Virus: Difference between revisions
→Antiviral drugs: use SVG. Also for some reason the frame is too wide... |
Snowmanradio (talk | contribs) →Structure: details - viruses and bacteria vary in size - the largest virus is about the size of the smallest bacteria, the micoplasma |
||
| Line 210: | Line 210: | ||
[[File:Hexon.svg|thumb|255px|Diagram of how a virus capsid can be constructed using multiple copies of just two [[protein molecule]]s|alt=A cartoon showing several identical molecules of protein forming a hexigon]] |
[[File:Hexon.svg|thumb|255px|Diagram of how a virus capsid can be constructed using multiple copies of just two [[protein molecule]]s|alt=A cartoon showing several identical molecules of protein forming a hexigon]] |
||
Viruses display a wide diversity of shapes and sizes, called ''[[morphology (biology)|morphologies]]''. |
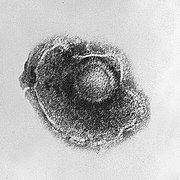
Viruses display a wide diversity of shapes and sizes, called ''[[morphology (biology)|morphologies]]''. Generally viruses are much smaller than bacteria. Most viruses that have been studied have a diameter between 10 and 300 [[nanometres]]. Some [[filovirus]]es have a total length of up to 1400 nm, however their diameters are only about 80 nm.<ref name="Collier3355"/> Most viruses cannot be seen with a [[light microscope]] so scanning and transmission [[electron microscope]]s are used to visualise virus particles.<ref>Collier pp. 33–37</ref> To increase the contrast between viruses and the background, electron-dense "stains" are used. These are solutions of salts of heavy metals such as [[tungsten]], that scatter the electrons from regions covered with the stain. When virus particles are coated with stain (positive staining), fine detail is obscured. [[Negative staining]] overcomes this problem by staining the background only.<ref name="pmid1715774">{{cite journal |
||
|author=Kiselev NA, Sherman MB, Tsuprun VL |
|author=Kiselev NA, Sherman MB, Tsuprun VL |
||
|title=Negative staining of proteins |
|title=Negative staining of proteins |
||
Revision as of 11:04, 5 October 2009
| Viruses | |
|---|---|
| |
| Rotavirus | |
| Virus classification | |
| Group: | I–VII
|
| Groups | |
|
I: dsDNA viruses | |
A virus (from the Latin virus meaning toxin or poison) is a microscopic infectious agent that can reproduce only inside a host cell. Viruses infect all types of organisms: from animals and plants, to bacteria and archaea.[1] Since the initial discovery of tobacco mosaic virus by Martinus Beijerinck in 1898,[2] more than 5,000 types of virus have been described in detail,[3] although most types of virus remain undiscovered.[4] Viruses are found in almost every ecosystem on Earth and these minute structures are numerically the most abundant type of biological entity.[5][6] The study of viruses is known as virology, and is a branch of microbiology.
Viruses consist of two or three parts: all viruses have genes made from either DNA or RNA, long molecules that carry genetic information; all have a protein coat that protects these genes; and some have an envelope of fat that surrounds them when they are outside a cell. Viruses vary in shape from simple helical and icosahedral shapes, to more complex structures. They are about 1/100th the size of bacteria.[7] The origins of viruses in the evolutionary history of life are unclear: some may have evolved from plasmids—pieces of DNA that can move between cells—while others may have evolved from bacteria. In evolution, viruses are an important means of horizontal gene transfer, which increases genetic diversity.[8]
Viruses spread in many ways; plant viruses are often transmitted from plant to plant by insects that feed on sap, such as aphids, while animal viruses can be carried by blood-sucking insects. These disease-bearing organisms are known as vectors. Influenza viruses are spread by coughing and sneezing, while others, such as norovirus, are transmitted by the faecal-oral route, when they contaminate hands, food, or water. Rotaviruses are often spread by direct contact with infected children. HIV is one of several viruses that are transmitted through sexual contact.
Not all viruses cause disease, as many viruses reproduce without causing any obvious harm to the infected organism. Some viruses including several causing viral hepatitis can cause life-long or chronic infections, and the viruses continue to replicate in the body despite the hosts' defence mechanisms. However, in most cases viral infections in animals cause an immune response that eliminates the infecting virus. The immune responses can also be directed against vaccines producing useful immunity to specific viral infections. Microorganisms such as bacteria also have defenses against viral infection, such as restriction modification systems. Antiviral drugs have been developed for both life-threatening and minor viral infections. Clinically useful antiviral drugs can only target a few specific steps in the synthesis of viral structures, as viruses utilize vital metabolism within the host's cells. This contrasts to the use of antibiotic drugs acting on infections caused by bacteria, which are larger than viruses and have a wider variety of their own metabolic pathways for drugs to interfere with.
Etymology
The word is from the Latin virus referring to poison and other noxious substances, first used in English in 1392.[9] Virulent, from Latin virulentus (poisonous) dates to 1400.[10] A meaning of "agent that causes infectious disease" is first recorded in 1728,[9] before the discovery of viruses by Dmitry Ivanovsky in 1892. The adjective viral dates to 1948.[11] The term virion is also used to refer to a single infective viral particle. The plural of virus is "viruses".
History

In 1884, the French microbiologist Charles Chamberland invented a filter (known today as the Chamberland filter or Chamberland-Pasteur filter), with pores smaller than bacteria. Thus, he could pass a solution containing bacteria through the filter and completely remove them from the solution.[12] In 1892 the Russian biologist Dmitry Ivanovsky used this filter to study what is now known as tobacco mosaic virus. His experiments showed that the crushed leaf extracts from infected tobacco plants are still infectious after filtration. Ivanovsky suggested the infection might be caused by a toxin produced by bacteria, but did not pursue the idea.[13] At the time it was thought that all infectious agents could be retained by filters and grown on a nutrient medium—this was part of the germ theory of disease.[2] In 1898 the Dutch microbiologist Martinus Beijerinck repeated the experiments and became convinced that this was a new form of infectious agent.[14] He went on to observe that the agent multiplied only in dividing cells, but as his experiments did not show that it was made of particles, he called it a contagium vivum fluidum (soluble living germ) and re-introduced the word virus.[13] Beijerinck maintained that viruses were liquid in nature, a theory later discredited by Wendell Stanley, who proved they were particulate.[13] In the same year, 1899, Friedrich Loeffler and Frosch passed the agent of foot-and-mouth disease (aphthovirus) through a similar filter and ruled out the possibility of a toxin because of the high dilution; they concluded that the agent could replicate.[13]
In the early 20th century, the English bacteriologist Frederick Twort discovered the viruses that infect bacteria, which are now called bacteriophages,[15] and the French-Canadian microbiologist Félix d'Herelle described viruses that, when added to bacteria on agar, would produce areas of dead bacteria. He accurately diluted a suspension of these viruses and discovered that the highest dilutions, rather than killing all the bacteria, formed discrete areas of dead organisms. Counting these areas and multiplying by the dilution factor allowed him to calculate the number of viruses in the suspension.[16]
By the end of the nineteenth century, viruses were defined in terms of their infectivity, filterability, and their requirement for living hosts. Viruses had been grown only in plants and animals. In 1906, Harrison invented a method for growing tissue in lymph, and, in 1913, E. Steinhardt, C. Israeli, and R. A. Lambert used this method to grow vaccinia virus in fragments of guinea pig corneal tissue.[17] In 1928, H. B. Maitland and M. C. Maitland grew vaccinia virus in suspensions of minced hens' kidneys. Their method was not widely adopted until the 1950s, when poliovirus was grown on a large scale for vaccine production.[18]
Another breakthrough came in 1931, when the American pathologist Ernest William Goodpasture grew influenza and several other viruses in fertilised chickens' eggs.[19] In 1949 John F. Enders, Thomas Weller, and Frederick Robbins grew polio virus in cultured human embryo cells, the first virus to be grown without using solid animal tissue or eggs. This work enabled Jonas Salk to make an effective polio vaccine.[20]
With the invention of electron microscopy in 1931 by the German engineers Ernst Ruska and Max Knoll came the first images of viruses.[21] In 1935 American biochemist and virologist Wendell Stanley examined the tobacco mosaic virus and found it to be mostly made from protein.[22] A short time later, this virus was separated into protein and RNA parts.[23] Tobacco mosaic virus was the first one to be crystallised and whose structure could therefore be elucidated in detail. The first X-ray diffraction pictures of the crystallised virus were obtained by Bernal and Fankuchen in 1941. On the basis of her pictures, Rosalind Franklin discovered the full structure of the virus in 1955.[24] In the same year, Heinz Fraenkel-Conrat and Robley Williams showed that purified tobacco mosaic virus RNA and its coat protein can assemble by themselves to form functional viruses, suggesting that this simple mechanism was probably how viruses assembled within their host cells.[25]
The second half of the twentieth century was the golden age of virus discovery and most of the 2,000 recognised species of animal, plant, and bacterial viruses were discovered during these years.[26][27] In 1957, equine arterivirus and the cause of Bovine virus diarrhea (a pestivirus) were discovered. In 1963, the hepatitis B virus was discovered by Baruch Blumberg,[28] and in 1965, Howard Temin described the first retrovirus. Reverse transcriptase, the key enzyme that retroviruses use to translate their RNA into DNA, was first described in 1970, independently by Howard Temin and David Baltimore.[29] In 1983 Luc Montagnier's team at the Pasteur Institute in France, first isolated the retrovirus now called HIV.[30]
Origins
Viruses are found wherever there is life and have probably existed since living cells first evolved.[31] The origin of viruses is unclear because they do not form fossils, so molecular techniques have been the most useful means of investigating how they arose.[32] These techniques rely on the availability of ancient viral DNA or RNA, but, unfortunately, most of the viruses that have been preserved and stored in laboratories are less than 90 years old.[33][34] There are three main hypotheses that try to explain the origins of viruses:[35][36]
- Regressive hypothesis
- Viruses may have once been small cells that parasitised larger cells. Over time, genes not required by their parasitism were lost. The bacteria rickettsia and chlamydia are living cells that, like viruses, can reproduce only inside host cells. They lend support to this hypothesis, as their dependence on parasitism is likely to have caused the loss of genes that enabled them to survive outside a cell. This is also called the degeneracy hypothesis.[37][38]
- Cellular origin hypothesis
- Some viruses may have evolved from bits of DNA or RNA that "escaped" from the genes of a larger organism. The escaped DNA could have come from plasmids (pieces of naked DNA that can move between cells) or transposons (molecules of DNA that replicate and move around to different positions within the genes of the cell).[39] Once called "jumping genes", transposons are examples of mobile genetic elements and could be the origin of some viruses. They were discovered in maize by Barbara McClintock in 1950.[40] This is sometimes called the vagrancy hypothesis.[37][41]
- Coevolution hypothesis
- Viruses may have evolved from complex molecules of protein and nucleic acid at the same time as cells first appeared on earth and would have been dependent on cellular life for many millions of years. Viroids are molecules of RNA that are not classified as viruses because they lack a protein coat. However, they have characteristics that are common to several viruses and are often called subviral agents.[42] Viroids are important pathogens of plants.[43] They do not code for proteins but interact with the host cell and use the host machinery for their replication.[44] The hepatitis delta virus of humans has an RNA genome similar to viroids but has protein coat derived from hepatitis B virus and cannot produce one of its own. It is therefore a defective virus and cannot replicate without the help of hepatitis B virus.[45]
The virophage 'sputnik' infects the mimivirus and the related mamavirus, which in turn infect the protozooan Acanthamoeba castellanii.[46] These viruses that are dependent on other virus species are called satellites and may represent evolutionary intermediates of viroids and viruses.[47][48] Prions are infectious protein molecules that do not contain DNA or RNA.[49] They cause an infection in sheep called scrapie and cattle bovine spongiform encephalopathy ("mad cow" disease). In humans they cause kuru and Creutzfeldt-Jakob disease.[50] They are able to replicate because some proteins can exist in two different shapes and the prion changes the normal shape of a host protein into the prion shape. This starts a chain reaction where each prion protein converts many host proteins into more prions, and these new prions then go on to convert even more protein into prions. Although they are fundamentally different from viruses and viroids, their discovery gives credence to the idea that viruses could have evolved from self-replicating molecules.[51]
Computer analysis of viral and host DNA sequences is giving a better understanding of the evolutionary relationships between different viruses and may help identify the ancestors of modern viruses. To date, such analyses have not helped to decide on which of these hypotheses are correct. However, it seems unlikely that all currently known viruses have a common ancestor and viruses have probably arisen numerous times in the past by one or more mechanisms.[52]
Microbiology
Life properties
Opinions differ on whether viruses are a form of life, or organic structures that interact with living organisms. They have been described as "organisms at the edge of life",[53] since they resemble organisms in that they possess genes and evolve by natural selection,[54] and reproduce by creating multiple copies of themselves through self-assembly. However, although they have genes, they do not have a cellular structure, which is often seen as the basic unit of life. Additionally, viruses do not have their own metabolism, and require a host cell to make new products. They therefore cannot reproduce outside a host cell (although bacterial species such as rickettsia and chlamydia are considered living organisms despite the same limitation). Accepted forms of life use cell division to reproduce, whereas viruses spontaneously assemble within cells, which is analogous to the autonomous growth of crystals. Virus self-assembly within host cells has implications for the study of the origin of life, as it lends further credence to the hypothesis that life could have started as self-assembling organic molecules.[1]
Structure

Viruses display a wide diversity of shapes and sizes, called morphologies. Generally viruses are much smaller than bacteria. Most viruses that have been studied have a diameter between 10 and 300 nanometres. Some filoviruses have a total length of up to 1400 nm, however their diameters are only about 80 nm.[7] Most viruses cannot be seen with a light microscope so scanning and transmission electron microscopes are used to visualise virus particles.[55] To increase the contrast between viruses and the background, electron-dense "stains" are used. These are solutions of salts of heavy metals such as tungsten, that scatter the electrons from regions covered with the stain. When virus particles are coated with stain (positive staining), fine detail is obscured. Negative staining overcomes this problem by staining the background only.[56]
A complete virus particle, known as a virion, consists of nucleic acid surrounded by a protective coat of protein called a capsid. These are formed from identical protein subunits called capsomers.[57] Viruses can have a lipid "envelope" derived from the host cell membrane. The capsid is made from proteins encoded by the viral genome and its shape serves as the basis for morphological distinction.[58][59] Virally coded protein subunits will self-assemble to form a capsid, generally requiring the presence of the virus genome. However, complex viruses code for proteins that assist in the construction of their capsid. Proteins associated with nucleic acid are known as nucleoproteins, and the association of viral capsid proteins with viral nucleic acid is called a nucleocapsid. The capsid and entire virus structure can be mechanically (physically) probed through atomic force microscopy. [60][61] In general, there are four main morphological virus types:
- Helical
- These viruses are composed of a single type of capsomer stacked around a central axis to form a helical structure, which may have a central cavity, or hollow tube. This arrangement results in rod-shaped or filamentous virions: these can be short and highly rigid, or long and very flexible. The genetic material, generally single-stranded RNA, but ssDNA in some cases, is bound into the protein helix, by interactions between the negatively charged nucleic acid and positive charges on the protein. Overall, the length of a helical capsid is related to the length of the nucleic acid contained within it and the diameter is dependent on the size and arrangement of capsomers. The well-studied Tobacco mosaic virus is an example of a helical virus.[62]
- Icosahedral
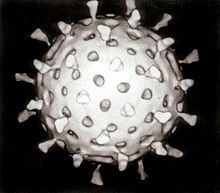
- Most animal viruses are icosahedral or near-spherical with icosahedral symmetry. A regular icosahedron is the optimum way of forming a closed shell from identical sub-units. The minimum number of identical capsomers required is twelve, each composed of five identical sub-units. Many viruses, such as rotavirus, have more than twelve capsomers and appear spherical but they retain this symmetry. Capsomers at the apices are surrounded by five other capsomers and are called pentons. Capsomers on the triangular faces are surround by six others and are call hexons.[63]
- Envelope
- Some species of virus envelope themselves in a modified form of one of the cell membranes, either the outer membrane surrounding an infected host cell, or internal membranes such as nuclear membrane or endoplasmic reticulum, thus gaining an outer lipid bilayer known as a viral envelope. This membrane is studded with proteins coded for by the viral genome and host genome; the lipid membrane itself and any carbohydrates present originate entirely from the host. The influenza virus and HIV use this strategy. Most enveloped viruses are dependent on the envelope for their infectivity.[64]
- Complex
These viruses possess a capsid that is neither purely helical, nor purely icosahedral, and that may possess extra structures such as protein tails or a complex outer wall. Some bacteriophages, such as Enterobacteria phage T4 have a complex structure consisting of an icosahedral head bound to a helical tail, which may have a hexagonal base plate with protruding protein tail fibres. This tail structure acts like a molecular syringe, attaching to the bacterial host and then injecting the viral genome into the cell.[65]
The poxviruses are large, complex viruses that have an unusual morphology. The viral genome is associated with proteins within a central disk structure known as a nucleoid. The nucleoid is surrounded by a membrane and two lateral bodies of unknown function. The virus has an outer envelope with a thick layer of protein studded over its surface. The whole particle is slightly pleiomorphic, ranging from ovoid to brick shape.[66] Mimivirus is the largest known virus, with a capsid diameter of 400 nm. Protein filaments measuring 100 nm project from the surface. The capsid appears hexagonal under an electron microscope, therefore the capsid is probably icosahedral.[67]
Some viruses that infect Archaea have complex structures that are unrelated to any other form of virus, with a wide variety of unusual shapes, ranging from spindle-shaped structures, to viruses that resemble hooked rods, teardrops or even bottles. Other archaeal viruses resemble the tailed bacteriophages, although they can have multiple tail structures.[68]
Genome
| Property | Parameters |
|---|---|
| Nucleic acid |
|
| Shape |
|
| Strandedness |
|
| Sense |
|
An enormous variety of genomic structures can be seen among viral species; as a group they contain more structural genomic diversity than plants, animals, archaea, or bacteria. A virus has either DNA or RNA genes and are called DNA viruses and RNA viruses respectively. By far most viruses have RNA. Plant viruses tend to have single-stranded RNA and bacteriophages tend to have double-stranded DNA.[69]
Viral genomes are circular, such as polyomaviruses, or linear, such as adenoviruses. The type of nucleic acid is irrelevant to the shape of the genome. Among RNA viruses, the genome is often divided up into separate parts within the virion and is called segmented. Each segment often codes for one protein and they are usually found together in one capsid. Every segment is not required to be in the same virion for the overall virus to be infectious, as demonstrated by the brome mosaic virus.[7]
A viral genome, irrespective of nucleic acid type, is either single-stranded or double-stranded. Single-stranded genomes consist of an unpaired nucleic acid, analogous to one-half of a ladder split down the middle. Double-stranded genomes consist of two complementary paired nucleic acids, analogous to a ladder. Some viruses, such as those belonging to the Hepadnaviridae, contain a genome that is partially double-stranded and partially single-stranded.[69]
For viruses with RNA or single-stranded DNA, the strands are said to be either positive-sense (called the plus-strand) or negative-sense (called the minus-strand), depending on whether it is complementary to the viral messenger RNA (mRNA). Positive-sense viral RNA is identical to viral mRNA and thus can be immediately translated by the host cell. Negative-sense viral RNA is complementary to mRNA and thus must be converted to positive-sense RNA by an RNA polymerase before translation. DNA nomenclature is similar to RNA nomenclature, in that the coding strand for the viral mRNA is complementary to it (−), and the non-coding strand is a copy of it (+).[69]
Genome size varies greatly between species. The smallest viral genomes code for only four proteins and have a mass of about 106 Daltons; the largest have a mass of about 108 Daltons and code for over one hundred proteins.[69] RNA viruses generally have smaller genome sizes than DNA viruses because of a higher error-rate when replicating, and have a maximum upper size limit. Beyond this limit, errors in the genome when replicating render the virus useless or uncompetitive. To compensate for this, RNA viruses often have segmented genomes where the genome is split into smaller molecules, thus reducing the chance of error. In contrast, DNA viruses generally have larger genomes because of the high fidelity of their replication enzymes.[70]

Viruses undergo genetic change by several mechanisms. These include a process called genetic drift where individual bases in the DNA or RNA mutate to other bases. Most of these point mutations are "silent"—they do not change the protein that the gene encodes—but others can confer evolutionary advantages such as resistance to antiviral drugs.[71] Antigenic shift occurs when there is a major change in the genome of the virus. This can be a result of recombination or reassortment. When this happens with influenza viruses, pandemics might result.[72] RNA viruses often exist as quasispecies or swarms of viruses of the same species but with slightly different genome nucleoside sequences. Such quasispecies are a prime target for natural selection.[73]
Segmented genomes confer evolutionary advantages; different strains of a virus with a segmented genome can shuffle and combine genes and produce progeny viruses or (offspring) that have unique characteristics. This is called reassortment or viral sex.[74]
Genetic recombination is the process by which a strand of DNA is broken and then joined to the end of a different DNA molecule. This can occur when viruses infect cells simultaneously and studies of viral evolution have shown that recombination has been rampant in the species studied.[75] Recombination is common to both RNA and DNA viruses.[76][77]
Replication cycle
Viral populations do not grow through cell division, because they are acellular; instead, they use the machinery and metabolism of a host cell to produce multiple copies of themselves, and they assemble in the cell.
The life cycle of viruses differs greatly between species but there are six basic stages in the life cycle of viruses:[78]
- Attachment is a specific binding between viral capsid proteins and specific receptors on the host cellular surface. This specificity determines the host range of a virus. For example, HIV infects only human T cells, because its surface protein, gp120, can interact with CD4 and receptors on the T cell's surface. This mechanism has evolved to favour those viruses that only infect cells in which they are capable of replication. Attachment to the receptor can induce the viral-envelope protein to undergo changes that results in the fusion of viral and cellular membranes.
- Penetration follows attachment; viruses enter the host cell through receptor mediated endocytosis or membrane fusion. This is often called viral entry. The infection of plant cells is different to that of animal cells. Plants have a rigid cell wall made of cellulose and viruses can only get inside the cells following trauma to the cell wall.[79] Viruses such as tobacco mosaic virus can also move directly in plants, from cell-to-cell, through pores called plasmodesmata.[80] Bacteria, like plants, have strong cell walls that a virus must breach to infect the cell. Some viruses have evolved mechanisms that inject their genome into the bacterial cell while the viral capsid remains outside.[81]
- Uncoating is a process in which the viral capsid is degraded by viral enzymes or host enzymes thus releasing the viral genomic nucleic acid.
- Replication involves synthesis of viral messenger RNA (mRNA) for viruses except positive sense RNA viruses (see above), viral protein synthesis and assembly of viral proteins and viral genome replication.
- Following the assembly of the virus particles, post-translational modification of the viral proteins often occurs. In viruses such as HIV, this modification, (sometimes called maturation), occurs after the virus has been released from the host cell.[82]
- Viruses are released from the host cell by lysis—a process that kills the cell by bursting its membrane. Enveloped viruses (e.g., HIV) typically are released from the host cell by budding. During this process the virus acquires its envelope, which is a modified piece of the host's plasma membrane.
The genetic material within viruses, and the method by which the material is replicated, vary between different types of viruses.
- DNA viruses
- The genome replication of most DNA viruses takes place in the cell's nucleus. If the cell has the appropriate receptor on its surface, these viruses enter the cell by fusion with the cell membrane or by endocytosis. Most DNA viruses are entirely dependent on the host cell's DNA and RNA synthesising machinery, and RNA processing machinery. The viral genome must cross the cell's nuclear membrane to access this machinery.[83]
- RNA viruses
- These viruses are unique because their genetic information is encoded in RNA. Replication usually takes place in the cytoplasm. RNA viruses can be placed into about four different groups depending on their modes of replication. The polarity (whether or not it can be used directly to make proteins) of the RNA largely determines the replicative mechanism, and whether the genetic material is single-stranded or double-stranded. RNA viruses use their own RNA replicase enzymes to create copies of their genomes.[84]
- Reverse transcribing viruses
- These replicate using reverse transcription, which is the formation of DNA from an RNA template. Reverse transcribing viruses containing RNA genomes use a DNA intermediate to replicate, whereas those containing DNA genomes use an RNA intermediate during genome replication. Both types use the reverse transcriptase enzyme to carry out the nucleic acid conversion. Retroviruses often integrate the DNA produced by reverse transcription into the host genome. They are susceptible to antiviral drugs that inhibit the reverse transcriptase enzyme, e.g. zidovudine and lamivudine. An example of the first type is HIV, which is a retrovirus. Examples of the second type are the Hepadnaviridae, which includes Hepatitis B virus.[85]
Effects on the host cell
The range of structural and biochemical effects that viruses have on the hosts cell is extensive.[86] These are called cytopathic effects.[87] Most virus infections eventually result in the death of the host cell. The causes of death include cell lysis, alterations to the cell's surface membrane and apoptosis.[88] Often cell death is caused by cessation of its normal activities because of suppression by virus-specific proteins, not all of which are components of the virus particle.[89]
Some viruses cause no apparent changes to the infected cell. Cells in which the virus is latent and inactive show few signs of infection and often function normally.[90] This causes persistent infections and the virus is often dormant for many months or years. This is often the case with herpes viruses.[91][92] Some viruses, such as Epstein-Barr virus, can cause cells to proliferate without causing malignancy,[93] while others, such as papillomaviruses, are established causes of cancer.[94]
Classification
Classification seeks to describe the diversity of viruses by naming and grouping them on the basis of similarities. In 1962, André Lwoff, Robert Horne, and Paul Tournier were the first to develop a means of virus classification, based on the Linnaean hierarchical system.[95] This system bases classification on phylum, class, order, family, genus, and species. Viruses were grouped according to their shared properties (not of their hosts) and the type of nucleic acid forming their genomes.[96] later the International Committee on Taxonomy of Viruses was formed.
ICTV classification
The International Committee on Taxonomy of Viruses (ICTV) developed the current classification system and wrote guidelines that put a greater weight on certain virus properties to maintain family uniformity. A universal system for classifying viruses, and a unified taxonomy, has been established since 1966. The 7th lCTV Report formalised for the first time the concept of the virus species as the lowest taxon (group) in a branching hierarchy of viral taxa.[97] However, at present only a small part of the total diversity of viruses has been studied, with analyses of samples from humans finding that about 20% of the virus sequences recovered have not been seen before, and samples from the environment, such as from seawater and ocean sediments, finding that the large majority of sequences are completely novel.[98]
The general taxonomic structure is as follows:
In the current (2008) ICTV taxonomy, five orders have been established, the Caudovirales, Herpesvirales, Mononegavirales, Nidovirales, and Picornavirales. The committee does not formally distinguish between subspecies, strains, and isolates. In total there are 5 orders, 82 families, 11 subfamilies, 307 genera, 2,083 species and about 3,000 types yet unclassified.[99][100]
Baltimore classification

The Nobel Prize-winning biologist David Baltimore devised the Baltimore classification system.[29][101] The ICTV classification system is used in conjunction with the Baltimore classification system in modern virus classification.[102][103][104]
The Baltimore classification of viruses is based on the mechanism of mRNA production. Viruses must generate mRNAs from their genomes to produce proteins and replicate themselves, but different mechanisms are used to achieve this in each virus family. Viral genomes may be single-stranded (ss) or double-stranded (ds), RNA or DNA, and may or may not use reverse transcriptase (RT). Additionally, ssRNA viruses may be either sense (+) or antisense (−). This classification places viruses into seven groups:
- I: dsDNA viruses (e.g. Adenoviruses, Herpesviruses, Poxviruses)
- II: ssDNA viruses (+ strand or "sense") DNA (e.g. Parvoviruses)
- III: dsRNA viruses (e.g. Reoviruses)
- IV:(+)ssRNA viruses (+ strand or sense) RNA (e.g. Coronaviruses, Picornaviruses, Togaviruses)
- V: (−)ssRNA viruses (− strand or antisense) RNA (e.g. Orthomyxoviruses, Rhabdoviruses)
- VI: ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle (e.g. Retroviruses)
- VII: dsDNA-RT viruses DNA with RNA intermediate in life-cycle (e.g. Hepadnaviruses)
As an example of viral classification, the chicken pox virus, varicella zoster (VZV), belongs to the order Herpesvirales, family Herpesviridae, subfamily Alphaherpesvirinae, and genus Varicellovirus. VZV is in Group I of the Baltimore Classification because it is a dsDNA virus that does not use reverse transcriptase.
Viruses and human disease

Examples of common human diseases caused by viruses include the common cold, influenza, chickenpox and cold sores. Many serious diseases such as ebola, AIDS, avian influenza and SARS are caused by viruses. The relative ability of viruses to cause disease is described in terms of virulence. Other diseases are under investigation as to whether they too have a virus as the causative agent, such as the possible connection between human herpes virus six (HHV6) and neurological diseases such as multiple sclerosis and chronic fatigue syndrome. There is controversy over whether the borna virus, previously thought to cause neurological diseases in horses, could be responsible for psychiatric illnesses in humans.[107]
Viruses have different mechanisms by which they produce disease in an organism, which largely depends on the viral species. Mechanisms at the cellular level primarily include cell lysis, the breaking open and subsequent death of the cell. In multicellular organisms, if enough cells die the whole organism will start to suffer the effects. Although viruses cause disruption of healthy homeostasis, resulting in disease, they may exist relatively harmlessly within an organism. An example would include the ability of the herpes simplex virus, which cause cold sores, to remain in a dormant state within the human body. This is called latency[108] and is a characteristic of the all herpes viruses including the Epstein-Barr virus, which causes glandular fever, and the varicella zoster virus, which causes chicken pox. Most people have been infected with at least one of these types of herpes virus.[109] However, these latent viruses might sometimes be beneficial, as the presence of the virus can increase immunity against bacterial pathogens, such as Yersinia pestis.[110] On the other hand, latent chickenpox infections return in later life as the disease called shingles.
Some viruses can cause life-long or chronic infections, where the viruses continue to replicate in the body despite the hosts' defence mechanisms.[111] This is common in hepatitis B virus and hepatitis C virus infections. People chronically infected are known as carriers, as they serve as reservoirs of infectious virus.[112] In populations with a high proportion of carriers, the disease is said to be endemic.[113] In contrast to acute lytic viral infections this persistence implies compatible interactions with the host organism. Persistent viruses may even broaden the evolutionary potential of host species.[114]
Epidemiology
Viral epidemiology is the branch of medical science that deals with the transmission and control of virus infections in humans. Transmission of viruses can be vertical, that is from mother to child, or horizontal, which means from person to person. Examples of vertical transmission include hepatitis B virus and HIV where the baby is born already infected with the virus.[115] Another, more rare, example is the varicella zoster virus, which although causing relatively mild infections in humans, can be fatal to the foetus and newly born baby.[116] Horizontal transmission is the most common mechanism of spread of viruses in populations. Transmission can be exchange of blood by sexual activity, e.g. HIV, hepatitis B and hepatitis C; by mouth by exchange of saliva, e.g. Epstein-Barr virus, or from contaminated food or water, e.g. norovirus; by breathing in viruses in the form of aerosols, e.g. influenza virus; and by insect vectors such as mosquitoes, e.g. dengue. The rate or speed of transmission of viral infections depends on factors that include population density, the number of susceptible individuals, (i.e. those who are not immune),[117] the quality of health care and the weather.[118]
Epidemiology is used to break the chain of infection in populations during outbreaks of viral diseases.[119] Control measures are used that are based on knowledge of how the virus is transmitted. It is important to find the source, or sources, of the outbreak and to identify the virus. Once the virus has been identified, the chain of transmission can sometimes be broken by vaccines. When vaccines are not available sanitation and disinfection can be effective. Often infected people are isolated from the rest of the community and those that have been exposed to the virus placed in quarantine.[120] To control the outbreak of foot and mouth disease in cattle in Britain in 2001, thousands of cattle were slaughtered.[121] Most viral infections of humans and other animals have incubation periods during which the infection causes no signs or symptoms.[122] Incubation periods for viral diseases range from a few days to weeks but are known for most infections.[123] Somewhat overlapping, but mainly following the incubation period, there is a period of communicability; a time when an infected individual or animal is contagious and can infect another person or animal.[124] This too is known for many viral infections and knowledge the length of both periods is important in the control of outbreaks.[125] When outbreaks cause an unusually high proportion of cases in a population, community or region they are called epidemics. If outbreaks spread worldwide they are called pandemics.[126]
Epidemics and pandemics
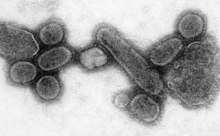
Native American populations were devastated by contagious diseases, particularly smallpox, brought to the Americas by European colonists. It is unclear how many Native Americans were killed by foreign diseases after the arrival of Columbus in the Americas, but the numbers have been estimated to be close to 70% of the indigenous population. The damage done by this disease significantly aided European attempts to displace and conquer the native population.[127]
A pandemic is a worldwide epidemic. The 1918 flu pandemic, commonly referred to as the Spanish flu, was a category 5 influenza pandemic caused by an unusually severe and deadly influenza A virus. The victims were often healthy young adults, in contrast to most influenza outbreaks, which predominantly affect juvenile, elderly, or otherwise weakened patients.[128]
The Spanish flu pandemic lasted from 1918 to 1919. Older estimates say it killed 40–50 million people,[129] while more recent research suggests that it may have killed as many as 100 million people, or 5% of the world's population in 1918.[130] Most researchers believe that HIV originated in sub-Saharan Africa during the twentieth century;[131] it is now a pandemic, with an estimated 38.6 million people now living with the disease worldwide.[132] The Joint United Nations Programme on HIV/AIDS (UNAIDS) and the World Health Organization (WHO) estimate that AIDS has killed more than 25 million people since it was first recognised on June 5, 1981, making it one of the most destructive epidemics in recorded history.[133] In 2007 there were 2.7 million new HIV infections and 2 million HIV-related deaths.[134]

Several highly lethal viral pathogens are members of the Filoviridae. Filoviruses are filament-like viruses that cause viral hemorrhagic fever, and include the ebola and marburg viruses. The Marburg virus attracted widespread press attention in April 2005 for an outbreak in Angola. Beginning in October 2004 and continuing into 2005, the outbreak was the world's worst epidemic of any kind of viral hemorrhagic fever.[135]
Cancer
Viruses are an established cause of cancer in humans and other species. Viral cancers only occur in a minority of infected persons (or animals). Cancer viruses come from a range of virus families, including both RNA and DNA viruses, and so there is no single type of "oncovirus" (an obsolete term originally used for acutely transforming retroviruses). The development of cancer is determined by a variety of factors such as host immunity[136] and mutations in the host.[137] Viruses accepted to cause human cancers include some genotypes of human papillomavirus, hepatitis B virus, hepatitis C virus, Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus and human T-lymphotropic virus. The most recently discovered human cancer virus is a polyomavirus (Merkel cell polyomavirus) that causes most cases of a rare form of skin cancer called Merkel cell carcinoma.[138] Hepatitis viruses can develop into a chronic viral infection that leads to liver cancer.[139][140] Infection by human T-lymphotropic virus can lead to tropical spastic paraparesis and adult T-cell leukemia.[141] Human papillomaviruses are an established cause of cancers of cervix, skin, anus, and penis.[142] Within the Herpesviridae, Kaposi's sarcoma-associated herpesvirus causes Kaposi's sarcoma and body cavity lymphoma, and Epstein–Barr virus causes Burkitt's lymphoma, Hodgkin’s lymphoma, B lymphoproliferative disorder and nasopharyngeal carcinoma.[143] Merkel cell polyomavirus closely related to SV40 and mouse polyomaviruses that have been used as animal models for cancer viruses for over 50 years.[144]
Host defence mechanisms
The body's first line of defence against viruses is the innate immune system. This comprises cells and other mechanisms that defend the host from infection in a non-specific manner. This means that the cells of the innate system recognise, and respond to, pathogens in a generic way, but unlike the adaptive immune system, it does not confer long-lasting or protective immunity to the host.[145]
RNA interference is an important innate defence against viruses.[146] Many viruses have a replication strategy that involves double-stranded RNA (dsRNA). When such a virus infects a cell, it releases its RNA molecule or molecules, which immediately bind to a protein complex called dicer that cuts the RNA into smaller pieces. A biochemical pathway called the RISC complex is activated, which degrades the viral mRNA and the cell survives the infection. Rotaviruses avoid this mechanism by not uncoating fully inside the cell and by releasing newly produced mRNA through pores in the particle's inner capsid. The genomic dsRNA remains protected inside the core of the virion.[147][148]
When the adaptive immune system of a vertebrate encounters a virus, it produces specific antibodies that bind to the virus and render it non-infectious. This is called humoral immunity. Two types of antibodies are important. The first called IgM is highly effective at neutralizing viruses but is only produced by the cells of the immune system for a few weeks. The second, called, IgG is produced indefinitely. The presence of IgM in the blood of the host is used to test for acute infection, whereas IgG indicates an infection sometime in the past.[149] IgG antibody is measured when tests for immunity are carried out.[150]
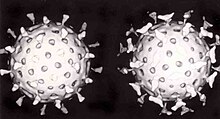
A second defence of vertebrates against viruses is called cell-mediated immunity and involves immune cells known as T cells. The body's cells constantly display short fragments of their proteins on the cell's surface, and if a T cell recognises a suspicious viral fragment there, the host cell is destroyed by killer T cells and the virus-specific T-cells proliferate. Cells such as the macrophage are specialists at this antigen presentation.[151] The production of interferon is an important host defence mechanism. This is a hormone produced by the body when viruses are present. Its role in immunity is complex, but it eventually stops the viruses from reproducing by killing the infected cell and its close neighbours.[152]
Not all virus infections produce a protective immune response in this way. HIV evades the immune system by constantly changing the amino acid sequence of the proteins on the surface of the virion. These persistent viruses evade immune control by sequestration, blockade of antigen presentation, cytokine resistance, evasion of natural killer cell activities, escape from apoptosis, and antigenic shift.[153] Other viruses, called neurotropic viruses, are disseminated by neural spread where the immune system may be unable to reach them.
Prevention and treatment
Because viruses use the machinery of a host cell to reproduce and reside within them, they are difficult to eliminate without killing the host cell. The most effective medical approaches to viral diseases so far are vaccinations to provide resistance to infection, and antiviral drugs.
Vaccines
Vaccination is a cheap and effective way of preventing infections by viruses. Vaccines were used to prevent viral infections long before the discovery of the actual viruses. Their use has resulted in a dramatic decline in morbidity (illness) and mortality (death) associated with viral infections such as polio, measles, mumps and rubella.[154] Smallpox infections have been eradicated.[155] Vaccines are available to prevent over thirteen viral infections of humans,[156] and more are used to prevent viral infections of animals.[157] Vaccines can consist of live-attenuated or killed viruses, or viral proteins (antigens).[158] Live vaccines contain weakened forms of the virus that causes the disease. Such viruses are called attenuated. Live vaccines can be dangerous when given to people with a weak immunity, (who are described as immunocompromised), because in these people, the weakened virus can cause the original disease.[159] Biotechnology and genetic engineering techniques are used to produce subunit vaccines. These vaccines use only the capsid proteins of the virus. Hepatitis B vaccine is an example of this type of vaccine.[160] Subunit vaccines are safe for immunocompromised patients because they cannot cause the disease.[161] However, the yellow fever virus vaccine, a live-attenuated strain called 17D, is probably the safest and most effective vaccine ever generated.[162]
Antiviral drugs
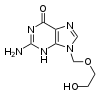
Over the past twenty years, the development of antiviral drugs has increased rapidly. This has been driven by the AIDS pandemic. Antiviral drugs are often nucleoside analogues, (fake DNA building blocks), which viruses incorporate into their genomes during replication. The life-cycle of the virus is then halted because the newly synthesised DNA is inactive. This is because these analogues lack the hydroxyl groups, which, along with phosphorus atoms, link together to form the strong "backbone" of the DNA molecule. This is called DNA chain termination.[163] Examples of nucleoside analogues are aciclovir for Herpes simplex virus infections and lamivudine for HIV and Hepatitis B virus infections. Aciclovir is one of the oldest and most frequently prescribed antiviral drugs.[164] Other antiviral drugs in use target different stages of the viral life cycle. HIV is dependent on a proteolytic enzyme called the HIV-1 protease for it to become fully infectious. There is a large class of drugs called protease inhibitors that inactivate this enzyme.
Hepatitis C is caused by an RNA virus. In 80% of people infected, the disease is chronic, and without treatment, they are infected for the remainder of their lives. However, there is now an effective treatment that uses the nucleoside analogue drug ribavirin combined with interferon.[165] The treatment of chronic carriers of the hepatitis B virus by using a similar strategy using lamivudine has been developed.[166]
Infection in other species
Viruses infect all cellular life and, although viruses occur universally, each cellular species has its own specific range that often infect only that species.[167] Viruses are important pathogens of livestock. Diseases such as Foot and Mouth Disease and bluetongue are caused by viruses.[168] Companion animals such as cats, dogs, and horses, if not vaccinated, are susceptible to serious viral infections. Canine parvovirus is caused by a small DNA virus and infections are often fatal in pups.[169] Like all invertebrates, the honey bee is susceptible to many viral infections.[170] Fortunately, most viruses co-exist harmlessly in their host and cause no signs or symptoms of disease.[2]
Plants

There are many types of plant virus, but often they cause only a loss of yield, and it is not economically viable to try to control them. Plant viruses are often spread from plant to plant by organisms, known as vectors. These are normally insects, but some fungi, nematode worms and single-celled organisms have been shown to be vectors. When control of plant virus infections is considered economical, for perennial fruits for example, efforts are concentrated on killing the vectors and removing alternate hosts such as weeds.[171] Plant viruses are harmless to humans and other animals because they can reproduce only in living plant cells.[172]
Plants have elaborate and effective defence mechanisms against viruses. One of the most effective is the presence of so-called resistance (R) genes. Each R gene confers resistance to a particular virus by triggering localised areas of cell death around the infected cell, which can often be seen with the unaided eye as large spots. This stops the infection from spreading.[173] RNA interference is also an effective defence in plants.[174] When they are infected, plants often produce natural disinfectants that kill viruses, such as salicylic acid, nitric oxide, and reactive oxygen molecules.[175]
Bacteria
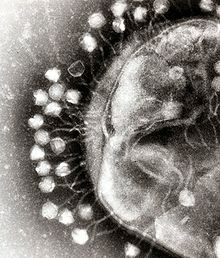
Bacteriophages are a common and diverse group of viruses and are the most abundant form of biological entity in aquatic environments—there are up to ten times more of these viruses in the oceans than there are bacteria,[176] reaching levels of 250,000,000 bacteriophages per millilitre of seawater.[177] These viruses infect specific bacteria by binding to surface receptor molecules and then entering the cell. Within a short amount of time, in some cases just minutes, bacterial polymerase starts translating viral mRNA into protein. These proteins go on to become either new virions within the cell, helper proteins, which help assembly of new virions, or proteins involved in cell lysis. Viral enzymes aid in the breakdown of the cell membrane, and, in the case of the T4 phage, in just over twenty minutes after injection over three hundred phages could be released.[178]
The major way bacteria defend themselves from bacteriophages is by producing enzymes that destroy foreign DNA. These enzymes, called restriction endonucleases, cut up the viral DNA that bacteriophages inject into bacterial cells.[179] Bacteria also contain a system that uses CRISPR sequences to retain fragments of the genomes of viruses that the bacteria have come into contact with in the past, which allows them to block the virus's replication through a form of RNA interference.[180][181] This genetic system provides bacteria with acquired immunity to infection.
Archaea
Some viruses replicate within archaea: these are double-stranded DNA viruses with unusual and sometimes unique shapes.[5][68] These viruses have been studied in most detail in the thermophilic archaea, particularly the orders Sulfolobales and Thermoproteales.[182] Defences against these viruses may involve RNA interference from repetitive DNA sequences within archaean genomes that are related to the genes of the viruses.[183][184]
Applications
Life sciences and medicine
Viruses are important to the study of molecular and cellular biology as they provide simple systems that can be used to manipulate and investigate the functions of cells.[185] The study and use of viruses have provided valuable information about aspects of cell biology.[186] For example, viruses have been useful in the study of genetics and helped our understanding of the basic mechanisms of molecular genetics, such as DNA replication, transcription, RNA processing, translation, protein transport, and immunology.
Geneticists often use viruses as vectors to introduce genes into cells that they are studying. This is useful for making the cell produce a foreign substance, or to study the effect of introducing a new gene into the genome. In similar fashion, virotherapy uses viruses as vectors to treat various diseases, as they can specifically target cells and DNA. It shows promising use in the treatment of cancer and in gene therapy. Eastern European scientists have used phage therapy as an alternative to antibiotics for some time, and interest in this approach is increasing, because of the high level of antibiotic resistance now found in some pathogenic bacteria.[187]
Materials science and nanotechnology
Current trends in nanotechnology promise to make much more versatile use of viruses. From the viewpoint of a materials scientist, viruses can be regarded as organic nanoparticles. Their surface carries specific tools designed to cross the barriers of their host cells. The size and shape of viruses, and the number and nature of the functional groups on their surface, is precisely defined. As such, viruses are commonly used in materials science as scaffolds for covalently linked surface modifications. A particular quality of viruses is that they can be tailored by directed evolution. The powerful techniques developed by life sciences are becoming the basis of engineering approaches towards nanomaterials, opening a wide range of applications far beyond biology and medicine.[188]
Because of their size, shape, and well-defined chemical structures, viruses have been used as templates for organizing materials on the nanoscale. Recent examples include work at the Naval Research Laboratory in Washington, DC, using Cowpea Mosaic Virus (CPMV) particles to amplify signals in DNA microarray based sensors. In this application, the virus particles separate the fluorescent dyes used for signalling to prevent the formation of non-fluorescent dimers that act as quenchers.[189] Another example is the use of CPMV as a nanoscale breadboard for molecular electronics.[190]
Weapons
The ability of viruses to cause devastating epidemics in human societies has led to the concern that viruses could be weaponised for biological warfare. Further concern was raised by the successful recreation of the infamous 1918 influenza virus in a laboratory.[191] The smallpox virus devastated numerous societies throughout history before its eradication. There are officially only two centers in the world which keep stocks of smallpox virus - the Russian Vector laboratory, and the United States Centers for Disease Control.[192] But fears that it may be used as a weapon are not totally unfounded;[192] the vaccine for smallpox is not safe—during the years before the eradication of smallpox disease more people became seriously ill as a result of vaccination than did people from smallpox[193] — and smallpox vaccination is no longer universally practiced.[194] Thus, much of the modern human population has almost no established resistance to smallpox.[192]
References
Notes
- ^ a b Koonin EV, Senkevich TG, Dolja VV (2006). "The ancient Virus World and evolution of cells". Biol. Direct. 1: 29. doi:10.1186/1745-6150-1-29. PMC 1594570. PMID 16984643. Retrieved 14 September 2008.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ a b c Dimmock p. 4
- ^ Dimmock p. 49
- ^ Breitbart M, Rohwer F (2005). "Here a virus, there a virus, everywhere the same virus?". Trends Microbiol. 13 (6): 278–84. doi:10.1016/j.tim.2005.04.003. PMID 15936660.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b Lawrence CM, Menon S, Eilers BJ; et al. (2009). "Structural and functional studies of archaeal viruses". J. Biol. Chem. 284 (19): 12599–603. doi:10.1074/jbc.R800078200. PMID 19158076.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Edwards RA, Rohwer F (2005). "Viral metagenomics". Nat. Rev. Microbiol. 3 (6): 504–10. doi:10.1038/nrmicro1163. PMID 15886693.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ a b c Collier pp. 33–55
- ^ Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H (2003). "Phage as agents of lateral gene transfer". Curr. Opin. Microbiol. 6 (4): 417–24. doi:10.1016/S1369-5274(03)00086-9. PMID 12941415.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ a b "virus". The Online Etymology Dictionary. Retrieved 12 September 2008.
- ^ "virulent, a." The Oxford English Dictionary — Online. Retrieved 12 September 2008.
- ^ "viral, a." The Oxford English Dictionary — Online. Retrieved 12 September 2008.
- ^ Shors pp. 76–77
- ^ a b c d Collier p. 3
- ^ Dimmock p.4–5
- ^ Shors p. 589
- ^ D'Herelle F (September 2007). "On an invisible microbe antagonistic toward dysenteric bacilli": brief note by Mr. F. D'Herelle, presented by Mr. Roux. 1917. Res. Microbiol. 158(7):553–4. Epub 2007 Jul 28. PMID 17855060
- ^ Steinhardt, E; Israeli, C; and Lambert, R.A. (1913) "Studies on the cultivation of the virus of vaccinia" J. Inf Dis. 13, 294–300
- ^ Collier p. 4
- ^ Goodpasture EW, Woodruff AM, Buddingh GJ (1931). "The cultivation of vaccine and other viruses in the chorioallantoic membrane of chick embryos". Science 74, pp. 371–372 PMID 17810781
- ^ Rosen FS (2004). "Isolation of poliovirus—John Enders and the Nobel Prize". New England Journal of Medicine, 351, pp. 1481–83 PMID 15470207
- ^ From Nobel Lectures, Physics 1981–1990, (1993) Editor-in-Charge Tore Frängsmyr, Editor Gösta Ekspång, World Scientific Publishing Co., Singapore.
- In 1887, Buist visualised one of the largest, Vaccinia virus, by optical microscopy after staining it. Vaccinia was not known to be a virus at that time. (Buist J.B. Vaccinia and Variola: a study of their life history Churchill, London)
- ^ Stanley WM, Loring HS (1936). "The isolation of crystalline tobacco mosaic virus protein from diseased tomato plants". Science, 83, p.85 PMID 17756690
- ^ Stanley WM, Lauffer MA (1939). "Disintegration of tobacco mosaic virus in urea solutions". Science 88, pp. 345–347 PMID 17788438
- ^ Creager AN, Morgan GJ (2008). "After the double helix: Rosalind Franklin's research on Tobacco mosaic virus". Isis. 99 (2): 239–72. doi:10.1086/588626. PMID 18702397.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - ^ Dimmock p. 12
- ^ Norrby E (2008). "Nobel Prizes and the emerging virus concept". Arch. Virol. 153 (6): 1109–23. doi:10.1007/s00705-008-0088-8. PMID 18446425.
- ^ ICTV list of virus discoveries and discoverers
- ^ Collier p. 745
- ^ a b Temin HM, Baltimore D (1972). "RNA-directed DNA synthesis and RNA tumor viruses". Adv. Virus Res. 17: 129–86. doi:10.1016/S0065-3527(08)60749-6. PMID 4348509.
{{cite journal}}:|access-date=requires|url=(help) - ^ Barré-Sinoussi, F., Chermann, J. C., Rey, F., Nugeyre, M. T., Chamaret, S., Gruest, J., Dauguet, C., Axler-Blin, C., Vezinet-Brun, F., Rouzioux, C., Rozenbaum, W., and Montagnier, L. (1983). "Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS)". Science. 220 (4599): 868–871. doi:10.1126/science.6189183. PMID 6189183.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Iyer LM, Balaji S, Koonin EV, Aravind L (2006). "Evolutionary genomics of nucleo-cytoplasmic large DNA viruses". Virus Res. 117 (1): 156–84. doi:10.1016/j.virusres.2006.01.009. PMID 16494962. Retrieved 14 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Liu Y, Nickle DC, Shriner D, et al. (2004). "Molecular clock-like evolution of human immunodeficiency virus type 1". Virology. 10;329(1):101–8, PMID 15476878
- ^ Shors p. 16
- ^ Collier pp. 18–19
- ^ Shors pp. 14–16
- ^ Collier pp. 11–21
- ^ a b Dimmock p. 16
- ^ Collier p. 11
- ^ Shors p. 574
- ^ McClintock, B. (1950). "The origin and behavior of mutable loci in maize". Proc Natl Acad Sci U S A. 36 (6): 344–55. doi:10.1073/pnas.36.6.344. PMID 15430309.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Collier pp. 11–12
- ^ Dimmock p. 55
- ^ Shors 551–3
- ^ Tsagris EM, de Alba AE, Gozmanova M, Kalantidis K (2008). "Viroids". Cell. Microbiol. 10 (11): 2168. doi:10.1111/j.1462-5822.2008.01231.x. PMID 18764915. Retrieved 19 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Shors p. 492–3
- ^ La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D (2008). "The virophage as a unique parasite of the giant mimivirus". Nature. 455 (7209): 100–4. doi:10.1038/nature07218. PMID 18690211.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Collier p. 777
- ^ Dimmock p. 55–7
- ^ Liberski PP (2008). "Prion diseases: a riddle wrapped in a mystery inside an enigma". Folia Neuropathol. 46 (2): 93–116. PMID 18587704. Retrieved 19 September 2008.
- ^ Dimmock pp. 57–58
- ^ Lupi O, Dadalti P, Cruz E, Goodheart C (2007). "Did the first virus self-assemble from self-replicating prion proteins and RNA?". Med. Hypotheses. 69 (4): 724–30. doi:10.1016/j.mehy.2007.03.031. PMID 17512677. Retrieved 19 September 2008.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Dimmock pp. 15–16
- ^ Rybicki EP (1990) "The classification of organisms at the edge of life, or problems with virus systematics." S Aft J Sci 86:182–186
- ^ Holmes EC (2007). "Viral evolution in the genomic age". PLoS Biol. 5 (10): e278. doi:10.1371/journal.pbio.0050278. PMC 1994994. PMID 17914905. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: unflagged free DOI (link) - ^ Collier pp. 33–37
- ^ Kiselev NA, Sherman MB, Tsuprun VL (1990). "Negative staining of proteins". Electron Microsc. Rev. 3 (1): 43–72. doi:10.1016/0892-0354(90)90013-I. PMID 1715774.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Collier p. 40
- ^ Caspar DL, Klug A (1962). "Physical principles in the construction of regular viruses". Cold Spring Harb. Symp. Quant. Biol. 27: 1–24. PMID 14019094.
- ^ Crick FH, Watson JD (1956). "Structure of small viruses". Nature. 177 (4506): 473–5. doi:10.1038/177473a0. PMID 13309339.
- ^ Falvo, M.R. (March 1997). "Manipulation of individual viruses: friction and mechanical properties". Biophysical Journal. 72 (3): 1396–1403. doi:10.1016/S0006-3495(97)78786-1. Retrieved 8 May 2009.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Kuznetsov, Yu. G. (1 September 2001). "Imaging of viruses by atomic force microscopy". J Gen Virol. 82 (9): 2025–2034. Retrieved 19 April 2009.
{{cite journal}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Collier p. 37
- ^ Collier pp. 40, 42
- ^ Collier pp. 42–43
- ^ Rossmann MG, Mesyanzhinov VV, Arisaka F, Leiman PG (2004). "The bacteriophage T4 DNA injection machine". Curr. Opin. Struct. Biol. 14 (2): 171–80. doi:10.1016/j.sbi.2004.02.001. PMID 15093831.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Long GW, Nobel J, Murphy FA, Herrmann KL, Lourie B (1970). "Experience with electron microscopy in the differential diagnosis of smallpox". Appl Microbiol. 20 (3): 497–504. PMC 376966. PMID 4322005.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Suzan-Monti M, La Scola B, Raoult D (2006). "Genomic and evolutionary aspects of Mimivirus". Virus Research. 117 (1): 145–155. doi:10.1016/j.virusres.2005.07.011. PMID 16181700.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b Prangishvili D, Forterre P, Garrett RA (2006). "Viruses of the Archaea: a unifying view". Nat. Rev. Microbiol. 4 (11): 837–48. doi:10.1038/nrmicro1527. PMID 17041631.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ a b c d Collier pp. 96–99
- ^ Pressing J, Reanney DC (1984). "Divided genomes and intrinsic noise". J Mol Evol. 20(2):135–46.
- ^ Pan XP, Li LJ, Du WB, Li MW, Cao HC, Sheng JF (2007). "Differences of YMDD mutational patterns, precore/core promoter mutations, serum HBV DNA levels in lamivudine-resistant hepatitis B genotypes B and C". J. Viral Hepat. 14 (11): 767–74. doi:10.1111/j.1365-2893.2007.00869.x. PMID 17927612. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Hampson AW, Mackenzie JS (2006). "The influenza viruses". Med. J. Aust. 185 (10 Suppl): S39–43. PMID 17115950. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Metzner KJ (2006). "Detection and significance of minority quasispecies of drug-resistant HIV-1". J HIV Ther. 11 (4): 74–81. PMID 17578210.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Goudsmit, Jaap. Viral Sex. Oxford Univ Press, 1998.ISBN 9780195124965 ISBN 0195124960
- ^ Worobey M, Holmes EC (1999). "Evolutionary aspects of recombination in RNA viruses". J. Gen. Virol. 80 ( Pt 10): 2535–43. PMID 10573145.
- ^ Lukashev AN (2005). "Role of recombination in evolution of enteroviruses". Rev. Med. Virol. 15 (3): 157–67. doi:10.1002/rmv.457. PMID 15578739.
- ^ Umene K (1999). "Mechanism and application of genetic recombination in herpesviruses". Rev. Med. Virol. 9 (3): 171–82. doi:10.1002/(SICI)1099-1654(199907/09)9:3<171::AID-RMV243>3.0.CO;2-A. PMID 10479778.
- ^ Collier pp. 75–91
- ^ Dimmock p. 70
- ^ Boevink P, Oparka KJ (2005). "Virus-host interactions during movement processes". Plant Physiol. 138 (4): 1815–21. doi:10.1104/pp.105.066761. PMC 1183373. PMID 16172094.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Dimmock p. 71
- ^ Barman S, Ali A, Hui EK, Adhikary L, Nayak DP (2001). "Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses". Virus Res. 77 (1): 61–9. doi:10.1016/S0168-1702(01)00266-0. PMID 11451488.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shors p. 54; Collier p. 78
- ^ Collier p. 79
- ^ Collier pp. 88–89
- ^ Collier pp. 115–146
- ^ Collier p. 115
- ^ Roulston A, Marcellus RC, Branton PE (1999). "Viruses and apoptosis". Annu. Rev. Microbiol. 53: 577–628. doi:10.1146/annurev.micro.53.1.577. PMID 10547702. Retrieved 20 December 2008.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Alwine JC (2008). "Modulation of host cell stress responses by human cytomegalovirus". Curr. Top. Microbiol. Immunol. 325: 263–79. doi:10.1007/978-3-540-77349-8_15. PMID 18637511.
{{cite journal}}:|access-date=requires|url=(help) - ^ Sinclair J (2008). "Human cytomegalovirus: Latency and reactivation in the myeloid lineage". J. Clin. Virol. 41 (3): 180–5. doi:10.1016/j.jcv.2007.11.014. PMID 18164651. Retrieved 20 December 2008.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Jordan MC, Jordan GW, Stevens JG, Miller G (1984). "Latent herpesviruses of humans". Ann. Intern. Med. 100 (6): 866–80. PMID 6326635.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Sissons JG, Bain M, Wills MR (2002). "Latency and reactivation of human cytomegalovirus". J. Infect. 44 (2): 73–7. doi:10.1053/jinf.2001.0948. PMID 12076064. Retrieved 20 December 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Barozzi P, Potenza L, Riva G, Vallerini D, Quadrelli C, Bosco R, Forghieri F, Torelli G, Luppi M (2007). "B cells and herpesviruses: a model of lymphoproliferation". Autoimmun Rev. 7 (2): 132–6. doi:10.1016/j.autrev.2007.02.018. PMID 18035323. Retrieved 20 December 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Subramanya D, Grivas PD (2008). "HPV and cervical cancer: updates on an established relationship". Postgrad Med. 120 (4): 7–13. doi:10.3810/pgm.2008.11.1928. PMID 19020360.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - ^ Lwoff A, Horne RW, Tournier P (1962). "A virus system". C. R. Hebd. Seances Acad. Sci. (in French). 254: 4225–7. PMID 14467544.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Lwoff A, Horne R, Tournier P (1962). "A system of viruses". Cold Spring Harb. Symp. Quant. Biol. 27: 51–5. PMID 13931895.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Fields p. 27
- As defined therein, "a virus species is a polythetic class of viruses that constitute a replicating lineage and occupy a particular ecological niche". A “polythetic" class is one whose members have several properties in common, although they do not necessarily all share a single common defining one. Members of a virus species are defined collectively by a consensus group of properties. Virus species thus differ from the higher viral taxa, which are “universal” classes and as such are defined by properties that are necessary for membership.
- ^ Delwart EL (2007). "Viral metagenomics". Rev. Med. Virol. 17 (2): 115–31. doi:10.1002/rmv.532. PMID 17295196.
- ^ Virus Taxonomy 2008. International Committee on Taxonomy of Viruses. Retrieved on September 15 2008.
- ^ ICTV Master Species List 2008
- This Excel file contains the official ICTV Master Species list for 2008. This spreadsheet lists all approved virus taxa and supersedes the previous taxonomy published as a part of the ICTV VIIIth Report. Produced by the International Committee on Taxonomy of Viruses. Retrieved on September 15 2008
- ^ Baltimore D (1974). "The strategy of RNA viruses". Harvey Lect. 70 Series: 57–74. PMID 4377923.
- ^ van Regenmortel MH, Mahy BW (2004). "Emerging issues in virus taxonomy". Emerging Infect. Dis. 10 (1): 8–13. PMID 15078590.
- ^ Mayo MA (1999). "Developments in plant virus taxonomy since the publication of the 6th ICTV Report. International Committee on Taxonomy of Viruses". Arch. Virol. 144 (8): 1659–66. doi:10.1007/s007050050620. PMID 10486120.
- ^ de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H (2004). "Classification of papillomaviruses". Virology. 324 (1): 17–27. doi:10.1016/j.virol.2004.03.033. PMID 15183049.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mainly Chapter 33 (Disease summaries), pages 367-392 in:Fisher, Bruce; Harvey, Richard P.; Champe, Pamela C. (2007). Lippincott's Illustrated Reviews: Microbiology (Lippincott's Illustrated Reviews Series). Hagerstwon, MD: Lippincott Williams & Wilkins. pp. pages 367–392. ISBN 0-7817-8215-5.
{{cite book}}:|pages=has extra text (help)CS1 maint: multiple names: authors list (link)|alt=A photograph of the upper body of a man labelled with the names of viruses that infect the different parts - ^ For common cold: National Institute of Allergy and Infectious Diseases (NIAID) > Common Cold. Last Updated December 10, 2007. Retrieved on 4 April, 2009
- ^ Chen C, Chiu Y, Wei F, Koong F, Liu H, Shaw C, Hwu H, Hsiao K (1999). "High seroprevalence of Borna virus infection in schizophrenic patients, family members and mental health workers in Taiwan". Mol Psychiatry. 4 (1): 33–8. doi:10.1038/sj.mp.4000484. PMID 10089006.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Margolis TP, Elfman FL, Leib D; et al. (2007). "Spontaneous reactivation of herpes simplex virus type 1 in latently infected murine sensory ganglia". J. Virol. 81 (20): 11069–74. doi:10.1128/JVI.00243-07. PMC 2045564. PMID 17686862. Retrieved 13 September 2008.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Whitley RJ, Roizman B (2001). "Herpes simplex virus infections". Lancet. 357 (9267): 1513–8. doi:10.1016/S0140-6736(00)04638-9. PMID 11377626.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Barton ES, White DW, Cathelyn JS; et al. (2007). "Herpesvirus latency confers symbiotic protection from bacterial infection". Nature. 447 (7142): 326–9. doi:10.1038/nature05762. PMID 17507983.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Bertoletti A, Gehring A (2007). "Immune response and tolerance during chronic hepatitis B virus infection". Hepatol. Res. 37 Suppl 3: S331–8. doi:10.1111/j.1872-034X.2007.00221.x. PMID 17931183.
- ^ Rodrigues C, Deshmukh M, Jacob T, Nukala R, Menon S, Mehta A (2001). "Significance of HBV DNA by PCR over serological markers of HBV in acute and chronic patients". Indian journal of medical microbiology. 19 (3): 141–4. PMID 17664817.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Nguyen VT, McLaws ML, Dore GJ (2007). "Highly endemic hepatitis B infection in rural Vietnam". Journal of Gastroenterology and Hepatology. 22 (12): 2093. doi:10.1111/j.1440-1746.2007.05010.x. PMID 17645465.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) - ^ Witzany G (2006) "Natural Genome-Editing Competences of Viruses'" Acta Biotheor 54: 235-253.
- ^ Fowler MG, Lampe MA, Jamieson DJ, Kourtis AP, Rogers MF (2007). "Reducing the risk of mother-to-child human immunodeficiency virus transmission: past successes, current progress and challenges, and future directions". Am. J. Obstet. Gynecol. 197 (3 Suppl): S3–9. doi:10.1016/j.ajog.2007.06.048. PMID 17825648.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Sauerbrei A, Wutzler P (2000). "The congenital varicella syndrome". Journal of perinatology : official journal of the California Perinatal Association. 20 (8 Pt 1): 548–54. PMID 11190597.
- ^ Garnett GP (2005). "Role of herd immunity in determining the effect of vaccines against sexually transmitted disease". J. Infect. Dis. 191 Suppl 1: S97–106. doi:10.1086/425271. PMID 15627236. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Platonov AE (2006). "(The influence of weather conditions on the epidemiology of vector-borne diseases by the example of West Nile fever in Russia)". Vestn. Akad. Med. Nauk SSSR (in Russian) (2): 25–9. PMID 16544901.
- ^ Shors p. 198
- ^ Shors pp. 199, 209
- ^ Shors p. 19
- ^ Shors p. 126
- ^ Shors pp. 193–194
- ^ Shors pp. 193–94
- ^ Shors p. 194
- ^ Shors pp. 192–193
- ^
- Ranlet P (2000). "The British, the Indians, and smallpox: what actually happened at Fort Pitt in 1763?". Pa Hist. 67 (3): 427–41. PMID 17216901.
{{cite journal}}:|access-date=requires|url=(help) - Van Rijn K (2006). ""Lo! The poor Indian!" colonial responses to the 1862–63 smallpox epidemic in British Columbia and Vancouver Island". Can Bull Med Hist. 23 (2): 541–60. PMID 17214129.
{{cite journal}}:|access-date=requires|url=(help) - Patterson KB, Runge T (2002). "Smallpox and the Native American". Am. J. Med. Sci. 323 (4): 216–22. doi:10.1097/00000441-200204000-00009. PMID 12003378.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - Sessa R, Palagiano C, Scifoni MG, di Pietro M, Del Piano M (1999). "The major epidemic infections: a gift from the Old World to the New?". Panminerva Med. 41 (1): 78–84. PMID 10230264.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - Bianchine PJ, Russo TA (1992). "The role of epidemic infectious diseases in the discovery of America". Allergy Proc. 13 (5): 225–32. doi:10.2500/108854192778817040. PMID 1483570. Retrieved 16 September 2008.
- Hauptman LM (1979). "Smallpox and American Indian; Depopulation in Colonial New York". N Y State J Med. 79 (12): 1945–9. PMID 390434.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - Fortuine R (1988). "Smallpox decimates the Tlingit (1787)". Alaska Med. 30 (3): 109. PMID 3041871.
{{cite journal}}:|access-date=requires|url=(help); More than one of|pages=and|page=specified (help)
- Ranlet P (2000). "The British, the Indians, and smallpox: what actually happened at Fort Pitt in 1763?". Pa Hist. 67 (3): 427–41. PMID 17216901.
- ^ Collier pp. 409–415
- ^ Patterson KD, Pyle GF (1991). "The geography and mortality of the 1918 influenza pandemic". Bull Hist Med. 65 (1): 4–21. PMID 2021692.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Johnson NP, Mueller J (2002). "Updating the accounts: global mortality of the 1918–1920 "Spanish" influenza pandemic". Bull Hist Med. 76 (1): 105–15. doi:10.1353/bhm.2002.0022. PMID 11875246. Retrieved 13 September 2008.
- ^
Gao F, Bailes E, Robertson DL; et al. (1999). "Origin of HIV-1 in the Chimpanzee Pan troglodytes troglodytes". Nature. 397 (6718): 436–441. doi:10.1038/17130. PMID doi:[https://doi.org/10.1038%2F17130 10.1038/17130 9989410 '"`UNIQ--templatestyles-0000013A-QINU`"'[[doi (identifier)|doi]]:[https://doi.org/10.1038%2F17130 10.1038/17130]].
{{cite journal}}: Check|pmid=value (help); Explicit use of et al. in:|author=(help); External link in|pmid=|pmid=at position 9 (help)CS1 maint: multiple names: authors list (link) - ^ Shors p. 447
- ^ Mawar N, Saha S, Pandit A, Mahajan U (2005). "The third phase of HIV pandemic: social consequences of HIV/AIDS stigma & discrimination & future needs" (PDF). Indian J. Med. Res. 122 (6): 471–84. PMID 16517997. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ "Status of the global HIV epidemic" (PDF). UNAIDS. 2008. Retrieved 15 September 2008.
- ^ Towner JS, Khristova ML, Sealy TK; et al. (2006). "Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola". J. Virol. 80 (13): 6497–516. doi:10.1128/JVI.00069-06. PMC 1488971. PMID 16775337. Retrieved 13 September 2008.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Einstein MH, Schiller JT, Viscidi RP, Strickler HD, Coursaget P, Tan T, Halsey N, Jenkins D (2009). "Clinician's guide to human papillomavirus immunology: knowns and unknowns". The Lancet Infectious Diseases. 9 (6): 347–56. doi:10.1016/S1473-3099(09)70108-2. PMID 19467474.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y (2008). "T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus". Proceedings of the National Academy of Sciences of the United States of America. 105 (42): 16272–7. doi:10.1073/pnas.0806526105. PMC 2551627. PMID 18812503.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Pulitzer MP, Amin BD, Busam KJ (2009). "Merkel cell carcinoma: review". Advances in Anatomic Pathology. 16 (3): 135–44. doi:10.1097/PAP.0b013e3181a12f5a. PMID 19395876.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Koike K (2007). "Hepatitis C virus contributes to hepatocarcinogenesis by modulating metabolic and intracellular signalling pathways". J. Gastroenterol. Hepatol. 22 Suppl 1: S108–11. doi:10.1111/j.1440-1746.2006.04669.x. PMID 17567457.
- ^ Hu J, Ludgate L (2007). "HIV-HBV and HIV-HCV coinfection and liver cancer development". Cancer Treat. Res. 133: 241–52. doi:10.1007/978-0-387-46816-7_9. PMID 17672044.
- ^ Bellon M, Nicot C (2007). "Telomerase: a crucial player in HTLV-I-induced human T-cell leukemia". Cancer genomics & proteomics. 4 (1): 21–5. PMID 17726237.
- ^ Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S (2007). "Human papillomavirus and cervical cancer". Lancet. 370 (9590): 890–907. doi:10.1016/S0140-6736(07)61416-0. PMID 17826171.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Klein E, Kis LL, Klein G (2007). "Epstein-Barr virus infection in humans: from harmless to life endangering virus-lymphocyte interactions". Oncogene. 26 (9): 1297–305. doi:10.1038/sj.onc.1210240. PMID 17322915.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ zur Hausen H (2008). "Novel human polyomaviruses--re-emergence of a well known virus family as possible human carcinogens". International Journal of Cancer. Journal International Du Cancer. 123 (2): 247–50. doi:10.1002/ijc.23620. PMID 18449881.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Alberts, Bruce (2002). Molecular Biology of the Cell; Fourth Edition. New York and London: Garland Science. ISBN 0-8153-3218-1. Retrieved 15 September 2008.
{{cite book}}: Unknown parameter|coauthors=ignored (|author=suggested) (help) - ^ Ding SW, Voinnet O (2007). "Antiviral immunity directed by small RNAs". Cell. 130 (3): 413–26. doi:10.1016/j.cell.2007.07.039. PMID 17693253. Retrieved 13 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Patton JT, Vasquez-Del Carpio R, Spencer E (2004). "Replication and transcription of the rotavirus genome". Curr. Pharm. Des. 10 (30): 3769–77. doi:10.2174/1381612043382620. PMID 15579070.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Jayaram H, Estes MK, Prasad BV (2004). "Emerging themes in rotavirus cell entry, genome organization, transcription and replication". Virus Res. 101 (1): 67–81. doi:10.1016/j.virusres.2003.12.007. PMID 15010218.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Greer S, Alexander GJ (1995). "Viral serology and detection". Baillieres Clin. Gastroenterol. 9 (4): 689–721. doi:10.1016/0950-3528(95)90057-8. PMID 8903801.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help) - ^ Matter L, Kogelschatz K, Germann D (1997). "Serum levels of rubella virus antibodies indicating immunity: response to vaccination of subjects with low or undetectable antibody concentrations". J. Infect. Dis. 175 (4): 749–55. doi:10.1086/513967. PMID 9086126.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Cascalho M, Platt JL (2007). "Novel functions of B cells". Crit. Rev. Immunol. 27 (2): 141–51. PMID 17725500. Retrieved 14 September 2008.
- ^ Le Page C, Génin P, Baines MG, Hiscott J (2000). "Interferon activation and innate immunity". Rev Immunogenet. 2 (3): 374–86. PMID 11256746.
{{cite journal}}:|access-date=requires|url=(help)CS1 maint: multiple names: authors list (link) - ^ Hilleman MR (2004). "Strategies and mechanisms for host and pathogen survival in acute and persistent viral infections". Proc. Natl. Acad. Sci. U.S.A. 101 Suppl 2: 14560–6. doi:10.1073/pnas.0404758101. PMC 521982. PMID 15297608. Retrieved 14 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Asaria P, MacMahon E (2006). "Measles in the United Kingdom: can we eradicate it by 2010?". BMJ. 333 (7574): 890–5. doi:10.1136/bmj.38989.445845.7C. PMID 17068034.
- ^ Lane JM (2006). "Mass vaccination and surveillance/containment in the eradication of smallpox". Curr. Top. Microbiol. Immunol. 304: 17–29. doi:10.1007/3-540-36583-4_2. PMID 16989262.
- ^ Arvin AM, Greenberg HB (2006). "New viral vaccines". Virology. 344 (1): 240–9. doi:10.1016/j.virol.2005.09.057. PMID 16364754.
- ^ Pastoret PP, Schudel AA, Lombard M (2007). "Conclusions--future trends in veterinary vaccinology". Rev. - Off. Int. Epizoot. 26 (2): 489–94, 495–501, 503–9. PMID 17892169.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Palese P (2006). "Making better influenza virus vaccines?". Emerging Infect. Dis. 12 (1): 61–5. PMID 16494719.
- ^ Thomssen R (1975). "Live attenuated versus killed virus vaccines". Monographs in allergy. 9: 155–76. PMID 1090805.
- ^ McLean AA (1986). "Development of vaccines against hepatitis A and hepatitis B". Rev. Infect. Dis. 8 (4): 591–8. PMID 3018891.
- ^ Casswall TH, Fischler B (2005). "Vaccination of the immunocompromised child". Expert review of vaccines. 4 (5): 725–38. doi:10.1586/14760584.4.5.725. PMID 16221073.
- ^ Barnett ED, Wilder-Smith A, Wilson ME (2008). "Yellow fever vaccines and international travelers". Expert Rev Vaccines. 7 (5): 579–87. doi:10.1586/14760584.7.5.579. PMID 18564013. Retrieved 16 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Magden J, Kääriäinen L, Ahola T (2005). "Inhibitors of virus replication: recent developments and prospects". Appl. Microbiol. Biotechnol. 66 (6): 612–21. doi:10.1007/s00253-004-1783-3. PMID 15592828.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Mindel A, Sutherland S (1983). "Genital herpes — the disease and its treatment including intravenous acyclovir". J. Antimicrob. Chemother. 12 Suppl B: 51–9. PMID 6355051.
- ^ Witthöft T, Möller B, Wiedmann KH; et al. (2007). "Safety, tolerability and efficacy of peginterferon alpha-2a and ribavirin in chronic hepatitis C in clinical practice: The German Open Safety Trial". J. Viral Hepat. 14 (11): 788–96. doi:10.1111/j.1365-2893.2007.00871.x. PMC 2156112. PMID 17927615.
{{cite journal}}:|access-date=requires|url=(help); Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Rudin D, Shah SM, Kiss A, Wetz RV, Sottile VM (2007). "Interferon and lamivudine vs. interferon for hepatitis B e antigen-positive hepatitis B treatment: meta-analysis of randomized controlled trials". Liver Int. 27 (9): 1185–93. doi:10.1111/j.1478-3231.2007.01580.x. PMC 2156150. PMID 17919229.
{{cite journal}}:|access-date=requires|url=(help); Unknown parameter|doi_brokendate=ignored (|doi-broken-date=suggested) (help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Dimmock p. 3
- ^ Goris N, Vandenbussche F, De Clercq K (2008). "Potential of antiviral therapy and prophylaxis for controlling RNA viral infections of livestock". Antiviral Res. 78 (1): 170–8. doi:10.1016/j.antiviral.2007.10.003. PMID 18035428. Retrieved 16 September 2008.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Carmichael L (2005). "An annotated historical account of canine parvovirus". J. Vet. Med. B Infect. Dis. Vet. Public Health. 52 (7–8): 303–11. doi:10.1111/j.1439-0450.2005.00868.x. PMID 16316389.
- ^ Chen YP, Zhao Y, Hammond J, Hsu H, Evans JD, Feldlaufer MF (October–November 2004). "Multiple virus infections in the honey bee and genome divergence of honey bee viruses". Journal of Invertebrate Pathology. 87 (2–3): 84–93. doi:10.1016/j.jip.2004.07.005. PMID 15579317.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Shors p. 584
- ^ Shors pp. 562–587
- ^ Dinesh-Kumar SP, Wai-Hong Tham, Baker BJ (2000). "Structure—function analysis of the tobacco mosaic virus resistance gene N". PNAS 97, 14789-94 PMID 11121079
- ^ Shors pp. 573–576
- ^ Soosaar JL, Burch-Smith TM, Dinesh-Kumar SP (2005). "Mechanisms of plant resistance to viruses". Nat. Rev. Microbiol. 3, pp. 789–98 PMID 16132037
- ^ Wommack KE, Colwell RR (2000). "Virioplankton: viruses in aquatic ecosystems". Microbiol. Mol. Biol. Rev. 64 (1): 69–114. doi:10.1128/MMBR.64.1.69-114.2000. PMC 98987. PMID 10704475.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Bergh O, Børsheim KY, Bratbak G, Heldal M (1989). "High abundance of viruses found in aquatic environments". Nature. 340 (6233): 467–8. doi:10.1038/340467a0. PMID 2755508.
{{cite journal}}: Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Shors pp. 595–97
- ^ Bickle TA, Krüger DH (1993). "Biology of DNA restriction". Microbiol. Rev. 57 (2): 434–50. PMC 372918. PMID 8336674.
{{cite journal}}: Unknown parameter|day=ignored (help); Unknown parameter|month=ignored (help) - ^ Barrangou R, Fremaux C, Deveau H; et al. (2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science (journal). 315 (5819): 1709–12. doi:10.1126/science.1138140. PMID 17379808.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Brouns SJ, Jore MM, Lundgren M; et al. (2008). "Small CRISPR RNAs guide antiviral defense in prokaryotes". Science (journal). 321 (5891): 960–4. doi:10.1126/science.1159689. PMID 18703739.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Prangishvili D, Garrett RA (2004). "Exceptionally diverse morphotypes and genomes of crenarchaeal hyperthermophilic viruses". Biochem. Soc. Trans. 32 (Pt 2): 204–8. doi:10.1042/BST0320204. PMID 15046572.
- ^ Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E (2005). "Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements". J. Mol. Evol. 60 (2): 174–82. doi:10.1007/s00239-004-0046-3. PMID 15791728.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006). "A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action". Biol. Direct. 1: 7. doi:10.1186/1745-6150-1-7. PMID 16545108.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Collier p.8
- ^ Lodish, Harvey; Berk, Arnold; Zipursky, S. Lawrence; Matsudaira, Paul; Baltimore, David; Darnell, James.Viruses:Structure, Function, and Uses Retrieved on September 16 2008
- ^ Matsuzaki S, Rashel M, Uchiyama J; et al. (2005). "Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases". J. Infect. Chemother. 11 (5): 211–9. doi:10.1007/s10156-005-0408-9. PMID 16258815.
{{cite journal}}: Explicit use of et al. in:|author=(help)CS1 maint: multiple names: authors list (link) - ^ Fischlechner, M; Donath, E (2007). "Viruses as Building Blocks for Materials and Devices". Angewandte Chemie International Edition. 46 (18): 3184. doi:10.1002/anie.200603445. PMID 17348058.
{{cite journal}}: More than one of|author=and|last1=specified (help); More than one of|pages=and|page=specified (help) - ^ Soto CM, Blum AS, Vora GJ; et al. (2006). "Fluorescent signal amplification of carbocyanine dyes using engineered viral nanoparticles". J. Am. Chem. Soc. 128 (15): 5184–9. doi:10.1021/ja058574x. PMID 16608355.
{{cite journal}}: Explicit use of et al. in:|author=(help); Unknown parameter|month=ignored (help)CS1 maint: multiple names: authors list (link) - ^ Blum AS, Soto CM, Wilson CD, et al. (2005). "An Engineered Virus as a Scaffold for Three-Dimensional Self-Assembly on the Nanoscale". Small, 7, 702.
- ^ Shors p. 331
- ^ a b c Artenstein AW, Grabenstein JD (2008). "Smallpox vaccines for biodefense: need and feasibility". Expert Review of Vaccines. 7 (8): 1225–37. doi:10.1586/14760584.7.8.1225. PMID 18844596. Retrieved 12 July 2009.
{{cite journal}}: Unknown parameter|month=ignored (help) - ^ Aragón TJ, Ulrich S, Fernyak S, Rutherford GW (2003). "Risks of serious complications and death from smallpox vaccination: a systematic review of the United States experience, 1963–1968". BMC public health. 3: 26. doi:10.1186/1471-2458-3-26. PMID 12911836.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) CS1 maint: unflagged free DOI (link) - ^ Weiss MM, Weiss PD, Mathisen G, Guze P (2004). "Rethinking smallpox". Clin. Infect. Dis. 39 (11): 1668–73. doi:10.1086/425745. PMID 15578369.
{{cite journal}}: CS1 maint: multiple names: authors list (link)
Bibliography
- Collier, Leslie; Balows, Albert; Sussman, Max (1998) Topley and Wilson's Microbiology and Microbial Infections ninth edition, Volume 1, Virology, volume editors: Mahy, Brian and Collier, Leslie. Arnold. ISBN 0340663162.
- Dimmock, N.J; Easton, Andrew J; Leppard, Keith (2007) Introduction to Modern Virology sixth edition, Blackwell Publishing, ISBN 1405136456.
- Knipe, David M; Howley, Peter M; Griffin, Diane E; Lamb, Robert A; Martin, Malcolm A; Roizman, Bernard; Straus Stephen E. (2007) Fields Virology, Lippincott Williams & Wilkins. ISBN 0781760607.
- Shors, Teri (2008). Understanding Viruses. Jones and Bartlett Publishers. ISBN 0763729329.
External links
Template:Link FA Template:Link FA Template:Link FA Template:Link FA