Coronaviridae
| Coronaviridae | |
|---|---|

| |
| |
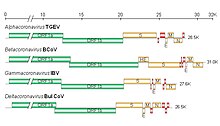
| Diagram, electron micrograph, and genome of coronavirus types. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Suborder: | Cornidovirineae |
| Family: | Coronaviridae |
| Subfamilies and genera | |
Coronaviridae is a family of enveloped, positive-sense, single-stranded RNA viruses which infects amphibians, birds, and mammals. The viral genome is 26–32 kilobases in length. The particles are typically decorated with large (~20 nm), club- or petal-shaped surface projections (the "peplomers" or "spikes"), which in electron micrographs of spherical particles create an image reminiscent of the solar corona.[1][2]
Virology

The 5' and 3' ends of the genome have a cap and poly(A) tract, respectively. The viral envelope, obtained by budding through membranes of the endoplasmic reticulum (ER) or Golgi apparatus, invariably contains two virus-specified (glyco)protein species, S and M. Glycoprotein S comprises the large surface projections, while M is a triple-spanning transmembrane protein. Toroviruses and a select subset of coronaviruses (in particular the members of subgroup A in the genus Betacoronavirus) possess, in addition to the peplomers composed of S, a second type of surface projections composed of the hemagglutinin-esterase protein. Another important structural protein is the phosphoprotein N, which is responsible for the helical symmetry of the nucleocapsid that encloses the genomic RNA.[3]
Genetic recombination can occur when at least two viral genomes are present in the same infected host cell. RNA recombination appears to be a major driving force in coronavirus evolution. Recombination can determine genetic variability within a CoV species, the capability of a CoV species to jump from one host to another and, infrequently, the emergence of a novel CoV.[4] The exact mechanism of recombination in CoVs is not known, but likely involves template switching during genome replication.[4]
Taxonomy

The family Coronaviridae is organized in 2 sub-families, 5 genera, 26 sub-genera, and 46 species.[5] Additional species are pending or tentative.[6]
Coronavirus
Coronavirus is the common name for Coronaviridae and Orthocoronavirinae, also called Coronavirinae.[8][9] Coronaviruses cause diseases in mammals and birds. In humans, the viruses cause respiratory infections, including the common cold, which are typically mild, though rarer forms such as SARS (including the one causing COVID-19) and MERS can be lethal.[10] Symptoms vary in other species: in chickens, they cause an upper respiratory disease, while in cows and pigs coronaviruses cause diarrhea. There are no vaccines or antiviral drugs to prevent or treat human coronavirus infections. They are enveloped viruses with a positive-sense single-stranded RNA genome and a nucleocapsid of helical symmetry. The genome size of coronaviruses ranges from approximately 26 to 32 kilobases, among the largest for an RNA virus (second only to a 41-kb nidovirus recently discovered in planaria).[11][12]
Orthocoronavirinae taxonomy
- Orthocoronavirinae
- Alphacoronavirus
- Betacoronavirus
- Gammacoronavirus
- Deltacoronavirus
References
- ^ King, Andrew M. Q.; Adams, Michael J.; Carstens, Eric B.; Lefkowitz, Elliot J., eds. (2012-01-01), "Order - Nidovirales", Virus Taxonomy, Elsevier, pp. 784–794, ISBN 978-0-12-384684-6, retrieved 2020-06-08
- ^ Bukhari, Khulud; Mulley, Geraldine; Gulyaeva, Anastasia A.; Zhao, Lanying; Shu, Guocheng; Jiang, Jianping; Neuman, Benjamin W. (2018-11-01). "Description and initial characterization of metatranscriptomic nidovirus-like genomes from the proposed new family Abyssoviridae, and from a sister group to the Coronavirinae, the proposed genus Alphaletovirus". Virology. 524: 160–171. doi:10.1016/j.virol.2018.08.010. ISSN 0042-6822. PMC 7112036. PMID 30199753.
- ^ McBride R, van Zyl M, Fielding BC (August 2014). "The coronavirus nucleocapsid is a multifunctional protein". Viruses. 6 (8): 2991–3018. doi:10.3390/v6082991. PMC 4147684. PMID 25105276.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ a b Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 2016 Jun;24(6):490-502. doi: 10.1016/j.tim.2016.03.003. Epub 2016 Mar 21. Review. PMID 27012512
- ^ "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 11 May 2020.
- ^ Gorbalenya A, Baker S, Baric R, de Groot R, Drosten C, Gulyaeva A, Haagmans B, Lauber C, Leontovich A, Neuman B, Penzar D, Perlman S, Poon L, Samborskiy D, Sidorov I, Sola I, Ziebuhr J (2020). "The species Severe acute respiratory syndrome related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2" (PDF). Nature Microbiology. 5 (4): 536–44. doi:10.1038/s41564-020-0695-z. PMC 7095448. PMID 32123347.
- ^ Fan Y, Zhao K, Shi ZL, Zhou P (March 2019). "Bat Coronaviruses in China". Viruses. 11 (3): 210. doi:10.3390/v11030210. PMC 6466186. PMID 30832341.
{{cite journal}}: CS1 maint: unflagged free DOI (link) - ^ "2017.012-015S" (xlsx). International Committee on Taxonomy of Viruses (ICTV). October 2018. Archived from the original on 14 May 2019. Retrieved 24 January 2020.
- ^ "The 2019–2020 Novel Coronavirus (Severe Acute Respiratory Syndrome Coronavirus 2) Pandemic: A Joint American College of Academic International Medicine‑World Academic Council of Emergency Medicine Multidisciplinary COVID‑19 Working Group Consensus Paper". ResearchGate. Retrieved May 16, 2020.
{{cite web}}: CS1 maint: url-status (link) - ^ "Coronavirus - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2020-04-02.
- ^ Saberi A, Gulyaeva AA, Brubacher JL, Newmark PA, Gorbalenya AE (November 2018). Stanley P (ed.). "A planarian nidovirus expands the limits of RNA genome size". PLOS Pathogens. 14 (11): e1007314. doi:10.1371/journal.ppat.1007314. PMC 6211748. PMID 30383829.
{{cite journal}}: CS1 maint: unflagged free DOI (link)
