DNA polymerase
| DNA-directed DNA polymerase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 3D structure of the DNA-binding helix-turn-helix motifs in human DNA polymerase beta (based on pdb file 7ICG) | |||||||||
| Identifiers | |||||||||
| EC no. | 2.7.7.7 | ||||||||
| CAS no. | 9012-90-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
A DNA polymerase is an enzyme (the suffix -ase is used to identify enzymes) that helps catalyze the polymerization of DNA bases (deoxyribonucleotides) into a DNA strand. DNA polymerases are best known for their negative feedback role[clarification needed] in DNA replication, in which the polymerase "reads" an intact DNA strand as a template and uses it to synthesize the new strand. This process forms a new DNA strand complementary to the template strand and identical to the template's original partner strand. DNA polymerases use magnesium ions as cofactors. Human DNA polymerases are 900-1000 amino acids long.
Function
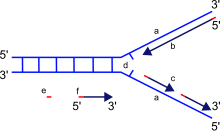

DNA polymerase can add free nucleotides only to the 3' end of the newly forming strand. This results in elongation of the newly forming strand in a 5'-3' direction. No known DNA polymerase is able to begin a new chain (de novo). DNA polymerase can add a nucleotide only on to a pre-existing 3'-OH group, and, therefore, needs a primer at which it can add the first nucleotide. Primers consist of RNA and/or DNA bases. In DNA replication, the first two bases are always RNA, and are synthesized by another enzyme called primase. An enzyme known as a helicase is required to unwind DNA from a double-strand structure to a single-strand structure to facilitate replication of each strand consistent with the semiconservative model of DNA replication.
It is important to note that the directionality of the newly forming strand (the daughter strand) is opposite to the direction in which DNA polymerase moves along the template strand. DNA polymerase moves along the template strand in a 3'-5' direction, and the daughter strand is formed in a 5'-3' direction. This difference enables the resultant double-stranded DNA formed to be composed of two DNA strands which are antiparallel to each other.
Error correction is a property of some, but not all, DNA polymerases. This process corrects mistakes in newly synthesized DNA. When an incorrect base pair is recognized, DNA polymerase moves backwards by one base pair of DNA. The 3'-5' exonuclease activity of the enzyme allows the incorrect base pair to be excised (this activity is known as proofreading). Following base excision, the polymerase can re-insert the correct base and replication can continue forwards.
Various DNA polymerases are extensively used in molecular biology experiments.
Variation across species
DNA polymerases have highly conserved structure, which means that their overall catalytic subunits vary, on a whole, very little from species to species. Conserved structures usually indicate important, irreplaceable functions of the cell, the maintenance of which provides evolutionary advantages.
Some viruses also encode special DNA polymerases, such as Hepatitis B virus DNA polymerase. These may selectively replicate viral DNA through a variety of mechanisms. Retroviruses encode an unusual DNA polymerase called reverse transcriptase, which is an RNA-dependent DNA polymerase (RdDp). It polymerizes DNA from a template of RNA.
DNA polymerase families
| DNA polymerase family A | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 c:o6-methyl-guanine pair in the polymerase-2 basepair position | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_A | ||||||||
| Pfam | PF00476 | ||||||||
| InterPro | IPR001098 | ||||||||
| PROSITE | PDOC00412 | ||||||||
| SCOP2 | 1dpi / SCOPe / SUPFAM | ||||||||
| |||||||||
| DNA polymerase family B | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of rb69 gp43 in complex with dna containing thymine glycol | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_B | ||||||||
| Pfam | PF00136 | ||||||||
| Pfam clan | CL0194 | ||||||||
| InterPro | IPR006134 | ||||||||
| PROSITE | PDOC00107 | ||||||||
| SCOP2 | 1noy / SCOPe / SUPFAM | ||||||||
| |||||||||
| DNA polymerase type B, organellar and viral | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 phi29 dna polymerase, orthorhombic crystal form, ssdna complex | |||||||||
| Identifiers | |||||||||
| Symbol | DNA_pol_B_2 | ||||||||
| Pfam | PF03175 | ||||||||
| Pfam clan | CL0194 | ||||||||
| InterPro | IPR004868 | ||||||||
| |||||||||
Based on sequence homology, DNA polymerases can be further subdivided into seven different families: A, B, C, D, X, Y, and RT.
Family A
Polymerases contain both replicative and repair polymerases. Replicative members from this family include the extensively studied T7 DNA polymerase, as well as the eukaryotic mitochondrial DNA Polymerase γ. Among the repair polymerases are Escherichia coli DNA pol I, Thermus aquaticus pol I, and Bacillus stearothermophilus pol I. These repair polymerases are involved in excision repair and processing of Okazaki fragments generated during lagging strand synthesis.
Family B
B family polymerases are involved in DNA repair and replication. These include; Pol II (bacterial), Pol B (archaebacterial), and Pol α, δ, and ε (eukaryotic). Pol ζ another B family polymerase, is made of two subunits Rev3 and Rev7 and is involved in translesion synthesis. Pol ζ is unique in that it can extend primers with terminal mismatches.
Family C
Polymerases are the primary bacterial chromosomal replicative enzymes. DNA Polymerase III alpha subunit from E. coli is the catalytic subunit [1] and possesses no known nuclease activity. A separate subunit, the epsilon subunit, possesses the 3'-5' exonuclease activity used for editing during chromosomal replication. Recent research has classified Family C polymerases as a subcategory of Family X[citation needed].
Family D
Polymerases are still not very well characterized. All known examples are found in the Euryarchaeota subdomain of Archaea and are thought to be replicative polymerases.
Family X
Contains the well-known eukaryotic polymerase pol β, as well as other eukaryotic polymerases such as pol σ, pol λ, pol μ, and terminal deoxynucleotidyl transferase (TdT). Pol β is required for short-patch base excision repair, a DNA repair pathway that is essential for repairing alkylated or oxidised bases as well as abasic sites. Pol λ and Pol μ are involved in non-homologous end-joining, a mechanism for rejoining DNA double-strand breaks. TdT is expressed only in lymphoid tissue, and adds "n nucleotides" to double-strand breaks formed during V(D)J recombination to promote immunological diversity. The yeast Saccharomyces cerevisiae has only one Pol X polymerase, Pol IV, which is involved in non-homologous end-joining.
Family Y
Y Polymerases differ from others in having a low fidelity on undamaged templates and in their ability to replicate through damaged DNA, known as translesion synthesis (TLS). Members of this family are hence called translesion synthesis polymerases. Depending on the lesion, TLS polymerases can bypass the damage in an error-free or error-prone fashion, the latter resulting in elevated mutagenesis. Xeroderma pigmentosum variant (XPV) patients for instance have mutations in the gene encoding Pol η (eta), which is error-free for UV-lesions. In XPV patients, alternative error-prone polymerases, e.g., Pol ζ (zeta) (polymerase ζ is a B Family polymerase a complex of the catalytic subunit REV3L with Rev7, which associates with Rev1[2]), are thought to be involved in mistakes that result in the cancer predisposition of these patients. Other members in humans are Pol ι (iota), Pol κ (kappa), and Rev1 (terminal deoxycytidyl transferase). In E. coli, two TLS polymerases, Pol IV (DINB) and Pol V (UmuD'2C), are known.
Family RT
Some viruses also encode special DNA polymerases, such as Hepatitis B virus DNA polymerase. These may selectively replicate viral DNA through a variety of mechanisms. Retroviruses encode an unusual DNA polymerase called reverse transcriptase, which is an RNA-dependent DNA polymerase (RdDp). It polymerizes DNA from a template of RNA. The reverse transcriptase family contains examples from both retroviruses and eukaryotic polymerases. The eukaryotic polymerases are usually restricted to telomerases. These polymerases use an RNA template to synthesize the DNA strand.
Prokaryotic DNA polymerases
Bacteria have 5 known DNA polymerases.
Eukaryotic DNA polymerases
Eukaryotes have at least 15 DNA Polymerases.[3]
See also
References
- ^ Attention: This template ({{cite doi}}) is deprecated. To cite the publication identified by doi:10.1016/j.cell.2006.07.028, please use {{cite journal}} (if it was published in a bona fide academic journal, otherwise {{cite report}} with
|doi=10.1016/j.cell.2006.07.028instead. - ^ Gan GN; Wittschieben JP; Wittschieben BØ; Wood RD (2008). "DNA polymerase zeta (pol zeta) in higher eukaryotes". Cell Research. 18 (1): 174–83. doi:10.1038/cr.2007.117. PMID 18157155.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link) - ^ I. Hubscher, U.; Maga, G.; Spadari, S. (2002). "Eukaryotic DNA polymerases". Annual Review of Biochemistry. 71: 133–63. doi:10.1146/annurev.biochem.71.090501.150041. PMID 12045093.
{{cite journal}}: More than one of|pages=and|page=specified (help)CS1 maint: multiple names: authors list (link)
External links
- Burgers P; Koonin E; Bruford E; et al. (2001). "Eukaryotic DNA polymerases: proposal for a revised nomenclature". J. Biol. Chem. 276 (47): 43487–90. doi:10.1074/jbc.R100056200. PMID 11579108.
{{cite journal}}: Unknown parameter|author-separator=ignored (help)CS1 maint: unflagged free DOI (link) - PDB Molecule of the Month DNA polymerase
- Unusual repair mechanism in DNA polymerase lambda, Ohio State University, July 25, 2006.
- DNA+polymerases at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
- EC 2.7.7.7
- A great animation of DNA Polymerase from WEHI at 1:45 minutes in
- 3D macromolecular structures of DNA polymerase from the EM Data Bank(EMDB)
