certain groups of chloroplasts.[8]
Phycobilins
Phycobilins are a third group of pigments found in cyanobacteria, and glaucophyte, red algal, and cryptophyte chloroplasts.[8][108] Phycobilins come in all colors, though phycoerytherin is one of the pigments that makes many red algae red.[109] Phycobilins often organize into relatively large protein complexes about 40 nanometers across called phycobilisomes.[8] Like photosystem I and ATP synthase, phycobilisomes jut into the stroma, preventing thylakoid stacking in red algal chloroplasts.[8] Cryptophyte chloroplasts and some cyanobacteria don't have their phycobilin pigments organized into phycobilisomes, and keep them in their thylakoid space instead.[8]
Specialized chloroplasts in C4 plants
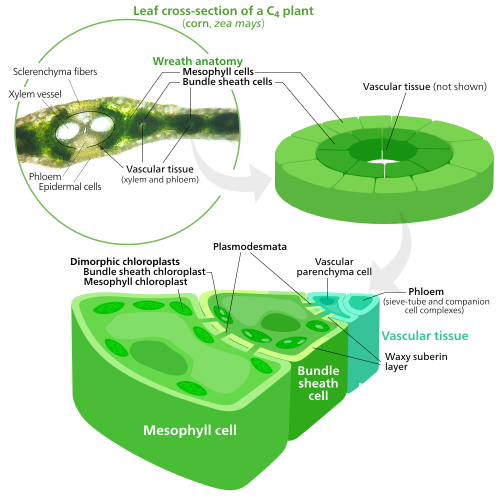
To fix carbon dioxide into sugar molecules in the process of photosynthesis, chloroplasts use an enzyme called rubisco. Rubisco has a problem—it has trouble distinguishing between carbon dioxide and oxygen, so at high oxygen concentrations, rubisco starts accidentally adding oxygen to sugar precursors. This has the end result of ATP energy being wasted and CO2 being released, all with no sugar being produced. This is a big problem, since O2 is produced by the initial light reactions of photosynthesis, causing issues down the line in the Calvin cycle which uses rubisco.[110]
C4 plants evolved a way to solve this—by spatially separating the light reactions and the Calvin cycle. The light reactions, which store light energy in ATP and NADPH, are done in the mesophyll cells of a C4 leaf. The Calvin cycle, which uses the stored energy to make sugar using rubisco, is done in the bundle sheath cells, a layer of cells surrounding a vein in a leaf.[110]
As a result, chloroplasts in C4 mesophyll cells and bundle sheath cells are specialized for each stage of photosynthesis. In mesophyll cells, chloroplasts are specialized for the light reactions, so they lack rubisco, and have normal grana and thylakoids,[95] which they use to make ATP and NADPH, as well as oxygen. They store CO2 in a four-carbon compound, which is why the process is called C4 photosynthesis. The four-carbon compound is then transported to the bundle sheath chloroplasts, where it drops off CO2 and returns to the mesophyll. Bundle sheath chloroplasts do not carry out the light reactions, preventing oxygen from building up in them and disrupting rubisco activity.[110] Because of this, they lack thylakoids organized into grana stacks—though bundle sheath chloroplasts still have free-floating thylakoids in the stroma where they still carry out cyclic electron flow, a light-driven method of synthesizing ATP to power the Calvin cycle without generating oxygen. They lack photosystem II, and only have photosystem I—the only protein complex needed for cyclic electron flow.[95][110] Because the job of bundle sheath chloroplasts is to carry out the Calvin cycle and make sugar, they often contain large starch grains.[95]
Both types of chloroplast contain large amounts of chloroplast peripheral reticulum,[95] which they use to get more surface area to transport stuff in and out of them.[88][89] Mesophyll chloroplasts have a little more peripheral reticulum than bundle sheath chloroplasts.[111]
Location
Distribution in a plant
Not all cells in a multicellular plant contain chloroplasts. All green parts of a plant contain chloroplasts—the chloroplasts, or more specifically, the chlorophyll in them are what make the photosynthetic parts of a plant green.[7] The plant cells which contain chloroplasts are usually parenchyma cells, though chloroplasts can also be found in collenchyma tissue.[112] A plant cell which contains chloroplasts is known as a chlorenchyma cell. A typical chlorenchyma cell of a land plant contains about 10 to 100 chloroplasts.

In some plants such as cacti, chloroplasts are found in the stems,[113] though in most plants, chloroplasts are concentrated in the leaves. One square millimeter of leaf tissue can contain half a million chloroplasts.[7] Within a leaf, chloroplasts are mainly found in the mesophyll layers of a leaf, and the guard cells of stomata. Palisade mesophyll cells can contain 30–70 chloroplasts per cell, while stomatal guard cells contain only around 8–15 per cell, as well as much less chlorophyll. Chloroplasts can also be found in the bundle sheath cells of a leaf, especially in C4 plants, which carry out the Calvin cycle in their bundle sheath cells. They are often absent from the epidermis of a leaf.[114]
Cellular location
Chloroplast movement
The chloroplasts of plant and algal cells can orient themselves to best suit the available light. In low-light conditions, they will spread out in a sheet—maximizing the surface area to absorb light. Under intense light, they will seek shelter by aligning in vertical columns along the plant cell's cell wall or turning sideways so that light strikes them edge-on. This reduces exposure and protects them from photooxidative damage.[115] This ability to distribute chloroplasts so that they can take shelter behind each other or spread out may be the reason why land plants evolved to have many small chloroplasts instead of a few big ones.[116] Chloroplast movement is considered one of the most closely regulated stimulus-response systems that can be found in plants.[117] Mitochondria have also been observed to follow chloroplasts as they move.[118]
In higher plants, chloroplast movement is run by phototropins, blue light photoreceptors also responsible for plant phototropism. In some algae, mosses, ferns, and flowering plants, chloroplast movement is influenced by red light in addition to blue light,[115] though very long red wavelengths inhibit movement rather than speeding it up. Blue light generally causes chloroplasts to seek shelter, while red light draws them out to maximize light absorption.[118]
Studies of Vallisneria gigantea, an aquatic flowering plant, have shown that chloroplasts can get moving within five minutes of light exposure, though they don't initially show any net directionality. They may move along microfilament tracks, and the fact that the microfilament mesh changes shape to form a honeycomb structure surrounding the chloroplasts after they have moved suggests that microfilaments may help to anchor chloroplasts in place.[117][118]
Function and chemistry
Plant innate immunity
Plants lack specialized immune cells—all plant cells participate in the plant immune response. Chloroplasts, along with the nucleus, cell membrane, and endoplasmic reticulum,[119] are key players in pathogen defense. Due to its role in a plant cell's immune response, pathogens frequently target the chloroplast.[119]
Plants have two main immune responses—the hypersensitive response, in which infected cells seal themselves off and undergo programmed cell death, and systemic acquired resistance, where infected cells release signals warning the rest of the plant of a pathogen's presence. Chloroplasts stimulate both responses by purposely damaging their photosynthetic system, producing reactive oxygen species. High levels of reactive oxygen species will cause the hypersensitive response. The reactive oxygen species also directly kill any pathogens within the cell. Lower levels of reactive oxygen species initiate systemic acquired resistance, triggering defense-molecule production in the rest of the plant.[119]
In some plants, chloroplasts are known to move closer to the infection site and the nucleus during an infection.[119]
Chloroplasts can serve as cellular sensors. After detecting stress in a cell, which might be due to a pathogen, chloroplasts begin producing molecules like salicylic acid, jasmonic acid, nitric oxide and reactive oxygen species which can serve as defense-signals. As cellular signals, reactive oxygen species are unstable molecules, so they probably don't leave the chloroplast, but instead pass on their signal to an unknown second messenger molecule. All these molecules initiate retrograde signaling—signals from the chloroplast that regulate gene expression in the nucleus.[119]
In addition to defense signaling, chloroplasts, with the help of the peroxisomes,[120] help synthesize an important defense molecule, jasmonate. Chloroplasts synthesize all the fatty acids in a plant cell[119][121]—linoleic acid, a fatty acid, is a precursor to jasmonate.[119]
Photosynthesis
One of the most important function of the chloroplast is carrying out photosynthesis to make food in the form of sugars for an alga or plant. Water (H2O) and carbon dioxide (CO2) are used in photosynthesis, and sugar and oxygen (O2) is made, using light energy. Photosynthesis is divided into two stages—the light reactions, where water is split to produce oxygen, and the dark reactions, or Calvin cycle, which builds sugar molecules from carbon dioxide. The two phases are linked by the energy carriers adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide phosphate (NADP+).[122]
Light reactions

The light reactions take place on the thylakoid membranes. They take light energy and store it in NADPH, a form of NADP+, and ATP to fuel the dark reactions.
Energy carriers
ATP is the phosphorylated version of adenosine diphosphate (ADP), which stores energy in a cell and powers most cellular activities. ATP is the energized form, while ADP is the (partially) depleted form. NADP+ is an electron carrier which ferries high energy electrons. In the light reactions, it gets reduced, meaning it picks up electrons, becoming NADPH.
Photophosphorylation
Like mitochondria, chloroplasts use the potential energy stored in an H+, or hydrogen ion gradient to generate ATP energy. The two photosystems capture light energy to energize electrons taken from water, and release them down an electron transport chain. The molecules between the photosystems harness the electrons' energy to pump hydrogen ions into the thylakoid space, creating a concentration gradient, with more hydrogen ions (up to a thousand times as many)[82] inside the thylakoid system than in the stroma. The hydrogen ions in the thylakoid space then diffuse back down their concentration gradient, flowing back out into the stroma through ATP synthase. ATP synthase uses the energy from the flowing hydrogen ions to phosphorylate adenosine diphosphate into adenosine triphosphate, or ATP.[82] Because chloroplast ATP synthase projects out into the stroma, the ATP is synthesized there, in position to be used in the dark reactions.[123]
NADP+ reduction
Electrons are often removed from the electron transport chains to charge NADP+ with electrons, reducing it to NADPH. Like ATP synthase, ferredoxin-NADP+ reductase, the enzyme that reduces NADP+, releases the NADPH it makes into the stroma, right where it's needed for the dark reactions.[123]
Because NADP+ reduction removes electrons from the electron transport chains, they must be replaced—the job of photosystem II, which splits water molecules (H2O) to obtain the electrons from its hydrogen atoms.[82][122]
Cyclic photophosphorylation
While photosystem II photolyzes water to obtain and energize new electrons, photosystem I simply reenergizes depleted electrons at the end of an electron transport chain. Normally, the reenergized electrons are taken by NADP+, though sometimes they can flow back down more H+-pumping electron transport chains to transport more hydrogen ions into the thylakoid space to generate more ATP. This is termed cyclic photophosphorylation because the electrons are recycled. Cyclic photophosphorylation is common in C4 plants, which need more ATP than NADPH.[110]
Dark reactions
The Calvin cycle, also known as the dark reactions, is a series of biochemical reactions that fixes CO2 into G3P sugar molecules and uses the energy and electrons from the ATP and NADPH made in the light reactions. The Calvin cycle takes place in the stroma of the chloroplast.[110]
While named "the dark reactions", in most plants, they take place in the light, since the dark reactions are dependent on the products of the light reactions.[7]
Carbon fixation and G3P synthesis
The Calvin cycle starts by using the enzyme Rubisco to fix CO2 into five-carbon Ribulose bisphosphate (RuBP) molecules. The result is unstable six-carbon molecules that immediately break down into three-carbon molecules called 3-phosphoglyceric acid, or 3-PGA. The ATP and NADPH made in the light reactions is used to convert the 3-PGA into glyceraldehyde-3-phosphate, or G3P sugar molecules. Most of the G3P molecules are recycled back into RuBP using energy from more ATP, but one out of every six produced leaves the cycle—the end product of the dark reactions.[110]
Sugar synthesis
Glyceraldehyde-3-phosphate can double up and form glucose-1-phosphate, glucose-6-phosphate or fructose-6-phosphate molecules which each include a phosphate group. To synthesize sucrose, a disaccharide commonly known as table sugar, the G3P molecules are first transported into the cytoplasm by a translocon in the chloroplast membrane. In the cytoplasm, they double up to form fructose-6-phosphate, join with glucose monomers, and have their phosphate groups removed to become the disaccharide sucrose.[124]
Alternatively, glucose monomers in the chloroplast can be linked together to make starch, which accumulates into starch grains in the chloroplast.[124]
Under conditions such as high atmospheric CO2 concentrations, large starch grains may form in the chloroplasts, distorting the grana and thylakoids. The starch granules displace the thylakoids, but leave them intact.[125]
Waterlogged roots can also cause starch buildup in the chloroplasts, possibly due to less sugar being exported through the phloem. This depletes a plant's free phosphate supply, which indirectly stimulates chloroplast starch synthesis.[125] While linked to low photosynthesis rates, the starch grains themselves may not necessarily interfere significantly with the efficiency of photosynthesis,[126] and might simply be a side effect of another photosynthesis-depressing factor.[125]
Photorespiration
Photorespiration can occur when the oxygen concentration is too high. Rubisco cannot distinguish between oxygen and carbon dioxide very well, so it can accidentally add O2 instead of CO2 to RuBP. This process reduces the efficiency of photosynthesis—it consumes ATP and oxygen, releases CO2, and produces no sugar. It can waste up to half the carbon fixed by the Calvin cycle.[122] Several mechanisms have evolved in different lineages that raise the carbon dioxide concentration relative to oxygen within the chloroplast, increasing the efficiency of photosynthesis. These mechanisms are called carbon dioxide concentrating mechanisms, or CCMs. These include Crassulacean acid metabolism, C4 carbon fixation,[122] and pyrenoids. Chloroplasts in C4 plants are notable as they exhibit a distinct chloroplast dimorphism.
pH
Because of the H+ gradient across the thylakoid membrane, the interior of the thylakoid is acidic, with a pH around 4,[127] while the stroma is slightly basic, with a pH of around 8.[128] The optimal stroma pH for the Calvin cycle is 8.1, with the reaction nearly stopping when the pH falls below 7.3.[129]
CO2 in water can form carbonic acid, which can disturb the pH of isolated chloroplasts, interfering with photosynthesis, even though CO2 is used in photosynthesis. However, chloroplasts in living plant cells are not affected by this as much.[128]
Chloroplasts can pump K+ and H+ ions in and out of themselves using a poorly understood light-driven transport system.[128]
In the presence of light, the pH of the thylakoid lumen can drop up to 1.5 pH units, while the pH of the stroma can rise by nearly one pH unit.[129]
Other chemical products
This section needs expansion with: needs more about lipids, also paramylon. You can help by adding to it. (March 2013) |
Chloroplasts are the site of complex lipid metabolism.[130]
Differentiation, replication, and inheritance
Chloroplasts are a special type of plant cell organelle called a plastid, though the two terms are sometimes used interchangeably. There are many other types of plastids, which carry out various functions. All chloroplasts in a plant are descended from undifferentiated proplastids found in the zygote,[131] or fertilized egg. Proplastids are commonly found in an adult plant's apical meristems. Chloroplasts do not normally develop from proplastids in root tip meristems[132]—instead, the formation of starch-storing amyloplasts is more common.[131]
In shoots, proplastids from shoot apical meristems gradually develop into chloroplasts in photosynthetic leaf tissues as the leaf matures, if exposed to the required light. This process involves invaginations of the inner plastid membrane, forming sheets of membrane that project into the internal stroma. These membrane sheets then fold to form thylakoids and grana.[133]
If angiosperm shoots are not exposed to the required light for chloroplast formation, proplastids may develop into an etioplast stage before becoming chloroplasts. An etioplast is a plastid that lacks chlorophyll, and has inner membrane invaginations that form a lattice of tubes in their stroma, called a prolamellar body. Within a few minutes of light exposure, the prolamellar body begins to reorganize into stacks of thylakoids, and chlorophyll starts to be produced. This process, where the etioplast becomes a chloroplast, takes several hours.[133] Gymnosperms do not require light to form chloroplasts.[133]
Light, however, does not guarantee that a proplastid will develop into a chloroplast—what type of plastid a proplastid becomes is largely influenced by the kind of cell it resides in.[131]
Plastid interconversion
Plastid differentiation is not permanent, in fact many interconversions are possible. Chloroplasts may be converted to chromoplasts, which are pigment-filled plastids responsible for the bright colors you see in flowers and ripe fruit. Starch storing amyloplasts can also be converted to chromoplasts, and it's possible for proplastids to develop straight into chromoplasts. Chromoplasts and amyloplasts can also become chloroplasts, like what happens when you illuminate a carrot or a potato. If a plant is injured, or something else causes a plant cell to revert to a meristematic state, chloroplasts and other plastids can turn back into proplastids. Chloroplast, amyloplast, chromoplast, proplast, etc., are not absolute states—intermediate forms are common.[131]
Chloroplast division
This section needs expansion with: functions, Z-ring dynamic assembly, regulators such as Giant Chloroplast 1. You can help by adding to it. (February 2013) |
Most chloroplasts in a photosynthetic cell do not develop directly from proplastids or etioplasts. In fact, a typical shoot meristematic plant cell contains only 7–20 proplastids. These proplastids differentiate into chloroplasts, which divide to create the 30–70 chloroplasts found in a mature photosynthetic plant cell. If the cell divides, chloroplast division provides the additional chloroplasts to partition between the two daughter cells.[134]
In single-celled algae, chloroplast division is the only way new chloroplasts are formed. There is no proplastid differentiation—when an algal cell divides, its chloroplast divides along with it, and each daughter cell receives a mature chloroplast.[133]
Almost all chloroplasts in a cell divide, rather than a small group of rapidly dividing chloroplasts.[135] Chloroplasts have no definite S-phase—their DNA replication is not synchronized or limited to that of their host cells.[136] Much of what we know about chloroplast division comes from studying the alga Cyanidioschyzon merolæ.[116]
The division process starts when the proteins FtsZ1 and FtsZ2 assemble into filaments, and with the help of a protein ARC6, form a structure called a Z-ring within the chloroplast's stroma.[116][137] The Min system manages the placement of the Z-ring, ensuring that the chloroplast is cleaved more or less evenly. The protein MinD prevents FtsZ from linking up and forming filaments. Another protein ARC3 may also be involved, but it is not very well understood. These proteins are active at the poles of the chloroplast, preventing Z-ring formation there, but near the center of the chloroplast, MinE inhibits them, allowing the Z-ring to form.[116]
Next, the two plastid-dividing rings, or PD rings form. The inner plastid-dividing ring is located in the inner side of the chloroplast's inner membrane, and is formed first.[116] The outer plastid-dividing ring is found wrapped around the outer chloroplast membrane. It consists of filaments about 5 nanometers across,[116] arranged in rows 6.4 nanometers apart, and shrinks to squeeze the chloroplast. This is when chloroplast constriction begins.[137]
In a few species like Cyanidioschyzon merolæ, chloroplasts have a third plastid-dividing ring located in the chloroplast's intermembrane space.[116][137]
Late into the constriction phase, dynamin proteins assemble around the outer plastid-dividing ring,[137] helping provide force to squeeze the chloroplast.[116] Meanwhile, the Z-ring and the inner plastid-dividing ring break down.[137] During this stage, the many chloroplast DNA plasmids floating around in the stroma are partitioned and distributed to the two forming daughter chloroplasts.[138]
Later, the dynamins migrate under the outer plastid dividing ring, into direct contact with the chloroplast's outer membrane,[137] to cleave the chloroplast in two daughter chloroplasts.[116]
A remnant of the outer plastid dividing ring remains floating between the two daughter chloroplasts, and a remnant of the dynamin ring remains attached to one of the daughter chloroplasts.[137]
Of the five or six rings involved in chloroplast division, only the outer plastid-dividing ring is present for the entire constriction and division phase—while the Z-ring forms first, constriction does not begin until the outer plastid-dividing ring forms.[137]
Regulation
In species of algae which contain a single chloroplast, regulation of chloroplast division is extremely important to ensure that each daughter cell receives a chloroplast. In organisms like plants, whose cells contain multiple chloroplasts, coordination is looser and less important. It's likely that chloroplast and cell division are somewhat synchronized, though the mechanisms for it are mostly unknown.[116]
Light has been shown to be a requirement for chloroplast division. Chloroplasts can grow and progress through some of the constriction stages under poor quality green light, but are slow to complete division—they require exposure to bright white light to complete division. Spinach leaves grown under green light have been observed to contain many large dumbbell-shaped chloroplasts. Exposure to white light can stimulate these chloroplasts to divide and reduce the population of dumbbell-shaped chloroplasts.[135][138]
Chloroplast inheritance
Like mitochondria, chloroplasts are usually inherited from a single parent. Biparental chloroplast inheritance—where plastid genes are inherited from both parent plants—occurs in very low levels in some flowering plants.[139]
Many mechanisms prevent biparental chloroplast DNA inheritance including selective destruction of chloroplasts or their genes within the gamete or zygote, and chloroplasts from one parent being excluded from the embryo. Parental chloroplasts can be sorted so that only one type is present in each offspring.[140]
Gymnosperms, such as pine trees, mostly pass on chloroplasts paternally,[141] while flowering plants often inherit chloroplasts maternally.[142][143] Flowering plants were once thought to only inherit chloroplasts maternally. However, there are now many documented cases of angiosperms inheriting chloroplasts paternally.[139]
Angiosperms which pass on chloroplasts maternally have many ways to prevent paternal inheritance. Most of them produce sperm cells which do not contain any plastids. There are many other documented mechanisms that prevent paternal inheritance in these flowering plants, such as different rates of chloroplast replication within the embryo.[139]
Among angiosperms, paternal chloroplast inheritance is observed more often in hybrids than in offspring from parents of the same species. This suggests that incompatible hybrid genes might interfere with the mechanisms that prevent paternal inheritance.[139]
Transplastomic plants
Recently, chloroplasts have caught attention by developers of genetically modified crops. Since in most flowering plants, chloroplasts are not inherited from the male parent, transgenes in these plastids cannot be disseminated by pollen. This makes plastid transformation a valuable tool for the creation and cultivation of genetically modified plants that are biologically contained, thus posing significantly lower environmental risks. This biological containment strategy is therefore suitable for establishing the coexistence of conventional and organic agriculture. While the reliability of this mechanism has not yet been studied for all relevant crop species, recent results in tobacco plants are promising, showing a failed containment rate of transplastomic plants at 3 in 1,000,000.[143]
See also
References
- ^ Campbell, Neil A. (2006). Biology: Exploring Life. Boston, Massachusetts: Pearson Prentice Hall. ISBN 978-0-13-250882-7.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "chloroplast". Online Etymology Dictionary.
- ^ a b c d e f g h i j Biology 8th Edition Campbell & Reece. Benjamin Cummings (Pearson). 2009. p. 516.
- ^ Mereschkowsky C (1905). "Über Natur und Ursprung der Chromatophoren im Pflanzenreiche". Biol Centralbl. 25: 593–604.
- ^ Schimper AFW (1883). "Über die Entwicklung der Chlorophyllkörner und Farbkörper". Bot. Zeitung. 41: 105–14, 121–31, 137–46, 153–62.
- ^ a b Kumar, Krithika (2010). "Cyanobacterial Heterocysts". Cold Spring Harb Perspect Biol. 2 (4): a000315. doi:10.1101/cshperspect.a000315. PMC 2845205. PMID 20452939.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e f Biology 8th edition—Campbell&Reece. Benjamin Cummings. 2008. pp. 186–187. ISBN 978-0-321-54325-7.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av aw ax ay az ba bb Aronsson, edited by David G. Robinson, Anna Stina Sandelius, Henrik (2009). The Chloroplast Interactions with the Environment (PDF) ([Online-Ausg.] ed.). Berlin, Heidelberg: Springer Berlin Heidelberg. pp. 3–7. ISBN 9783540686965.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ Bryant, Donald A. (1 November 1979). "The structure of cyanobacterial phycobilisomes: a model". Archives of Microbiology. 123 (2): 113–127. doi:10.1007/BF00446810. PMID 10049814.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag Keeling, Patrick J (2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany. 91 (10): 1481–93. doi:10.3732/ajb.91.10.1481. PMID 21652304. Retrieved 27 January 2013.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d McFadden, Geoffrey I (NaN undefined NaN). "Evolution: Red Algal Genome Affirms a Common Origin of All Plastids". Current Biology. 14 (13): R514–R516. doi:10.1016/j.cub.2004.06.041. PMID 15242632. Retrieved 7 April 2013.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "Chloroplasts". Kimball's Biology Pages. Retrieved 27 December 2012.
- ^ Joyard J Block MA, Douce R (1991). "Molecular aspects of plastid envelope biochemistry". Eur J Biochem. 199 (3): 489–509. doi:10.1111/j.1432-1033.1991.tb16148.x. PMID 1868841.
- ^ "Chloroplast". Encyclopedia of Science. Retrieved 27 December 2012.
- ^ a b c d e f Nakayama, Takuro (2012). "Evolving a photosynthetic organelle". BMC Biology. 10: 35. doi:10.1186/1741-7007-10-35. PMC 3337241. PMID 22531210. Retrieved 27 January 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b c d e f McFadden, Geoffrey I (2001). "Chloroplast Origin and Integration". Plant Physiology. 125 (1): 50–3. doi:10.1104/pp.125.1.50. PMC 1539323. PMID 11154294. Retrieved 27 January 2013.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Archibald, J.M. (2009). "The Puzzle of Plastid Evolution". Current biology. 19 (2): R81–R88. doi:10.1016/j.cub.2008.11.067. PMID 19174147.
- ^ a b c d e f g h i j k l m n o p q r s t u v w x Wise, edited by Robert R. (2006). The structure and function of plastids. Dordrecht: Springer. pp. 3–21. ISBN 978-1-4020-4061-0.
{{cite book}}:|first=has generic name (help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Ball, S. (10 January 2011). "The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis". Journal of Experimental Botany. 62 (6): 1775–1801. doi:10.1093/jxb/erq411. PMID 21220783. Retrieved 6 June 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e Nair, Sethu C. (30). "What Do Human Parasites Do with a Chloroplast Anyway?". PLoS Biology. 9 (8): e1001137. doi:10.1371/journal.pbio.1001137. PMC 3166169. PMID 21912515. Retrieved 6 June 2013.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai Keeling, P. J. (2 February 2010). "The endosymbiotic origin, diversification and fate of plastids". Philosophical Transactions of the Royal Society B: Biological Sciences. 365 (1541): 729–748. doi:10.1098/rstb.2009.0103. Retrieved 8 June 2013.
- ^ a b c d e f g h i j Biology 8th Edition Campbell & Reece. Benjamin Cummings (Pearson). 2009. pp. 582–592.
- ^ "rhodo-". The Free Dictionary. Farlex. Retrieved 7 June 2013.
- ^ a b Lewis, L. A. (1 October 2004). "Green algae and the origin of land plants". American Journal of Botany. 91 (10): 1535–1556. doi:10.3732/ajb.91.10.1535. PMID 21652308. Retrieved 7 June 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Machida, M. (25 April 2006). "Genes for the peptidoglycan synthesis pathway are essential for chloroplast division in moss". Proceedings of the National Academy of Sciences. 103 (17): 6753–6758. doi:10.1073/pnas.0510693103.
{{cite journal}}:|access-date=requires|url=(help) - ^ Moroney, James V (1999). "How Do Algae Concentrate CO2 to Increase the Efficiency of Photosynthetic Carbon Fixation?". Plant Physiology. 119 (1): 9–16. doi:10.1104/pp.119.1.9. PMC 1539202. PMID 9880340. Retrieved 7 June 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Tartar, Aurélien (1 April 2004). "The non-photosynthetic, pathogenic green alga sp. has retained a modified, functional plastid genome". FEMS Microbiology Letters. 233 (1): 153–157. doi:10.1016/j.femsle.2004.02.006. PMID 15043882. Retrieved 9 June 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Chaal, Balbir K.; Green, Beverley R. (2005). "Protein import pathways in 'complex' chloroplasts derived from secondary endosymbiosis involving a red algal ancestor". Plant Molecular Biology. 57 (3): 333–42. doi:10.1007/s11103-004-7848-y. PMID 15830125.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Moustafa, A.; Beszteri, B.; Maier, U. G.; Bowler, C.; Valentin, K.; Bhattacharya, D. (2009). "Genomic Footprints of a Cryptic Plastid Endosymbiosis in Diatoms". Science. 324 (5935): 1724–6. doi:10.1126/science.1172983. PMID 19556510.
- ^ Rogers, M. B. (16 October 2006). "The Complete Chloroplast Genome of the Chlorarachniophyte Bigelowiella natans: Evidence for Independent Origins of Chlorarachniophyte and Euglenid Secondary Endosymbionts". Molecular Biology and Evolution. 24 (1): 54–62. doi:10.1093/molbev/msl129. Retrieved 11 June 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f g h i j k l m n Hackett, J. D.; Anderson, D. M.; Erdner, D. L.; Bhattacharya, D. (2004). "Dinoflagellates: A remarkable evolutionary experiment". American Journal of Botany. 91 (10): 1523–34. doi:10.3732/ajb.91.10.1523. PMID 21652307.
- ^ Dorrell, Richard G (2011). "Do Red and Green Make Brown?: Perspectives on Plastid Acquisitions within Chromalveolates". Eukaryot Cell. 10 (7): A SYMPHONY OF RED, GREEN, AND BROWN–THE DIVERSITY OF ALGAE. doi:10.1128/EC.00326-10. PMC 3147421. PMID 21622904.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Meeson, B. W. (1 January 1982). "Characterization of Peridinin-Chlorophyll α-Proteins from the Marine Dinoflagellate Ceratium furca". Botanica Marina. 25 (8). doi:10.1515/botm.1982.25.8.347.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Tengs, T; Dahlberg, OJ; Shalchian-Tabrizi, K; Klaveness, D; Rudi, K; Delwiche, CF; Jakobsen, KS (2000). "Phylogenetic analyses indicate that the 19'Hexanoyloxy-fucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin". Molecular biology and evolution. 17 (5): 718–29. doi:10.1093/oxfordjournals.molbev.a026350. PMID 10779532.
- ^ a b c Schnepf, Eberhard (1 June 1999). "Dinophyte chloroplasts and phylogeny - A review". Grana. 38 (2–3): 81–97. doi:10.1080/00173139908559217.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Patrick J. Keeling (2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany. 91 (10): 1481–1493. doi:10.3732/ajb.91.10.1481. PMID 21652304.[dead link]
- ^ a b c d Nowack, EC (2011). "Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora". Molecular Biology Evol. 28 (1): 407–22. doi:10.1093/molbev/msq209. PMID 20702568.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e Dann, Leighton (2002). Bioscience—Explained (PDF). Green DNA: BIOSCIENCE EXPLAINED.
- ^ a b c d e Clegg, MT (1994). "Rates and patterns of chloroplast DNA evolution". Proceedings of the National Academy of Sciences. 91 (15): 6795–6801. doi:10.1073/pnas.91.15.6795. PMC 44285. PMID 8041699.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f Shaw, Joey (2006). "Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III". American Journal of Botany. 94 (3): 275–88. doi:10.3732/ajb.94.3.275. PMID 21636401. Retrieved 2 January 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Skovgaard A (1998). "Role of chloroplast retention in a marine dinoflagellate". Aquatic Microbial Ecology. 15: 293–301. doi:10.3354/ame015293. Archived from the original (PDF) on 18 November 2010.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help) - ^ C.Michael Hogan. 2010. Deoxyribonucleic acid. Encyclopedia of Earth. National Council for Science and the Environment. eds. S.Draggan and C.Cleveland. Washington DC
- ^ "ctDNA — chloroplast DNA". AllAcronyms.com. Retrieved 2 January 2013.
- ^ The Oxford Dictionary of Abbreviations. ctDNA—Dictionary definition. 1998.
{{cite book}}: CS1 maint: location missing publisher (link) - ^ "Chloroplasts and Other Plastids". University of Hamburg. Retrieved 27 December 2012.
- ^ a b c d e f g h i j Sandelius, Anna Stina (2009). The Chloroplast: Interactions with the Environment. Springer. p. 18. ISBN 9783540686965.
- ^ a b Burgess,, Jeremy (1989). An introduction to plant cell development ([Pbk. ed.). Cambridge: Cambridge university press. p. 62. ISBN 0521316111.
{{cite book}}: CS1 maint: extra punctuation (link) - ^ a b c Kolodner, Richard (1979). "Inverted repeats in chloroplast DNA from higher plants". Proceedings of the National Academy of Sciences. 76 (1): 41–45. doi:10.1073/pnas.76.1.41. PMC 382872. PMID 16592612.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Palmer, JD (1982). "Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost". Cell. 29 (2): 537–50. doi:10.1016/0092-8674(82)90170-2. PMID 6288261. Retrieved 2 January 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Bendich, Arnold J (2004). "Circular Chloroplast Chromosomes: The Grand Illusion". The Plant Cell. 16 (7): 1661–1666. doi:10.1105/tpc.160771. Retrieved 12 June 2013.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Plant Biochemistry, 3rd edition. Academic Press. 2005. p. 517. ISBN 9780120883912.
- ^ a b Kobayashi, Tamaki (2002). "Detection and Localization of a Chloroplast-Encoded HU-Like Protein That Organizes Chloroplast Nucleoids". The Plant Cell. 14 (7): 1579–1589. doi:10.1105/tpc.002717. PMC 150708. PMID 12119376. Retrieved 21 April 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d Harris, EH (1994). "Chloroplast ribosomes and protein synthesis". American Society for Microbiology—Mol. Biol. Rev. 58 (4): 700–754. PMC 372988. PMID 7854253. Retrieved 27 January 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Wakasugi, T (1998). "Updated Gene Map of Tobacco Chloroplast DNA". Plant Molecular Biology Reporter. 16 (3): 231. doi:10.1023/A:1007564209282. Retrieved 2 January 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Krause K (2008). "From chloroplasts to "cryptic" plastids: evolution of plastid genomes in parasitic plants". Curr. Genet. 54 (3): 111–21. doi:10.1007/s00294-008-0208-8. PMID 18696071.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Peng, Lianwei (24). "Multistep Assembly of Chloroplast NADH Dehydrogenase-Like Subcomplex A Requires Several Nucleus-Encoded Proteins, Including CRR41 and CRR42, in Arabidopsis". Plant Cell. 24 (1): 202–214. doi:10.1105/tpc.111.090597. PMC 3289569. PMID 22274627.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Huang CY, Ayliffe MA, Timmis JN (2003). "Direct measurement of the transfer rate of chloroplast DNA into the nucleus". Nature. 422 (6927): 72–6. doi:10.1038/nature01435. PMID 12594458.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b c Martin W, Rujan T, Richly E (2002). "Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus". Proc. Natl. Acad. Sci. U.S.A. 99 (19): 12246–51. doi:10.1073/pnas.182432999. PMC 129430. PMID 12218172. Archived from the original on 18 November 2010.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Archibald, John M. (1 December 2006). "Algal Genomics: Exploring the Imprint of Endosymbiosis". Current Biology. 16 (24): R1033–R1035. doi:10.1016/j.cub.2006.11.008. Retrieved 12 June 2013.
- ^ Koussevitzky, Shai (4). "Signals from Chloroplasts Converge to Regulate Nuclear Gene Expression". Science. 316 (5825): 715–719. doi:10.1126/science.+1140516. PMID 17395793. Retrieved 27 January 2013.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Hedtke, Boris (8). "Mitochondrial and Chloroplast Phage-Type RNA Polymerases in Arabidopsis". Science. 277 (5327): 809–811. doi:10.1126/science.277.5327.809. PMID 9242608. Retrieved 27 January 2013.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an Soll, Jürgen (2004). "Plant cell biology: Protein import into chloroplasts". Nature Reviews Molecular Cell Biology. 5 (3): 198–208. doi:10.1038/nrm1333.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b Biology 8th edition—Campbell & Reece. Benjamin Cummings. 2008. p. 340. ISBN 978-0-321-54325-7.
- ^ a b c d Wise, Robert R. (2007). Structure and function of plastids. Berlin: Springer. pp. 53–74. ISBN 9781402065705.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d Lee, Dong Wook (2006). "Functional Characterization of Sequence Motifs in the Transit Peptide of Arabidopsis Small Subunit of Rubisco". Plant Physiology. 140 (2): 466–483. doi:10.1104/pp.105.074575.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e May, Timo (2000). "14-3-3 Proteins Form a Guidance Complex with Chloroplast Precursor Proteins in Plants". The Plant Cell. 12 (1): 53–64. doi:10.1105/tpc.12.1.53.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - ^ Lung, S.-C. (18 April 2012). "A Transit Peptide-Like Sorting Signal at the C Terminus Directs the Bienertia sinuspersici Preprotein Receptor Toc159 to the Chloroplast Outer Membrane". The Plant Cell. 24 (4): 1560–1578. doi:10.1105/tpc.112.096248.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d Soll, Jr. (15). "Phosphorylation of the Transit Sequence of Chloroplast Precursor Proteins". Journal of Biological Chemistry. 271 (11): 6545–6554. doi:10.1074/jbc.271.11.6545.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ a b c d e f g h i Jarvis, Paul (1 December 2001). "Toc, Tic, and chloroplast protein import". Biochimica et Biophysica Acta (BBA) — Molecular Cell Research. 1541 (1–2): 64–79. doi:10.1016/S0167-4889(01)00147-1.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Sun, Yuh-Ju (2 January 2001). "Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon". Nature Structural Biology. 9 (2): 95–100. doi:10.1038/nsb744.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e Agne, B. (10 May 2010). "The Acidic A-Domain of Arabidopsis Toc159 Occurs as a Hyperphosphorylated Protein". Plant Physiology. 153 (3): 1016–1030. doi:10.1104/pp.110.158048.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d Shingo Kikuchi; et al. ("2013). "Uncovering the Protein Translocon at the Chloroplast Inner Envelope Membrane". Science. 339 (571): 571–573. doi:10.1126/science.1229262.
{{cite journal}}: Check date values in:|year=(help); Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help) - ^ Mitochondrial Function and Biogenesis. Springer. 2004. p. 59. ISBN 9783540214892.
{{cite book}}: Cite uses deprecated parameter|authors=(help) - ^ Wise, R.R.; Hoober, J.K. (2007). The Structure and Function of Plastids. Springer. pp. 32–33. ISBN 9781402065705.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ "Oedogonium Link ex Hirn, 1900: 17". algaeBASE. Retrieved 19 May 2013.
- ^ "Chlamydomonas Ehrenberg, 1833: 288". algaeBASE. Retrieved 19 May 2013.
- ^ "Spirogyra Link, 1820: 5". algaeBASE. Retrieved 19 May 2013.
- ^ "Sirogonium Kützing, 1843: 278". algaeBASE. Retrieved 19 May 2013.
- ^ "Zygnema C.Agardh, 1817: xxxii, 98". algaeBASE. Retrieved 19 May 2013.
- ^ "Micrasterias C.Agardh ex Ralfs, 1848: 68". algaeBASE. Retrieved 19 May 2013.
- ^ John, D.M. (2002). The freshwater algal flora of the British Isles : an identification guide to freshwater and terrestrial algae (Repr. with corrections. ed.). Cambridge: Cambridge University Press. p. 335. ISBN 9780521770514.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f g Biology 8th edition—Campbell & Reece. Benjamin Cummings. 2008. pp. 196–197. ISBN 978-0-321-54325-7.
- ^ Koike, H (1998 May). "Polypeptide composition of envelopes of spinach chloroplasts: two major proteins occupy 90% of outer envelope membranes" (PDF). Plant & cell physiology. 39 (5): 526–32. doi:10.1093/oxfordjournals.pcp.a029400. PMID 9664716. Retrieved 18 May 2013.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Soll, Jürgen (2004). "Plant cell biology: Protein import into chloroplasts". Nature Reviews Molecular Cell Biology. 5 (3): 198–208. doi:10.1038/nrm1333. PMID 14991000. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Köhler RH, Hanson MR (1 January 2000). "Plastid tubules of higher plants are tissue-specific and developmentally regulated". J. Cell. Sci. 113 (Pt 1): 81–9. PMID 10591627. Archived from the original on 18 November 2010.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help) - ^ Gray JC, Sullivan JA, Hibberd JM, Hansen MR (2001). "Stromules: mobile protrusions and interconnections between plastids". Plant Biology. 3 (3): 223–33. doi:10.1055/s-2001-15204.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e f g h i j k Burgess,, Jeremy (1989). An introduction to plant cell development ([Pbk. ed.). Cambridge: Cambridge university press. p. 46. ISBN 0521316111.
{{cite book}}: CS1 maint: extra punctuation (link) - ^ a b Whatley, Jean M (5). "The occurrence of a peripheral reticulum in plastids of the gymnosperm Welwitschia mirabilis". New Phytologist. 74 (2): 215–220. doi:10.1111/j.1469-8137.1975.tb02608.x.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|month=ignored (help) - ^ a b c Wise, Robert R (2007). The Structure and Function of Plastids. Springer. pp. 17–18. ISBN 9781402065705.
- ^ Manuell, Andrea L. (1 January 2007). "Structure of the Chloroplast Ribosome: Novel Domains for Translation Regulation". PLoS Biology. 5 (8): e209. doi:10.1371/journal.pbio.0050209. PMC 1939882. PMID 17683199. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help)CS1 maint: unflagged free DOI (link) - ^ a b c d Austin II, Jotham R (2006). "Plastoglobules Are Lipoprotein Subcompartments of the Chloroplast That Are Permanently Coupled to Thylakoid Membranes and Contain Biosynthetic Enzymes". The Plant Cell. 18 (7): 1693–1703. doi:10.1105/tpc.105.039859. PMC 1488921. PMID 16731586. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b Crumpton-Taylor, Matilda (2011). "Control of Starch Granule Numbers in Arabidopsis Chloroplasts". Plant Physiology. 158 (2): 905–916. doi:10.1104/pp.111.186957. PMC 3271777. PMID 22135430. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Zeeman, Samuel C. (1 January 2007). "Starch breakdown: recent discoveries suggest distinct pathways and novel mechanisms" (PDF). Functional Plant Biology. 34 (6): 465. doi:10.1071/FP06313. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ J D, Rochaix (1998). The molecular biology of chloroplasts and mitochondria in Chlamydomonas. Dordrecht [u.a.]: Kluwer Acad. Publ. pp. 550–565. ISBN 9780792351740.
- ^ a b c d e f Steer, Brian E.S. Gunning, Martin W. (1996). Plant cell biology : structure and function. Boston, Mass.: Jones and Bartlett Publishers. p. 24. ISBN 0867205040.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ Hanson, David (2002). "Variability of the pyrenoid-based CO2 concentrating mechanism in hornworts (Anthocerotophyta)". Functional Plant Biology. 29 (3): 407. doi:10.1071/PP01210. Retrieved 31 December 2012.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Ma, Yunbing (2011). "Identification of a Novel Gene, CIA6, Required for Normal Pyrenoid Formation in Chlamydomonas reinhardtii". Plant Physiology. 156 (2): 884–96. doi:10.1104/pp.111.173922. PMC 3177283. PMID 21527423. Retrieved 31 December 2012.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Retallack, B (1970). "The Development and Structure of Pyrenoids in Bulbochaete hiloensis". Journal of Cell Science. 6 (1): 229–240. PMID 5417694. Retrieved 2 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Brown, Malcolm R (1970). "Structure and Function of the Algal Pyrenoid" (PDF). Journal of Phycology. Retrieved 31 December 2012.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c d e f Mustardy, L. (24 October 2008). "The Three-Dimensional Network of the Thylakoid Membranes in Plants: Quasihelical Model of the Granum-Stroma Assembly". The Plant Cell. 20 (10): 2552–2557. doi:10.1105/tpc.108.059147. PMC 2590735. PMID 18952780. Retrieved 19 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Austin II, Jotham R (2011). "Three-Dimensional Architecture of Grana and Stroma Thylakoids of Higher Plants as Determined by Electron Tomography". Plant Physiology. 155 (4): 1601–1611. doi:10.1104/pp.110.170647. PMC 3091084. PMID 21224341. Retrieved 18 May 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "The Chloroplast Import Receptor Toc90 Partially Restores the Accumulation of Toc159 Client Proteins in the Arabidopsis thaliana ppi2 Mutant". Molecular Plant. Retrieved 19 May 2013.
- ^ "thylakoid". Merriam-Webster Dictionary. Merriam-Webster. Retrieved 19 May 2013.
- ^ a b c Biology 8th Edition Campbell & Reece. Benjamin Cummings (Pearson). 2009. pp. 190–193.
- ^ "Australian scientists discover first new chlorophyll in 60 years". University of Sydney. 20 August 2010.
- ^ a b c Takaichi, Shinichi (15 June 2011). "Carotenoids in Algae: Distributions, Biosyntheses and Functions". Marine Drugs. 9 (12): 1101–1118. doi:10.3390/md9061101. PMC 3131562.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ Shapley, Dan. "Why Do Leaves Change Color in Fall?". News Articles. The Daily Green. Retrieved 21 May 2013.
- ^ a b Howe, C.J (27 August 2008). "The origin of plastids". Philosophical Transactions of the Royal Society B: Biological Sciences. 363 (1504): 2675–2685. doi:10.1098/rstb.2008.0050.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ "Introduction to the Rhodophyta". University of California Museum of Paleontology. Retrieved 20 May 2013.
- ^ a b c d e f g Biology 8th Edition Campbell & Reece. Benjamin Cummings (Pearson). 2009. pp. 200–201.
- ^ Lawton, June R (1988). "Ultrastructure of Chloroplast Membranes in Leaves of Maize and Ryegrass as Revealed by Selective Staining Methods". New Phytologist. 108 (3): 277–283. doi:10.1111/j.1469-8137.1988.tb04163.x. JSTOR 2433294.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Roberts, editor, Keith (2007). Handbook of plant science. Chichester, West Sussex, England: Wiley. p. 16. ISBN 978-0-470-05723-0.
{{cite book}}:|first=has generic name (help)CS1 maint: multiple names: authors list (link) - ^ Biology 8th edition—Campbell&Reece. Benjamin Cummings. 2008. p. 742. ISBN 978-0-321-54325-7.
- ^ "Guard Cell Photosynthesis". Plant Physiology Online. Sinauer. Retrieved 14 April 2013.
- ^ a b Eckardt, Nancy A (2003). "Controlling Organelle Positioning: A Novel Chloroplast Movement Protein". The Plant Cell. 15 (12): 2755–2757. doi:10.1105/tpc.151210. Retrieved 6 March 2013.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c d e f g h i j Glynn, Jonathan M. (1 May 2007). "Chloroplast Division". Traffic. 8 (5): 451–461. doi:10.1111/j.1600-0854.2007.00545.x. PMID 17451550. Retrieved 2 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b Dong, Xia-Jing (1998). "Microfilaments Anchor Chloroplasts along the Outer Periclinal Wall in Vallisneria Epidermal Cells through Cooperation of PFR and Photosynthesis" (PDF). Plant Cell Physiology. 39 (12): 1299–1306. doi:10.1093/oxfordjournals.pcp.a029334. Retrieved 6 March 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Takagi, Shingo (NaN undefined NaN). "Actin-based photo-orientation movement of chloroplasts in plant cells". Journal of Experimental Biology (206): 1963–1969. Retrieved 6 March 2013.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|month=ignored (help) - ^ a b c d e f g Padmanabhan, Meenu S. (1 November 2010). "All Hands on Deck—The Role of Chloroplasts, Endoplasmic Reticulum, and the Nucleus in Driving Plant Innate Immunity". Molecular Plant-Microbe Interactions. 23 (11): 1368–1380. doi:10.1094/MPMI-05-10-0113. PMID 20923348. Retrieved 4 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ Katsir, L. (2008). "Jasmonate signaling: a conserved mechanism of hormone sensing". Curr Bio. 11 (4): 428–435. doi:10.1016/j.pbi.2008.05.004.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Schnurr, Judy (2002). "Fatty Acid Export from the Chloroplast. Molecular Characterization of a Major Plastidial Acyl-Coenzyme A Synthetase from Arabidopsis". Plant Physiology. 129 (4): 1700–1709. doi:10.1104/pp.003251. PMC 166758. PMID 12177483. Retrieved 4 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d Biology—Concepts and Connections. Pearson. 2009. pp. 108–118.
- ^ a b Clarke, Jeremy M. Berg; John L. Tymoczko; Lubert Stryer. Web content by Neil D. (2002). Biochemistry (5. ed., 4. print. ed.). New York, NY [u.a.]: W. H. Freeman. pp. Section 19.4. ISBN 0-7167-3051-0.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b Clarke, Jeremy M. Berg; John L. Tymoczko; Lubert Stryer. Web content by Neil D. (2002). Biochemistry (5. ed., 4. print. ed.). New York, NY [u.a.]: W. H. Freeman. pp. Section 20.1. ISBN 0-7167-3051-0.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b c Wample, Robert L (1983 September). "Effect of Flooding on Starch Accumulation in Chloroplasts of Sunflower (Helianthus annuus L.)" (PDF). Plant Physiology. 73 (1): 195–198. doi:10.1104/pp.73.1.195. PMC 1066435. PMID 16663176. Retrieved 5 March 2013.
{{cite journal}}: Check date values in:|date=(help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Carmi, A.; Shomer, I. (1979). "Starch Accumulation and Photosynthetic Activity in Primary Leaves of Bean (Phaseolus vulgaris L.)". Annals of Botany. 44 (4): 479–484.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Berg JM (2002). Biochemistry (5th ed.). W H Freeman. pp. Section 19.4. Retrieved 30 October 2012.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ a b c Hauser, Matthias (1995). "Stimulation by Light of Rapid pH Regulation in the Chloroplast Stroma in Vivo as lndicated by CO, Solubilization in Leaves". Plant Physiology. 108 (3): 1059–1066. doi:10.1104/pp.108.3.1059. PMC 157457. PMID 12228527. Retrieved 2 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b Werdan, Karl (1975). "The role of pH in the regulation of carbon fixation in the chloroplast stroma. Studies on CO2 fixation in the light and dark". Science Direct. 396 (2): 276. doi:10.1016/0005-2728(75)90041-9. Retrieved 9 December 2012.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Joyard, Jacques (23). "Chloroplast proteomics highlights the subcellular compartmentation of lipid metabolism" (PDF). Science Direct: 1. Retrieved 28 December 2012.
{{cite journal}}: Check date values in:|date=and|year=/|date=mismatch (help); Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d Burgess,, Jeremy (1989). An introduction to plant cell development ([Pbk. ed.). Cambridge: Cambridge university press. p. 56. ISBN 0521316111.
{{cite book}}: CS1 maint: extra punctuation (link) - ^ Steer, Brian E.S. Gunning, Martin W. (1996). Plant cell biology : structure and function. Boston, Mass.: Jones and Bartlett Publishers. p. 20. ISBN 0867205040.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ^ a b c d e Burgess,, Jeremy (1989). An introduction to plant cell development ([Pbk. ed.). Cambridge: Cambridge university press. pp. 54–55. ISBN 0521316111.
{{cite book}}: CS1 maint: extra punctuation (link) - ^ Burgess,, Jeremy (1989). An introduction to plant cell development ([Pbk. ed.). Cambridge: Cambridge university press. p. 57. ISBN 0521316111.
{{cite book}}: CS1 maint: extra punctuation (link) - ^ a b Possingham, J. V. (18 May 1976). "Chloroplast Replication and Chloroplast DNA Synthesis in Spinach Leaves" (PDF). Proceedings of the Royal Society B: Biological Sciences. 193 (1112): 295–305. doi:10.1098/rspb.1976.0047. Retrieved 2 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Heinhorst, S (1993). "DNA replication in chloroplasts". Journal of Cell Science. 104 (104): 1–9. Retrieved 3 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d e f g h Miyagishima, Shin-ya (2003). "A Plant-Specific Dynamin-Related Protein Forms a Ring at the Chloroplast Division Site". The Plant Cell. 15 (3): 655–665. doi:10.1105/tpc.009373. PMC 150020. PMID 12615939. Retrieved 2 February 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b Hashimoto, Haruki (1989). "Effect of Light on the Chloroplast Division Cycle and DNA Synthesis in Cultured Leaf Discs of Spinach". Plant Physiology. 89 (4): 1178–1183. doi:10.1104/pp.89.4.1178. PMC 1055993. PMID 16666681.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help); Unknown parameter|month=ignored (help) - ^ a b c d Hansen, A. K. (1 January 2007). "Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): implications for phylogenetic studies". American Journal of Botany. 94 (1): 42–46. doi:10.3732/ajb.94.1.42. PMID 21642206. Retrieved 21 April 2013.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Birky Jr, C William (1995). "Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution" (PDF). Proceedings of the National Academy of Science. 92 (25): 11331–11338. Bibcode:1995PNAS...9211331B. doi:10.1073/pnas.92.25.11331. Retrieved 21 April 2013.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Powell W, Morgante M, McDevitt R, Vendramin GG, Rafalski JA (1995). "Polymorphic simple sequence repeat regions in chloroplast genomes: applications to the population genetics of pines". Proc. Natl. Acad. Sci. U.S.A. 92 (17): 7759–63. doi:10.1073/pnas.92.17.7759. PMC 41225. PMID 7644491.
In the pines, the chloroplast genome is transmitted through pollen
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Stegemann S, Hartmann S, Ruf S, Bock R (2003). "High-frequency gene transfer from the chloroplast genome to the nucleus". Proc. Natl. Acad. Sci. U.S.A. 100 (15): 8828–33. doi:10.1073/pnas.1430924100. PMC 166398. PMID 12817081. Archived from the original on 18 November 2010.
most angiosperm species inherit their chloroplasts maternally
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Ruf S, Karcher D, Bock R (2007). "Determining the transgene containment level provided by chloroplast transformation". Proc. Natl. Acad. Sci. U.S.A. 104 (17): 6998–7002. doi:10.1073/pnas.0700008104. PMC 1849964. PMID 17420459. Archived from the original on 18 November 2010.
{{cite journal}}: Unknown parameter|deadurl=ignored (|url-status=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link)
External links
- Chloroplast - Cell Centered Database
- http://www.ncbi.nlm.nih.gov/nuccore/7525012?report=graphChloroplasts and Photosynthesis: The Role of Light from Kimball's Biology Pages
- Clegg MT, Gaut BS, Learn GH, Morton BR (1994). "Rates and patterns of chloroplast DNA evolution". Proc. Natl. Acad. Sci. U.S.A. 91 (15): 6795–801. doi:10.1073/pnas.91.15.6795. PMC 44285. PMID 8041699.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - 3D structures of proteins associated with thylakoid membrane
- Co-Extra research on chloroplast transformation
- NCBI full chloroplast genome










![Toc34 from a pea plant. Toc34 has three almost identical molecules (shown in slightly different shades of green), each of which forms a dimer with one of its adjacent molecules. Part of a GDP molecule binding site is highlighted in pink.[70]](http://upload.wikimedia.org/wikipedia/commons/thumb/e/e4/TOC34.png/350px-TOC34.png)




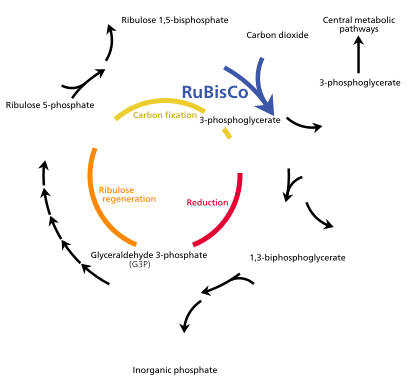




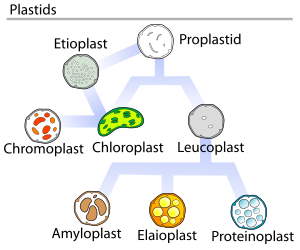

![Most chloroplasts in plant cells, and all chloroplasts in algae arise from chloroplast division.[133]](http://upload.wikimedia.org/wikipedia/commons/thumb/d/df/Chloroplast_division.svg/800px-Chloroplast_division.svg.png)