Laminin: Difference between revisions
Amkilpatrick (talk | contribs) Reflinks: Converting bare references |
Undid revision 567808845 by Miguelferig (talk) This is one reason why Laminin is notable in popular culture and therefore merits inclusion in this article. |
||
| Line 211: | Line 211: | ||
[[LAMA1]]; [[LAMA2]]; [[LAMA3]]; [[LAMA5]]; [[LAMB1]]; [[LAMB2]]; [[LAMB3]]; [[LAMB4]]; |
[[LAMA1]]; [[LAMA2]]; [[LAMA3]]; [[LAMA5]]; [[LAMB1]]; [[LAMB2]]; [[LAMB3]]; [[LAMB4]]; |
||
[[LAMC1]]; [[LAMC3]]; [[NTN1]]; [[NTN2L]]; [[NTN4]]; [[NTNG1]]; [[NTNG2]]; [[USH2A]]; |
[[LAMC1]]; [[LAMC3]]; [[NTN1]]; [[NTN2L]]; [[NTN4]]; [[NTNG1]]; [[NTNG2]]; [[USH2A]]; |
||
==Role in culture== |
|||
Many [[Christian apologists]], most notably [[Louie Giglio]], have written about the fact that laminin is shaped like a [[Christian cross]], comparing the role of the cross in [[Christian theology|theology]] and laminin's role in the human body. For example, David D. Swanson states: "Our knowledge of truth is most clearly revealed on the cross of Christ. And what holds the human body together? Laminin. And what does it look like? A cross. Coincidence? Some would say yes, but I think it is yet another way God reveals his glory to us. I think God is the one who holds all things together-our bodies-our world-our lives."<ref>{{cite book|last=Swanson|first=David D.|title=Learning to Be You|year=2012|publisher=Baker Books|isbn=1441238492}}</ref> Fazale Rana, a biochemist and apologist, disagrees with Giglio and instead argues that "Instead of pointing to superficial features of biomolecules such as the “cross-shaped” architecture of laminin, there are many more substantive ways to use biochemistry to argue for the necessity of a Creator."<ref>{{cite web|last=Rana|first=Fazale|title=Crossed off the List: Is the Cross Shape of Laminin Evidence for the Creator?|url=http://www.reasons.org/articles/crossed-off-the-list-is-the-cross-shape-of-laminin-evidence-for-the-creator|publisher=RTB|accessdate=26 July 2013}}</ref> |
|||
==See also== |
==See also== |
||
Revision as of 20:55, 11 August 2013
Laminins are major proteins in the basal lamina (one of the layers of the basement membrane), a protein network foundation for most cells and organs. The laminins are an important and biologically active part of the basal lamina, influencing cell differentiation, migration, and adhesion, as well as phenotype and survival.[1]
Laminins are trimeric proteins that contain an α-chain, a β-chain, and a γ-chain, found in five, four, and three genetic variants, respectively. The laminin molecules are named according to their chain composition. Thus, laminin-511 contains α5, β1, and γ1 chains.[2] Fourteen other chain combinations have been identified in vivo. The trimeric proteins intersect to form a cross-like structure that can bind to other cell membrane and extracellular matrix molecules.[3] The three shorter arms are particularly good at binding to other laminin molecules, which allows them to form sheets. The long arm is capable of binding to cells, which helps anchor organized tissue cells to the membrane.
The laminin family of glycoproteins are an integral part of the structural scaffolding in almost every tissue of an organism. They are secreted and incorporated into cell-associated extracellular matrices. Laminin is vital for the maintenance and survival of tissues. Defective laminins can cause muscles to form improperly, leading to a form of muscular dystrophy, lethal skin blistering disease (junctional epidermolysis bullosa) and defects of the kidney filter (nephrotic syndrome). [4]
Types
Fifteen laminin trimers have been identified. The laminins are combinations of different alpha-, beta-, and gamma-chains.[5]
- The five forms of alpha-chains are: LAMA1, LAMA2, LAMA3, LAMA4, LAMA5
- The beta-chains include: LAMB1, LAMB2, LAMB3, LAMB4
- The gamma-chains are: LAMC1, LAMC2, LAMC3
Laminins were previously numbered - e.g. laminin-1, laminin-2, laminin-3 - but the nomenclature was recently[when?] changed to describe which chains are present in each isoform. For example, laminin-511 contains an α5-chain, a β1-chain and a γ1 chain.[2]
Networks
Laminins form independent networks and are associated with type IV collagen networks via entactin,[6] fibronectin,[7] and perlecan. They also bind to cell membranes through integrin receptors and other plasma membrane molecules, such as the dystroglycan glycoprotein complex and Lutheran blood group glycoprotein.[3] Through these interactions, laminins critically contribute to cell attachment and differentiation, cell shape and movement, maintenance of tissue phenotype, and promotion of tissue survival.[3][5] Some of these biological functions of laminin have been associated with specific amino-acid sequences or fragments of laminin.[3] For example, the peptide sequence [GTFALRGDNGDNGQ], which is located on the alpha-chain of laminin, promotes adhesion of endothelial cells.[8]
Pathology
Dysfunctional structure of one particular laminin, laminin-211, is the cause of one form of congenital muscular dystrophy.[9] Laminin-211 is composed of an α2, a β1 and a γ1 chains. This laminin's distribution includes the brain and muscle fibers. In muscle, it binds to alpha dystroglycan and integrin alpha7—beta1 via the G domain, and via the other end binds to the extracellular matrix. Abnormal laminin-332, which is essential for epithelial cell adhesion to the basement membrane, leads to a condition called junctional epidermolysis bullosa, characterized by generalized blisters, exuberant granulation tissue of skin and mucosa, and pitted teeth. Malfunctional laminin-521 in the kidney filter causes leakage of protein into the urine and nephrotic syndrome.[4]
Laminins in cell culture
Recently, several publications have reported that laminins can be used to culture cells, such as pluripotent stem cells, that are difficult to culture on other substrates. Mostly, two types of laminins have been used. Laminin-111 extracted from mouse sarcomas is one popular laminin type, as well as a mixture of laminins 511 and 521 from human placenta.[10] Various laminin isoforms are practically impossible to isolate from tissues in pure form due to extensive cross-linking and the need for harsh extraction conditions, such as proteolytic enzymes or low pH, that cause degradation. However, professor Tryggvason's group at the Karolinska Institute in Sweden showed how to produce recombinant laminins using HEK293 cells in 2000. Kortesmaa et al. 2000. This made it possible to test if laminins could have a significant role in vitro as they have in the human body. In 2008, two groups independently showed that mouse embryonic stem cells can be grown for months on top of recombinant laminin-511.[11][12] Later, Rodin et al. showed that recombinant laminin 511 can be used to create a totally xeno-free and defined cell culture environment to culture human pluripotent ES cells and human iPS cells.[13]
Role in neural development
Laminin-111 is a major substrate along which nerve axons will grow, both in vivo and in vitro. For example, it lays down a path that developing retinal ganglion cells follow on their way from the retina to the tectum. It is also often used as a substrate in cell culture experiments. Interestingly, the presence of laminin-1 can influence how the growth cone responds to other cues. For example, growth cones are repelled by netrin when grown on laminin-111, but are attracted to netrin when grown on fibronectin. This effect of laminin-111 probably occurs through a lowering of intracellular cyclic AMP.
Role in cancer
This section's tone or style may not reflect the encyclopedic tone used on Wikipedia. (July 2012) |
The majority of transcripts that harbor an internal ribosome entry site (IRES) are involved in cancer development via corresponding proteins. A crucial event in tumor progression referred to as epithelial to mesenchymal transition (EMT) allows carcinoma cells to acquire invasive properties. The translational activation of the extracellular matrix component laminin B1 (LamB1) during EMT has been recently reported suggesting an IRES-mediated mechanism. In this study, the IRES activity of LamB1 was determined by independent bicistronic reporter assays. Strong evidences exclude an impact of cryptic promoter or splice sites on IRES-driven translation of LamB1. Furthermore, no other LamB1 mRNA species arising from alternative transcription start sites or polyadenylation signals were detected that account for its translational control. Mapping of the LamB1 5'-untranslated region (UTR) revealed the minimal LamB1 IRES motif between -293 and -1 upstream of the start codon. Notably, RNA affinity purification showed that the La protein interacts with the LamB1 IRES. This interaction and its regulation during EMT were confirmed by ribonucleoprotein immunoprecipitation. In addition, La was able to positively modulate LamB1 IRES translation. In summary, these data indicate that the LamB1 IRES is activated by binding to La which leads to translational upregulation during hepatocellular EMT.[14]
Laminin domains
| Laminin Domain I | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Laminin_I | ||||||||
| Pfam | PF06008 | ||||||||
| InterPro | IPR009254 | ||||||||
| |||||||||
| Laminin Domain II | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Laminin_II | ||||||||
| Pfam | PF06009 | ||||||||
| InterPro | IPR010307 | ||||||||
| |||||||||
| Laminin B (Domain IV) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Laminin_B | ||||||||
| Pfam | PF00052 | ||||||||
| Pfam clan | CL0001 | ||||||||
| InterPro | IPR000034 | ||||||||
| |||||||||
| Laminin EGF-like (Domains III and V) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of three consecutive laminin-type epidermal growth factor-like (le) modules of laminin gamma1 chain harboring the nidogen binding site | |||||||||
| Identifiers | |||||||||
| Symbol | Laminin_EGF | ||||||||
| Pfam | PF00053 | ||||||||
| Pfam clan | CL0001 | ||||||||
| InterPro | IPR002049 | ||||||||
| PROSITE | PDOC00021 | ||||||||
| SCOP2 | 1tle / SCOPe / SUPFAM | ||||||||
| |||||||||
| Laminin G domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
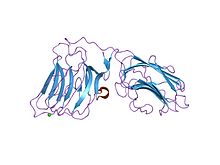 laminin alpha 2 chain lg4-5 domain pair, ca1 site mutant | |||||||||
| Identifiers | |||||||||
| Symbol | Laminin_G_1 | ||||||||
| Pfam | PF00054 | ||||||||
| Pfam clan | CL0004 | ||||||||
| InterPro | IPR012679 | ||||||||
| SCOP2 | 1qu0 / SCOPe / SUPFAM | ||||||||
| |||||||||
| Laminin G domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 the structure of the ligand-binding domain of neurexin 1beta: regulation of lns domain function by alternative splicing | |||||||||
| Identifiers | |||||||||
| Symbol | Laminin_G_2 | ||||||||
| Pfam | PF02210 | ||||||||
| Pfam clan | CL0004 | ||||||||
| InterPro | IPR012680 | ||||||||
| SMART | TSPN | ||||||||
| |||||||||
| Laminin N-terminal (Domain VI) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | Laminin_N | ||||||||
| Pfam | PF00055 | ||||||||
| Pfam clan | CL0202 | ||||||||
| InterPro | IPR008211 | ||||||||
| SMART | LamNT | ||||||||
| SCOP2 | 1klo / SCOPe / SUPFAM | ||||||||
| |||||||||
Laminins contain several conserved protein domains.
Laminin I and Laminin II
Laminins are trimeric molecules; laminin-1 is an alpha1 beta1 gamma1 trimer. It has been suggested that the domains I and II from laminin A, B1 and B2 may come together to form a triple helical coiled-coil structure.[15]
Laminin B
The laminin B domain (also known as domain IV) is an extracellular module of unknown function. It is found in a number of different proteins that include, heparan sulphate proteoglycan from basement membrane, a laminin-like protein from Caenorhabditis elegans and laminin. Laminin IV domain is not found in short laminin chains (alpha4 or beta3).
Laminin EGF-like
Beside different types of globular domains each laminin subunit contains, in its first half, consecutive repeats of about 60 amino acids in length that include eight conserved cysteines.[16] The tertiary structure of this domain is remotely similar in its N-terminus to that of the EGF-like module.[17][18] It is also known as a 'LE' or 'laminin-type EGF-like' domain. The number of copies of the laminin EGF-like domain in the different forms of laminins is highly variable; from 3 up to 22 copies have been found. In mouse laminin gamma-1 chain, the seventh LE domain has been shown to be the only one that binds with a high affinity to nidogen.[19] The binding-sites are located on the surface within the loops C1-C3 and C5-C6.[17][18] Long consecutive arrays of laminin EGF-like domains in laminins form rod-like elements of limited flexibility, which determine the spacing in the formation of laminin networks of basement membranes.[20][21]
Laminin G
The laminin globular (G) domain can be found in one to several copies in various laminin family members, which includes a large number of extracellular proteins. The C terminus of laminin alpha chain contains a tandem repeat of five laminin G domains, which are critical for heparin-binding and cell attachment activity.[22] Laminin alpha4 is distributed in a variety of tissues including peripheral nerves, dorsal root ganglion, skeletal muscle and capillaries; in the neuromuscular junction, it is required for synaptic specialisation.[23] The structure of the laminin-G domain has been predicted to resemble that of pentraxin.[24]
Laminin G domains can vary in their function, and a variety of binding functions has been ascribed to different LamG modules. For example, the laminin alpha1 and alpha2 chains each has five C-terminal laminin G domains, where only domains LG4 and LG5 contain binding sites for heparin, sulphatides and the cell surface receptor dystroglycan.[22] Laminin G-containing proteins appear to have a wide variety of roles in cell adhesion, signalling, migration, assembly and differentiation.
Laminin N-terminal
Basement membrane assembly is a cooperative process in which laminins polymerise through their N-terminal domain (LN or domain VI) and anchor to the cell surface through their G domains. Netrins may also associate with this network through heterotypic LN domain interactions.[21] This leads to cell signalling through integrins and dystroglycan (and possibly other receptors) recruited to the adherent laminin. This LN domain dependent self-assembly is considered to be crucial for the integrity of basement membranes, as highlighted by genetic forms of muscular dystrophy containing the deletion of the LN module from the alpha 2 laminin chain.[25] The laminin N-terminal domain is found in all laminin and netrin subunits except laminin alpha 3A, alpha 4 and gamma 2.
Human proteins containing laminin domains
Laminin Domain I
LAMA1; LAMA2; LAMA3; LAMA4; LAMA5;
Laminin Domain II
LAMA1; LAMA2; LAMA3; LAMA4; LAMA5;
Laminin B (Domain IV)
HSPG2; LAMA1; LAMA2; LAMA3; LAMA5; LAMC1; LAMC2; LAMC3;
Laminin EGF-like (Domains III and V)
AGRIN; ATRN; ATRNL1; CELSR1; CELSR2; CELSR3; CRELD1; HSPG2; LAMA1; LAMA2; LAMA3; LAMA4; LAMA5; LAMB1; LAMB2; LAMB3; LAMB4; LAMC1; LAMC2; LAMC3; MEGF10; MEGF12; MEGF6; MEGF8; MEGF9; NSR1; NTN1; NTN2L; NTN4; NTNG1; NTNG2; RESDA1; SCARF1; SCARF2; SREC; STAB1; USH2A;
Laminin G domain
AGRIN; CASPR4; CELSR1; CELSR2; CELSR3; CNTNAP1; CNTNAP2; CNTNAP3; CNTNAP4; CNTNAP5; COL11A1; COL11A2; COL24A1; COL5A1; COL5A3; CRB1; CRB2; CSPG4; EGFLAM; FAT; FAT2; FAT4; GAS6; HSPG2; LAMA1; LAMA2; LAMA3; LAMA4; LAMA5; NELL2; NRXN1; NRXN2; NRXN3; PROS1; RESDA1; SLIT1; SLIT2; SLIT3; USH2A;
Laminin N-terminal (Domain VI)
LAMA1; LAMA2; LAMA3; LAMA5; LAMB1; LAMB2; LAMB3; LAMB4; LAMC1; LAMC3; NTN1; NTN2L; NTN4; NTNG1; NTNG2; USH2A;
Role in culture
Many Christian apologists, most notably Louie Giglio, have written about the fact that laminin is shaped like a Christian cross, comparing the role of the cross in theology and laminin's role in the human body. For example, David D. Swanson states: "Our knowledge of truth is most clearly revealed on the cross of Christ. And what holds the human body together? Laminin. And what does it look like? A cross. Coincidence? Some would say yes, but I think it is yet another way God reveals his glory to us. I think God is the one who holds all things together-our bodies-our world-our lives."[26] Fazale Rana, a biochemist and apologist, disagrees with Giglio and instead argues that "Instead of pointing to superficial features of biomolecules such as the “cross-shaped” architecture of laminin, there are many more substantive ways to use biochemistry to argue for the necessity of a Creator."[27]
See also
References
- ^ Timpl R; et al. (1979). "Laminin – a glycoprotein from basement membranes". J Biol Chem. 254 (19): 9933–7. PMID 114518.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - ^ a b Aumailley M; et al. (2005). "A simplified laminin nomenclature". Matrix Biol. 24 (5): 326–32. doi:10.1016/j.matbio.2005.05.006. PMID 15979864.
{{cite journal}}: Unknown parameter|author-separator=ignored (help) - ^ a b c d M. A. Haralson and John R. Hassell (1995). Extracellular matrix: a practical approach. Ithaca, N.Y: IRL Press. ISBN 0-19-963220-0.
- ^ a b Yurchenko P and Batton BL (2009). "Developmental and Pathogenic Mechanisms of Basement Membrane Assembly". Curr Pharm Des. 15 (12): 1277–94. doi:10.2174/138161209787846766. PMC 2978668. PMID 19355968.
- ^ a b Colognato H, Yurchenco P (2000). "Form and function: the laminin family of heterotrimers". Dev. Dyn. 218 (2): 213–34. doi:10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. PMID 10842354.
- ^ Smith J, Ockleford CD (1994). "Laser scanning confocal examination and comparison of nidogen (entactin) with laminin in term human amniochorion". Placenta. 15 (1): 95–106. doi:10.1016/S0143-4004(05)80240-1. PMID 8208674.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Ockleford CD, Bright N, Hubbard A, D'Lacey C , Smith J, Gardiner L, Sheikh T, Albentosa, M, Turtle K (1993). "Micro-Trabeculae, Macro-Plaques or Mini-Basement Membranes in Human Term Fetal Membranes?". Phil. Trans. R. Soc. Lond. B. 342 (1300): 121–136. doi:10.1098/rstb.1993.0142.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Beck et al., 1999.[specify]
- ^ Hall, T. E.; Bryson-Richardson, RJ; et al. (2007). "The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin α2-deficient congenital muscular dystrophy". PNAS. 104 (17): 7092–7097. doi:10.1073/pnas.0700942104. PMC 1855385. PMID 17438294.
- ^ Wewer; et al. (1983). "Human laminin isolated in a nearly intact, biologically active form from placenta by limited proteolysis". J Biol Chem. 258 (20): 12654–60. PMID 6415055.
- ^ Domogatskaya; et al. (2008). "Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro". Stem Cells. 26 (11): 2800–9. doi:10.1634/stemcells.2007-0389. PMID 18757303.
- ^ Miyakzaki; et al. (2008). "Recombinant human laminin isoforms can support the undifferentiated growth of human embryonic stem cells". Biochem. Biophys. Res. Commun. 375 (1): 27–32. doi:10.1016/j.bbrc.2008.07.111. PMID 18675790.
- ^ http://www.nature.com/nbt/journal/v28/n6/full/nbt.1620.html
- ^ Petz M, Them N, Huber H, Beug H, Mikulits W. (2011). "La enhances IRES-mediated translation of laminin B1 during malignant epithelial to mesenchymal transition". Nucleic Acids Rsearch. 39 (18): 01–13. doi:10.1093/nar/gkr717. PMC 3245933. PMID 21896617.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Sasaki M, Kleinman HK, Huber H, Deutzmann R, Yamada Y (1988). "Laminin, a multidomain protein. The A chain has a unique globular domain and homology with the basement membrane proteoglycan and the laminin B chains". J. Biol. Chem. 263 (32): 16536–44. PMID 3182802.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Engel J (1989). "EGF-like domains in extracellular matrix proteins: localized signals for growth and differentiation?". FEBS Lett. 251 (1–2): 1–7. doi:10.1016/0014-5793(89)81417-6. PMID 2666164.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Stetefeld J, Mayer U, Timpl R, Huber R (1996). "Crystal structure of three consecutive laminin-type epidermal growth factor-like (LE) modules of laminin gamma1 chain harboring the nidogen binding site". J. Mol. Biol. 257 (3): 644–57. doi:10.1006/jmbi.1996.0191. PMID 8648630.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Baumgartner R, Czisch M, Mayer U, Poschl E, Huber R, Timpl R, Holak TA (1996). "Structure of the nidogen binding LE module of the laminin gamma1 chain in solution". J. Mol. Biol. 257 (3): 658–68. doi:10.1006/jmbi.1996.0192. PMID 8648631.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Mayer U, Poschl E, Gerecke DR, Wagman DW, Burgeson RE, Timpl R (1995). "Low nidogen affinity of laminin-5 can be attributed to two serine residues in EGF-like motif gamma 2III4". FEBS Lett. 365 (2–3): 129–32. doi:10.1016/0014-5793(95)00438-F. PMID 7781764.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Beck K, Hunter I, Engel J (1990). "Structure and function of laminin: anatomy of a multidomain glycoprotein". FASEB J. 4 (2): 148–60. PMID 2404817.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b Yurchenco PD, Cheng YS (1993). "Self-assembly and calcium-binding sites in laminin. A three-arm interaction model". J. Biol. Chem. 268 (23): 17286–99. PMID 8349613.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Tisi D, Talts JF, Timpl R, Hohenester E (2000). "Structure of the C-terminal laminin G-like domain pair of the laminin alpha2 chain harbouring binding sites for alpha-dystroglycan and heparin". EMBO J. 19 (7): 1432–40. doi:10.1093/emboj/19.7.1432. PMC 310212. PMID 10747011.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Ichikawa N, Kasai S, Suzuki N, Nishi N, Oishi S, Fujii N, Kadoya Y, Hatori K, Mizuno Y, Nomizu M, Arikawa-Hirasawa E (2005). "Identification of neurite outgrowth active sites on the laminin alpha4 chain G domain". Biochemistry. 44 (15): 5755–62. doi:10.1021/bi0476228. PMID 15823034.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Beckmann G, Hanke J, Bork P, Reich JG (1998). "Merging extracellular domains: fold prediction for laminin G-like and amino-terminal thrombospondin-like modules based on homology to pentraxins". J. Mol. Biol. 275 (5): 725–30. doi:10.1006/jmbi.1997.1510. PMID 9480764.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Xu H, Wu XR, Wewer UM, Engvall E (1994). "Murine muscular dystrophy caused by a mutation in the laminin alpha 2 (Lama2) gene". Nat. Genet. 8 (3): 297–302. doi:10.1038/ng1194-297. PMID 7874173.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Swanson, David D. (2012). Learning to Be You. Baker Books. ISBN 1441238492.
- ^ Rana, Fazale. "Crossed off the List: Is the Cross Shape of Laminin Evidence for the Creator?". RTB. Retrieved 26 July 2013.
External links
- The Laminin Protein
- Laminin Precursors as nutritional supplements
- BioLamina - The Laminin Experts
- Laminin at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
